Breaking Bad Proteins—Discovery Approaches and the Road to Clinic for Degraders
Abstract
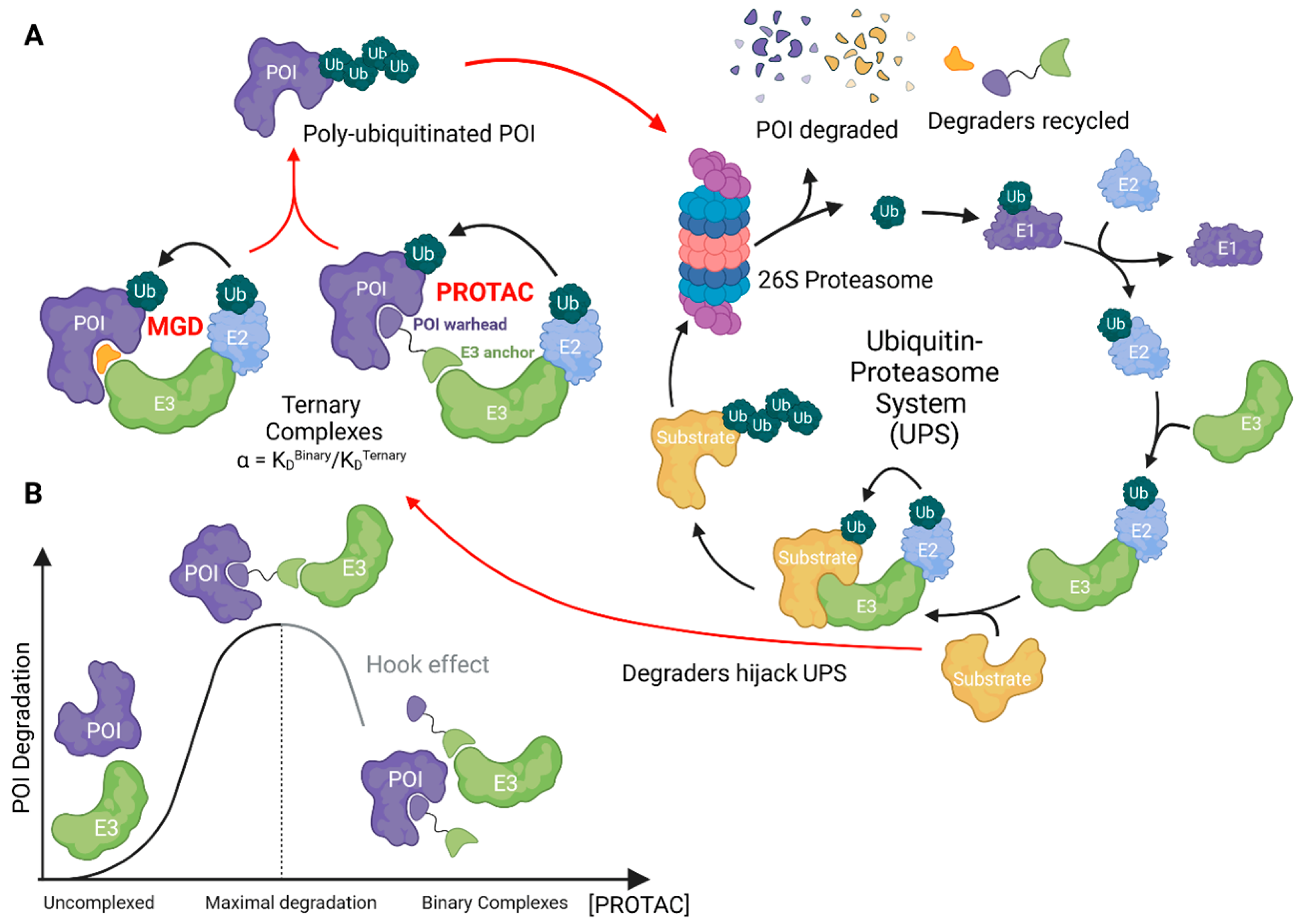
:1. General Concepts in TPD Drug Discovery
2. Ubiquitin Ligases in TPD Drug Discovery
3. Ternary Complex Assessment for Compound Development
4. Directly Recruiting E3s for TPD
5. Recruiting Alternative UPS Components for TPD
6. TPD as a Therapeutic Strategy
6.1. Selectivity
6.2. Degraders for Non-Enzymatic Functions
6.3. Resistance Mutations
7. Discovery Approaches for Bifunctional Degraders
7.1. Aspects to Consider for POI Ligand Development
7.2. Aspects to Consider for E3 Ligand Development
7.3. Linker Optimization
7.4. Discovery Approaches for Molecular Glue Degraders
7.4.1. Serendipity
7.4.2. Cellular Screening Approaches
7.4.3. Biophysical Screening Approaches
7.5. Target Validation via Degraders
7.6. Assays for Validation of Degraders
Confirmation of Mechanism of Action
7.7. Therapeutic Areas and Clinical Application
7.7.1. Heterobifunctional Degraders in Oncology
- The aforementioned low bioavailability and, therefore, potentially subtherapeutic exposure at target
- Emergent resistance due to loss of function, or decreased expression of the cognate E3 ligase
- Elevated rates of protein re-expression in response to treatment, counteracting active degradation by the heterobifunctional agent.
7.7.2. Molecular Glue Degraders in Oncology
7.7.3. Heterobifunctional Degraders for Inflammatory Indications
7.7.4. Molecular Glue Degraders for Inflammatory Indications
7.7.5. Heterobifunctional Degraders for CNS Disorders
AD
PD
HD
FTD and ALS
7.7.6. Molecular Glue Degraders for CNS Disorders
8. Concluding Remarks and Future Perspectives
Funding
Conflicts of Interest
References
- Sakamoto, K.M.; Kim, K.B.; Kumagai, A.; Mercurio, F.; Crews, C.M.; Deshaies, R.J. Protacs: Chimeric Molecules That Target Proteins to the Skp1–Cullin–F Box Complex for Ubiquitination and Degradation. Proc. Natl. Acad. Sci. USA 2001, 98, 8554–8559. [Google Scholar] [CrossRef] [PubMed]
- Słabicki, M.; Kozicka, Z.; Petzold, G.; Li, Y.-D.; Manojkumar, M.; Bunker, R.D.; Donovan, K.A.; Sievers, Q.L.; Koeppel, J.; Suchyta, D.; et al. The CDK Inhibitor CR8 Acts as a Molecular Glue Degrader That Depletes Cyclin K. Nature 2020, 585, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Chen, P.; Cao, L.; Li, Y.; Zeng, Z.; Cui, Y.; Wu, Q.; Li, J.; Wang, J.-H.; Dong, M.-Q.; et al. Discovery of a Molecular Glue Promoting CDK12-DDB1 Interaction to Trigger Cyclin K Degradation. eLife 2020, 9, e59994. [Google Scholar] [CrossRef] [PubMed]
- Mayor-Ruiz, C.; Bauer, S.; Brand, M.; Kozicka, Z.; Siklos, M.; Imrichova, H.; Kaltheuner, I.H.; Hahn, E.; Seiler, K.; Koren, A.; et al. Rational Discovery of Molecular Glue Degraders via Scalable Chemical Profiling. Nat. Chem. Biol. 2020, 16, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Thomas, K.L.; Bouguenina, H.; Miller, D.S.J.; Sialana, F.J.; Hayhow, T.G.; Choudhary, J.S.; Rossanese, O.W.; Bellenie, B.R. Degradation by Design: New Cyclin K Degraders from Old CDK Inhibitors. ACS Chem. Biol. 2024, 19, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Hsia, O.; Hinterndorfer, M.; Cowan, A.D.; Iso, K.; Ishida, T.; Sundaramoorthy, R.; Nakasone, M.A.; Imrichova, H.; Schätz, C.; Rukavina, A.; et al. Targeted Protein Degradation via Intramolecular Bivalent Glues. Nature 2024, 627, 204–211. [Google Scholar] [CrossRef]
- Hassan, M.M.; Li, Y.-D.; Ma, M.W.; Teng, M.; Byun, W.S.; Puvar, K.; Lumpkin, R.; Sandoval, B.; Rutter, J.C.; Jin, C.Y.; et al. Exploration of the Tunability of BRD4 Degradation by DCAF16 Trans-Labelling Covalent Glues. bioRxiv 2023. [Google Scholar] [CrossRef]
- Shergalis, A.G.; Marin, V.L.; Rhee, D.Y.; Senaweera, S.; McCloud, R.L.; Ronau, J.A.; Hutchins, C.W.; McLoughlin, S.; Woller, K.R.; Warder, S.E.; et al. CRISPR Screen Reveals BRD2/4 Molecular Glue-like Degrader via Recruitment of DCAF16. ACS Chem. Biol. 2023, 18, 331–339. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Ermondi, G.; Jimenez, D.G.; Rossi Sebastiano, M.; Kihlberg, J.; Caron, G. Conformational Sampling Deciphers the Chameleonic Properties of a VHL-Based Degrader. Pharmaceutics 2023, 15, 272. [Google Scholar] [CrossRef]
- Atilaw, Y.; Poongavanam, V.; Svensson Nilsson, C.; Nguyen, D.; Giese, A.; Meibom, D.; Erdelyi, M.; Kihlberg, J. Solution Conformations Shed Light on PROTAC Cell Permeability. ACS Med. Chem. Lett. 2021, 12, 107–114. [Google Scholar] [CrossRef]
- Hornberger, K.R.; Araujo, E.M.V. Physicochemical Property Determinants of Oral Absorption for PROTAC Protein Degraders. J. Med. Chem. 2023, 66, 8281–8287. [Google Scholar] [CrossRef] [PubMed]
- Bondeson, D.P.; Mares, A.; Smith, I.E.D.; Ko, E.; Campos, S.; Miah, A.H.; Mulholland, K.E.; Routly, N.; Buckley, D.L.; Gustafson, J.L.; et al. Catalytic in Vivo Protein Knockdown by Small-Molecule PROTACs. Nat. Chem. Biol. 2015, 11, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Belcher, B.P.; Ward, C.C.; Nomura, D.K. Ligandability of E3 Ligases for Targeted Protein Degradation Applications. Biochemistry 2023, 62, 588–600. [Google Scholar] [CrossRef]
- Du, G.; Jiang, J.; Wu, Q.; Henning, N.J.; Donovan, K.A.; Yue, H.; Che, J.; Lu, W.; Fischer, E.S.; Bardeesy, N.; et al. Discovery of a Potent Degrader for Fibroblast Growth Factor Receptor 1/2. Angew. Chem. Int. Ed. 2021, 60, 15905–15911. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.R.; De Vita, E.; Conole, D.; Sathyamurthi, P.S.; Zhang, X.; Fellows, E.; Fleites, C.M.; Queisser, M.A.; Harling, J.D.; Tate, E.W. Structure-Guided Design and Optimization of Covalent VHL-Targeted Sulfonyl Fluoride PROTACs. bioRxiv 2023. [Google Scholar] [CrossRef] [PubMed]
- Nowak, R.P.; Ragosta, L.; Huerta, F.; Liu, H.; Ficarro, S.B.; Cruite, J.T.; Metivier, R.J.; Donovan, K.A.; Marto, J.A.; Fischer, E.S.; et al. Development of a Covalent Cereblon-Based PROTAC Employing a Fluorosulfate Warhead. RSC Chem. Biol. 2023, 4, 906–912. [Google Scholar] [CrossRef] [PubMed]
- Johansson, H.; Isabella Tsai, Y.-C.; Fantom, K.; Chung, C.-W.; Kümper, S.; Martino, L.; Thomas, D.A.; Eberl, H.C.; Muelbaier, M.; House, D.; et al. Fragment-Based Covalent Ligand Screening Enables Rapid Discovery of Inhibitors for the RBR E3 Ubiquitin Ligase HOIP. J. Am. Chem. Soc. 2019, 141, 2703–2712. [Google Scholar] [CrossRef] [PubMed]
- Schiemer, J.; Maxwell, A.; Horst, R.; Liu, S.; Uccello, D.P.; Borzilleri, K.; Rajamohan, N.; Brown, M.F.; Calabrese, M.F. A Covalent BTK Ternary Complex Compatible with Targeted Protein Degradation. Nat. Commun. 2023, 14, 1189. [Google Scholar] [CrossRef]
- Gabizon, R.; Shraga, A.; Gehrtz, P.; Livnah, E.; Shorer, Y.; Gurwicz, N.; Avram, L.; Unger, T.; Aharoni, H.; Albeck, S.; et al. Efficient Targeted Degradation via Reversible and Irreversible Covalent PROTACs. J. Am. Chem. Soc. 2020, 142, 11734–11742. [Google Scholar] [CrossRef]
- Xue, G.; Chen, J.; Liu, L.; Zhou, D.; Zuo, Y.; Fu, T.; Pan, Z. Protein Degradation through Covalent Inhibitor-Based PROTACs. Chem. Commun. 2020, 56, 1521–1524. [Google Scholar] [CrossRef]
- Zeng, M.; Xiong, Y.; Safaee, N.; Nowak, R.P.; Donovan, K.A.; Yuan, C.J.; Nabet, B.; Gero, T.W.; Feru, F.; Li, L.; et al. Exploring Targeted Degradation Strategy for Oncogenic KRASG12C. Cell Chem. Biol. 2020, 27, 19–31.e6. [Google Scholar] [CrossRef]
- Bond, M.J.; Chu, L.; Nalawansha, D.A.; Li, K.; Crews, C.M. Targeted Degradation of Oncogenic KRAS G12C by VHL-Recruiting PROTACs. ACS Cent. Sci. 2020, 6, 1367–1375. [Google Scholar] [CrossRef]
- Boike, L.; Henning, N.J.; Nomura, D.K. Advances in Covalent Drug Discovery. Nat. Rev. Drug Discov. 2022, 21, 881–898. [Google Scholar] [CrossRef] [PubMed]
- Mares, A.; Miah, A.H.; Smith, I.E.D.; Rackham, M.; Thawani, A.R.; Cryan, J.; Haile, P.A.; Votta, B.J.; Beal, A.M.; Capriotti, C.; et al. Extended Pharmacodynamic Responses Observed upon PROTAC-Mediated Degradation of RIPK2. Commun. Biol. 2020, 3, 140. [Google Scholar] [CrossRef]
- Riching, K.M.; Caine, E.A.; Urh, M.; Daniels, D.L. The Importance of Cellular Degradation Kinetics for Understanding Mechanisms in Targeted Protein Degradation. Chem. Soc. Rev. 2022, 51, 6210–6221. [Google Scholar] [CrossRef]
- Dikic, I.; Schulman, B.A. An Expanded Lexicon for the Ubiquitin Code. Nat. Rev. Mol. Cell Biol. 2023, 24, 273–287. [Google Scholar] [CrossRef]
- Ishida, T.; Ciulli, A. E3 Ligase Ligands for PROTACs: How They Were Found and How to Discover New Ones. SLAS Discov. 2021, 26, 484–502. [Google Scholar] [CrossRef]
- Henning, N.J.; Manford, A.G.; Spradlin, J.N.; Brittain, S.M.; Zhang, E.; McKenna, J.M.; Tallarico, J.A.; Schirle, M.; Rape, M.; Nomura, D.K. Discovery of a Covalent FEM1B Recruiter for Targeted Protein Degradation Applications. J. Am. Chem. Soc. 2022, 144, 701–708. [Google Scholar] [CrossRef]
- Hickey, C.M.; Digianantonio, K.M.; Zimmermann, K.; Harbin, A.; Quinn, C.; Patel, A.; Gareiss, P.; Chapman, A.; Tiberi, B.; Dobrodziej, J.; et al. Co-Opting the E3 Ligase KLHDC2 for Targeted Protein Degradation by Small Molecules. Nat. Struct. Mol. Biol. 2024, 31, 311–322. [Google Scholar] [CrossRef]
- Zhang, X.; Luukkonen, L.M.; Eissler, C.L.; Crowley, V.M.; Yamashita, Y.; Schafroth, M.A.; Kikuchi, S.; Weinstein, D.S.; Symons, K.T.; Nordin, B.E.; et al. DCAF11 Supports Targeted Protein Degradation by Electrophilic Proteolysis-Targeting Chimeras. J. Am. Chem. Soc. 2021, 143, 5141–5149. [Google Scholar] [CrossRef] [PubMed]
- Schröder, M.; Renatus, M.; Liang, X.; Meili, F.; Zoller, T.; Ferrand, S.; Gauter, F.; Li, X.; Sigoillot, F.; Gleim, S.; et al. DCAF1-Based PROTACs with Activity against Clinically Validated Targets Overcoming Intrinsic- and Acquired-Degrader Resistance. Nat. Commun. 2024, 15, 275. [Google Scholar] [CrossRef] [PubMed]
- Taherbhoy, A.M.; Daniels, D.L. Harnessing UBR5 for Targeted Protein Degradation of Key Transcriptional Regulators. Trends Pharmacol. Sci. 2023, 44, 758–761. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, J.; Wang, T.; Luo, M.; Chen, Y.; Chen, C.; Ronai, Z.; Zhou, Y.; Ruppin, E.; Han, L. Expanding PROTACtable Genome Universe of E3 Ligases. Nat. Commun. 2023, 14, 6509. [Google Scholar] [CrossRef] [PubMed]
- Dragovich, P.S.; Pillow, T.H.; Blake, R.A.; Sadowsky, J.D.; Adaligil, E.; Adhikari, P.; Bhakta, S.; Blaquiere, N.; Chen, J.; Dela Cruz-Chuh, J.; et al. Antibody-Mediated Delivery of Chimeric BRD4 Degraders. Part 1: Exploration of Antibody Linker, Payload Loading, and Payload Molecular Properties. J. Med. Chem. 2021, 64, 2534–2575. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Zhang, X.; Lv, D.; Zhang, Q.; He, Y.; Zhang, P.; Liu, X.; Thummuri, D.; Yuan, Y.; Wiegand, J.S.; et al. A Selective BCL-XL PROTAC Degrader Achieves Safe and Potent Antitumor Activity. Nat. Med. 2019, 25, 1938–1947. [Google Scholar] [CrossRef] [PubMed]
- Simpson, L.M.; Glennie, L.; Brewer, A.; Zhao, J.-F.; Crooks, J.; Shpiro, N.; Sapkota, G.P. Target Protein Localization and Its Impact on PROTAC-Mediated Degradation. Cell Chem. Biol. 2022, 29, 1482–1504.e7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Crowley, V.M.; Wucherpfennig, T.G.; Dix, M.M.; Cravatt, B.F. Electrophilic PROTACs That Degrade Nuclear Proteins by Engaging DCAF16. Nat. Chem. Biol. 2019, 15, 737–746. [Google Scholar] [CrossRef]
- Marei, H.; Tsai, W.-T.K.; Kee, Y.-S.; Ruiz, K.; He, J.; Cox, C.; Sun, T.; Penikalapati, S.; Dwivedi, P.; Choi, M.; et al. Antibody Targeting of E3 Ubiquitin Ligases for Receptor Degradation. Nature 2022, 610, 182–189. [Google Scholar] [CrossRef]
- Cotton, A.D.; Nguyen, D.P.; Gramespacher, J.A.; Seiple, I.B.; Wells, J.A. Development of Antibody-Based PROTACs for the Degradation of the Cell-Surface Immune Checkpoint Protein PD-L1. J. Am. Chem. Soc. 2021, 143, 593–598. [Google Scholar] [CrossRef]
- Squair, D.R.; Virdee, S. A New Dawn beyond Lysine Ubiquitination. Nat. Chem. Biol. 2022, 18, 802–811. [Google Scholar] [CrossRef] [PubMed]
- Donovan, K.A.; Ferguson, F.M.; Bushman, J.W.; Eleuteri, N.A.; Bhunia, D.; Ryu, S.; Tan, L.; Shi, K.; Yue, H.; Liu, X.; et al. Mapping the Degradable Kinome Provides a Resource for Expedited Degrader Development. Cell 2020, 183, 1714–1731.e10. [Google Scholar] [CrossRef] [PubMed]
- Bondeson, D.P.; Smith, B.E.; Burslem, G.M.; Buhimschi, A.D.; Hines, J.; Jaime-Figueroa, S.; Wang, J.; Hamman, B.D.; Ishchenko, A.; Crews, C.M. Lessons in PROTAC Design from Selective Degradation with a Promiscuous Warhead. Cell Chem. Biol. 2018, 25, 78–87.e5. [Google Scholar] [CrossRef] [PubMed]
- Roy, M.J.; Winkler, S.; Hughes, S.J.; Whitworth, C.; Galant, M.; Farnaby, W.; Rumpel, K.; Ciulli, A. SPR-Measured Dissociation Kinetics of PROTAC Ternary Complexes Influence Target Degradation Rate. ACS Chem. Biol. 2019, 14, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Gadd, M.S.; Testa, A.; Lucas, X.; Chan, K.-H.; Chen, W.; Lamont, D.J.; Zengerle, M.; Ciulli, A. Structural Basis of PROTAC Cooperative Recognition for Selective Protein Degradation. Nat. Chem. Biol. 2017, 13, 514–521. [Google Scholar] [CrossRef] [PubMed]
- Geiger, T.M.; Walz, M.; Meyners, C.; Kuehn, A.; Dreizler, J.K.; Sugiarto, W.O.; Maciel, E.; Zheng, M.; Lermyte, F.; Hausch, F. Discovery of a Potent PROTAC Enables Targeting of FKBP51’s Scaffolding Functions. Angew. Chem. Int. Ed. 2023, 63, e202309706. [Google Scholar] [CrossRef] [PubMed]
- Farnaby, W.; Koegl, M.; Roy, M.J.; Whitworth, C.; Diers, E.; Trainor, N.; Zollman, D.; Steurer, S.; Karolyi-Oezguer, J.; Riedmueller, C.; et al. BAF Complex Vulnerabilities in Cancer Demonstrated via Structure-Based PROTAC Design. Nat. Chem. Biol. 2019, 15, 672–680. [Google Scholar] [CrossRef] [PubMed]
- Kaneshige, A.; Bai, L.; Wang, M.; McEachern, D.; Meagher, J.L.; Xu, R.; Wang, Y.; Jiang, W.; Metwally, H.; Kirchhoff, P.D.; et al. A Selective Small-Molecule STAT5 PROTAC Degrader Capable of Achieving Tumor Regression in Vivo. Nat. Chem. Biol. 2023, 19, 703–711. [Google Scholar] [CrossRef]
- Wurz, R.P.; Rui, H.; Dellamaggiore, K.; Ghimire-Rijal, S.; Choi, K.; Smither, K.; Amegadzie, A.; Chen, N.; Li, X.; Banerjee, A.; et al. Affinity and Cooperativity Modulate Ternary Complex Formation to Drive Targeted Protein Degradation. Nat. Commun. 2023, 14, 4177. [Google Scholar] [CrossRef]
- Chung, C.; Dai, H.; Fernandez, E.; Tinworth, C.P.; Churcher, I.; Cryan, J.; Denyer, J.; Harling, J.D.; Konopacka, A.; Queisser, M.A.; et al. Structural Insights into PROTAC-Mediated Degradation of Bcl-XL. ACS Chem. Biol. 2020, 15, 2316–2323. [Google Scholar] [CrossRef]
- Zorba, A.; Nguyen, C.; Xu, Y.; Starr, J.; Borzilleri, K.; Smith, J.; Zhu, H.; Farley, K.A.; Ding, W.; Schiemer, J.; et al. Delineating the Role of Cooperativity in the Design of Potent PROTACs for BTK. Proc. Natl. Acad. Sci. USA 2018, 115, E7285–E7292. [Google Scholar] [CrossRef] [PubMed]
- Metzger, M.B.; Hristova, V.A.; Weissman, A.M. HECT and RING Finger Families of E3 Ubiquitin Ligases at a Glance. J. Cell Sci. 2012, 125, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Budhidarmo, R.; Nakatani, Y.; Day, C.L. RINGs Hold the Key to Ubiquitin Transfer. Trends Biochem. Sci. 2012, 37, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Rotin, D.; Kumar, S. Physiological Functions of the HECT Family of Ubiquitin Ligases. Nat. Rev. Mol. Cell Biol. 2009, 10, 398–409. [Google Scholar] [CrossRef]
- Berndsen, C.E.; Wolberger, C. New Insights into Ubiquitin E3 Ligase Mechanism. Nat. Struct. Mol. Biol. 2014, 21, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Spratt, D.E.; Walden, H.; Shaw, G.S. RBR E3 Ubiquitin Ligases: New Structures, New Insights, New Questions. Biochem. J. 2014, 458, 421–437. [Google Scholar] [CrossRef] [PubMed]
- Cotton, T.R.; Lechtenberg, B.C. Chain Reactions: Molecular Mechanisms of RBR Ubiquitin Ligases. Biochem. Soc. Trans. 2020, 48, 1737–1750. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, E.S.; Schulman, B.A.; Zheng, N. Structural Assembly of Cullin-RING Ubiquitin Ligase Complexes. Curr. Opin. Struct. Biol. 2010, 20, 714–721. [Google Scholar] [CrossRef] [PubMed]
- Sarikas, A.; Hartmann, T.; Pan, Z.-Q. The Cullin Protein Family. Genome Biol. 2011, 12, 220. [Google Scholar] [CrossRef]
- Zheng, N.; Schulman, B.A.; Song, L.; Miller, J.J.; Jeffrey, P.D.; Wang, P.; Chu, C.; Koepp, D.M.; Elledge, S.J.; Pagano, M.; et al. Structure of the Cul1–Rbx1–Skp1–F BoxSkp2 SCF Ubiquitin Ligase Complex. Nature 2002, 416, 703–709. [Google Scholar] [CrossRef]
- Kamura, T.; Maenaka, K.; Kotoshiba, S.; Matsumoto, M.; Kohda, D.; Conaway, R.C.; Conaway, J.W.; Nakayama, K.I. VHL-Box and SOCS-Box Domains Determine Binding Specificity for Cul2-Rbx1 and Cul5-Rbx2 Modules of Ubiquitin Ligases. Genes. Dev. 2004, 18, 3055–3065. [Google Scholar] [CrossRef] [PubMed]
- Mahrour, N.; Redwine, W.B.; Florens, L.; Swanson, S.K.; Martin-Brown, S.; Bradford, W.D.; Staehling-Hampton, K.; Washburn, M.P.; Conaway, R.C.; Conaway, J.W. Characterization of Cullin-Box Sequences That Direct Recruitment of Cul2-Rbx1 and Cul5-Rbx2 Modules to Elongin BC-Based Ubiquitin Ligases. J. Biol. Chem. 2008, 283, 8005–8013. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Wei, Y.; Reboul, J.; Vaglio, P.; Shin, T.-H.; Vidal, M.; Elledge, S.J.; Harper, J.W. BTB Proteins Are Substrate-Specific Adaptors in an SCF-like Modular Ubiquitin Ligase Containing CUL-3. Nature 2003, 425, 316–321. [Google Scholar] [CrossRef] [PubMed]
- Geyer, R.; Wee, S.; Anderson, S.; Yates, J.; Wolf, D.A. BTB/POZ Domain Proteins Are Putative Substrate Adaptors for Cullin 3 Ubiquitin Ligases. Mol. Cell 2003, 12, 783–790. [Google Scholar] [CrossRef] [PubMed]
- Hannah, J.; Zhou, P. Distinct and Overlapping Functions of the Cullin E3 Ligase Scaffolding Proteins CUL4A and CUL4B. Gene 2015, 573, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Angers, S.; Li, T.; Yi, X.; MacCoss, M.J.; Moon, R.T.; Zheng, N. Molecular Architecture and Assembly of the DDB1–CUL4A Ubiquitin Ligase Machinery. Nature 2006, 443, 590–593. [Google Scholar] [CrossRef] [PubMed]
- Fischer, E.S.; Scrima, A.; Böhm, K.; Matsumoto, S.; Lingaraju, G.M.; Faty, M.; Yasuda, T.; Cavadini, S.; Wakasugi, M.; Hanaoka, F.; et al. The Molecular Basis of CRL4DDB2/CSA Ubiquitin Ligase Architecture, Targeting, and Activation. Cell 2011, 147, 1024–1039. [Google Scholar] [CrossRef] [PubMed]
- Skaar, J.R.; Pagan, J.K.; Pagano, M. Mechanisms and Function of Substrate Recruitment by F-Box Proteins. Nat. Rev. Mol. Cell Biol. 2013, 14, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Reitsma, J.M.; Mamrosh, J.L.; Zhang, Y.; Straube, R.; Deshaies, R.J. Cand1-Mediated Adaptive Exchange Mechanism Enables Variation in F-Box Protein Expression. Mol. Cell 2018, 69, 773–786.e6. [Google Scholar] [CrossRef]
- Metzger, M.B.; Pruneda, J.N.; Klevit, R.E.; Weissman, A.M. RING-Type E3 Ligases: Master Manipulators of E2 Ubiquitin-Conjugating Enzymes and Ubiquitination. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2014, 1843, 47–60. [Google Scholar] [CrossRef]
- Lucas, X.; Ciulli, A. Recognition of Substrate Degrons by E3 Ubiquitin Ligases and Modulation by Small-Molecule Mimicry Strategies. Curr. Opin. Struct. Biol. 2017, 44, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Schneekloth, J.S.; Fonseca, F.N.; Koldobskiy, M.; Mandal, A.; Deshaies, R.; Sakamoto, K.; Crews, C.M. Chemical Genetic Control of Protein Levels: Selective In Vivo Targeted Degradation. J. Am. Chem. Soc. 2004, 126, 3748–3754. [Google Scholar] [CrossRef] [PubMed]
- Galdeano, C.; Gadd, M.S.; Soares, P.; Scaffidi, S.; Van Molle, I.; Birced, I.; Hewitt, S.; Dias, D.M.; Ciulli, A. Structure-Guided Design and Optimization of Small Molecules Targeting the Protein–Protein Interaction between the von Hippel–Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities. J. Med. Chem. 2014, 57, 8657–8663. [Google Scholar] [CrossRef] [PubMed]
- Soares, P.; Gadd, M.S.; Frost, J.; Galdeano, C.; Ellis, L.; Epemolu, O.; Rocha, S.; Read, K.D.; Ciulli, A. Group-Based Optimization of Potent and Cell-Active Inhibitors of the von Hippel–Lindau (VHL) E3 Ubiquitin Ligase: Structure–Activity Relationships Leading to the Chemical Probe (2S,4R)-1-((S)-2-(1-Cyanocyclopropanecarboxamido)-3,3-Dimethylbutanoyl)-4-Hydroxy-N-(4-(4-Methylthiazol-5-Yl)Benzyl)Pyrrolidine-2-Carboxamide (VH298). J. Med. Chem. 2018, 61, 599–618. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Ando, H.; Suzuki, T.; Ogura, T.; Hotta, K.; Imamura, Y.; Yamaguchi, Y.; Handa, H. Identification of a Primary Target of Thalidomide Teratogenicity. Science 2010, 327, 1345–1350. [Google Scholar] [CrossRef] [PubMed]
- Chamberlain, P.P.; Lopez-Girona, A.; Miller, K.; Carmel, G.; Pagarigan, B.; Chie-Leon, B.; Rychak, E.; Corral, L.G.; Ren, Y.J.; Wang, M.; et al. Structure of the Human Cereblon–DDB1–Lenalidomide Complex Reveals Basis for Responsiveness to Thalidomide Analogs. Nat. Struct. Mol. Biol. 2014, 21, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Krönke, J.; Udeshi, N.D.; Narla, A.; Grauman, P.; Hurst, S.N.; McConkey, M.; Svinkina, T.; Heckl, D.; Comer, E.; Li, X.; et al. Lenalidomide Causes Selective Degradation of IKZF1 and IKZF3 in Multiple Myeloma Cells. Science 2014, 343, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Middleton, R.E.; Sun, H.; Naniong, M.; Ott, C.J.; Mitsiades, C.S.; Wong, K.-K.; Bradner, J.E.; Kaelin, W.G. The Myeloma Drug Lenalidomide Promotes the Cereblon-Dependent Destruction of Ikaros Proteins. Science 2014, 343, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Krönke, J.; Fink, E.C.; Hollenbach, P.W.; MacBeth, K.J.; Hurst, S.N.; Udeshi, N.D.; Chamberlain, P.P.; Mani, D.R.; Man, H.W.; Gandhi, A.K.; et al. Lenalidomide Induces Ubiquitination and Degradation of CK1α in Del(5q) MDS. Nature 2015, 523, 183–188. [Google Scholar] [CrossRef]
- Słabicki, M.; Yoon, H.; Koeppel, J.; Nitsch, L.; Roy Burman, S.S.; Di Genua, C.; Donovan, K.A.; Sperling, A.S.; Hunkeler, M.; Tsai, J.M.; et al. Small-Molecule-Induced Polymerization Triggers Degradation of BCL6. Nature 2020, 588, 164–168. [Google Scholar] [CrossRef]
- House, C.M.; Frew, I.J.; Huang, H.-L.; Wiche, G.; Traficante, N.; Nice, E.; Catimel, B.; Bowtell, D.D.L. A Binding Motif for Siah Ubiquitin Ligase. Proc. Natl. Acad. Sci. USA 2003, 100, 3101–3106. [Google Scholar] [CrossRef] [PubMed]
- McCoull, W.; Cheung, T.; Anderson, E.; Barton, P.; Burgess, J.; Byth, K.; Cao, Q.; Castaldi, M.P.; Chen, H.; Chiarparin, E.; et al. Development of a Novel B-Cell Lymphoma 6 (BCL6) PROTAC To Provide Insight into Small Molecule Targeting of BCL6. ACS Chem. Biol. 2018, 13, 3131–3141. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Qian, Y.; Altieri, M.; Dong, H.; Wang, J.; Raina, K.; Hines, J.; Winkler, J.D.; Crew, A.P.; Coleman, K.; et al. Hijacking the E3 Ubiquitin Ligase Cereblon to Efficiently Target BRD4. Chem. Biol. 2015, 22, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, Y.; Wu, Y.; Xing, D. Developments of CRBN-Based PROTACs as Potential Therapeutic Agents. Eur. J. Med. Chem. 2021, 225, 113749. [Google Scholar] [CrossRef] [PubMed]
- Cieślak, M.; Słowianek, M. Cereblon-Recruiting PROTACs: Will New Drugs Have to Face Old Challenges? Pharmaceutics 2023, 15, 812. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Robert, E.I.; Van Breugel, P.C.; Strubin, M.; Zheng, N. A Promiscuous α-Helical Motif Anchors Viral Hijackers and Substrate Receptors to the CUL4–DDB1 Ubiquitin Ligase Machinery. Nat. Struct. Mol. Biol. 2010, 17, 105–111. [Google Scholar] [CrossRef]
- Meyers, M.; Cismoski, S.; Panidapu, A.; Chie-Leon, B.; Nomura, D.K. Targeted Protein Degradation through Recruitment of the CUL4 Complex Adaptor Protein DDB1. ACS Chem. Biol. 2024, 19, 58–68. [Google Scholar] [CrossRef]
- Hong, S.H.; Divakaran, A.; Osa, A.; Huang, O.W.; Wertz, I.E.; Nomura, D.K. Exploiting the Cullin E3 Ligase Adaptor Protein SKP1 for Targeted Protein Degradation. ACS Chem. Biol. 2024, 19, 442–450. [Google Scholar] [CrossRef] [PubMed]
- King, E.A.; Cho, Y.; Hsu, N.S.; Dovala, D.; McKenna, J.M.; Tallarico, J.A.; Schirle, M.; Nomura, D.K. Chemoproteomics-Enabled Discovery of a Covalent Molecular Glue Degrader Targeting NF-κB. Cell Chem. Biol. 2023, 30, 394–402.e9. [Google Scholar] [CrossRef] [PubMed]
- Forte, N.; Dovala, D.; Hesse, M.J.; McKenna, J.M.; Tallarico, J.A.; Schirle, M.; Nomura, D.K. Targeted Protein Degradation through E2 Recruitment. ACS Chem. Biol. 2023, 18, 897–904. [Google Scholar] [CrossRef]
- Bashore, C.; Prakash, S.; Johnson, M.C.; Conrad, R.J.; Kekessie, I.A.; Scales, S.J.; Ishisoko, N.; Kleinheinz, T.; Liu, P.S.; Popovych, N.; et al. Targeted Degradation via Direct 26S Proteasome Recruitment. Nat. Chem. Biol. 2023, 19, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Ciulli, A.; Testa, A.; Hughes, S.; Butcher, S.P. Butcher Bifunctional Molecules for Targeting Uchl5. U.S. Patent No. 20210283139A1, 16 September 2021. [Google Scholar]
- Ciulli, A.; Testa, A.; Hughes, S.; Butcher, S.P. Butcher Bifunctional Molecules for Targeting Rpn11. U.S. Patent No. 20210260200A1, 26 August 2021. [Google Scholar]
- Testa, A.; Hughes, S.; Butcher, S.P.; Ciulli, A. Bifunctional Molecules for Targeting Usp14. WO Patent No. 2019238886A1, 19 December 2019. [Google Scholar]
- Ali, E.M.H.; Loy, C.A.; Trader, D.J. ByeTAC: Bypassing an E3 Ligase for Targeted Protein Degradation. bioRxiv 2024. [Google Scholar] [CrossRef]
- Miyamoto-Sato, E.; Imanishi, S.; Huang, L.; Itakura, S.; Iwasaki, Y.; Ishizaka, M. A First-Class Degrader Candidate Targeting Both KRAS G12D and G12V Mediated by CANDDY Technology Independent of Ubiquitination. Molecules 2023, 28, 5600. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Zhou, H.; Xu, R.; Zhao, Y.; Chinnaswamy, K.; McEachern, D.; Chen, J.; Yang, C.-Y.; Liu, Z.; Wang, M.; et al. A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo. Cancer Cell 2019, 36, 498–511.e17. [Google Scholar] [CrossRef] [PubMed]
- Tong, B.; Spradlin, J.N.; Novaes, L.F.T.; Zhang, E.; Hu, X.; Moeller, M.; Brittain, S.M.; McGregor, L.M.; McKenna, J.M.; Tallarico, J.A.; et al. A Nimbolide-Based Kinase Degrader Preferentially Degrades Oncogenic BCR-ABL. ACS Chem. Biol. 2020, 15, 1788–1794. [Google Scholar] [CrossRef]
- Luo, M.; Spradlin, J.N.; Boike, L.; Tong, B.; Brittain, S.M.; McKenna, J.M.; Tallarico, J.A.; Schirle, M.; Maimone, T.J.; Nomura, D.K. Chemoproteomics-Enabled Discovery of Covalent RNF114-Based Degraders That Mimic Natural Product Function. Cell Chem. Biol. 2021, 28, 559–566.e15. [Google Scholar] [CrossRef] [PubMed]
- De Nardo, D.; Balka, K.R.; Cardona Gloria, Y.; Rao, V.R.; Latz, E.; Masters, S.L. Interleukin-1 Receptor–Associated Kinase 4 (IRAK4) Plays a Dual Role in Myddosome Formation and Toll-like Receptor Signaling. J. Biol. Chem. 2018, 293, 15195–15207. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Sano, R.; Apatira, M.; DeAnda, F.; Gururaja, T.; Yang, M.; Lundgaard, G.; Pan, C.; Liu, J.; Zhai, Y.; et al. Bruton’s Tyrosine Kinase Inhibitors with Distinct Binding Modes Reveal Differential Functional Impact on B-Cell Receptor Signaling. Mol. Cancer Ther. 2024, 23, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Montoya, S.; Bourcier, J.; Noviski, M.; Lu, H.; Thompson, M.C.; Chirino, A.; Jahn, J.; Sondhi, A.K.; Gajewski, S.; Tan, Y.S.; et al. Kinase-Impaired BTK Mutations Are Susceptible to Clinical-Stage BTK and IKZF1/3 Degrader NX-2127. Science 2024, 383, eadi5798. [Google Scholar] [CrossRef]
- Yuan, H.; Zhu, Y.; Cheng, Y.; Hou, J.; Jin, F.; Li, M.; Jia, W.; Cheng, Z.; Xing, H.; Liu, M.; et al. BTK Kinase Activity Is Dispensable for the Survival of Diffuse Large B-Cell Lymphoma. J. Biol. Chem. 2022, 298, 102555. [Google Scholar] [CrossRef]
- Ackerman, L.; Acloque, G.; Bacchelli, S.; Schwartz, H.; Feinstein, B.J.; La Stella, P.; Alavi, A.; Gollerkeri, A.; Davis, J.; Campbell, V.; et al. IRAK4 Degrader in Hidradenitis Suppurativa and Atopic Dermatitis: A Phase 1 Trial. Nat. Med. 2023, 29, 3127–3136. [Google Scholar] [CrossRef]
- Lu, H.; Zhou, Q.; He, J.; Jiang, Z.; Peng, C.; Tong, R.; Shi, J. Recent Advances in the Development of Protein–Protein Interactions Modulators: Mechanisms and Clinical Trials. Signal Transduct. Target. Ther. 2020, 5, 213. [Google Scholar] [CrossRef]
- Wang, Z.; He, N.; Guo, Z.; Niu, C.; Song, T.; Guo, Y.; Cao, K.; Wang, A.; Zhu, J.; Zhang, X.; et al. Proteolysis Targeting Chimeras for the Selective Degradation of Mcl-1/Bcl-2 Derived from Nonselective Target Binding Ligands. J. Med. Chem. 2019, 62, 8152–8163. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Yang, Z.; Gao, H.; Yang, H.; Zhu, S.; An, Z.; Wang, J.; Li, Q.; Chandarlapaty, S.; Deng, H.; et al. Potent and Preferential Degradation of CDK6 via Proteolysis Targeting Chimera Degraders. J. Med. Chem. 2019, 62, 7575–7582. [Google Scholar] [CrossRef]
- Druker, B.J.; Tamura, S.; Buchdunger, E.; Ohno, S.; Segal, G.M.; Fanning, S.; Zimmermann, J.; Lydon, N.B. Effects of a Selective Inhibitor of the Abl Tyrosine Kinase on the Growth of Bcr–Abl Positive Cells. Nat. Med. 1996, 2, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Braun, T.P.; Eide, C.A.; Druker, B.J. Response and Resistance to BCR-ABL1-Targeted Therapies. Cancer Cell 2020, 37, 530–542. [Google Scholar] [CrossRef]
- Lai, A.C.; Toure, M.; Hellerschmied, D.; Salami, J.; Jaime-Figueroa, S.; Ko, E.; Hines, J.; Crews, C.M. Modular PROTAC Design for the Degradation of Oncogenic BCR-ABL. Angew. Chem. Int. Ed. 2016, 55, 807–810. [Google Scholar] [CrossRef]
- Liu, H.; Ding, X.; Liu, L.; Mi, Q.; Zhao, Q.; Shao, Y.; Ren, C.; Chen, J.; Kong, Y.; Qiu, X.; et al. Discovery of Novel BCR-ABL PROTACs Based on the Cereblon E3 Ligase Design, Synthesis, and Biological Evaluation. Eur. J. Med. Chem. 2021, 223, 113645. [Google Scholar] [CrossRef]
- Burslem, G.M.; Schultz, A.R.; Bondeson, D.P.; Eide, C.A.; Savage Stevens, S.L.; Druker, B.J.; Crews, C.M. Targeting BCR-ABL1 in Chronic Myeloid Leukemia by PROTAC-Mediated Targeted Protein Degradation. Cancer Res. 2019, 79, 4744–4753. [Google Scholar] [CrossRef] [PubMed]
- Lyczek, A.; Berger, B.-T.; Rangwala, A.M.; Paung, Y.; Tom, J.; Philipose, H.; Guo, J.; Albanese, S.K.; Robers, M.B.; Knapp, S.; et al. Mutation in Abl Kinase with Altered Drug-Binding Kinetics Indicates a Novel Mechanism of Imatinib Resistance. Proc. Natl. Acad. Sci. USA 2021, 118, e2111451118. [Google Scholar] [CrossRef]
- Heinlein, C.A.; Chang, C. Androgen Receptor in Prostate Cancer. Endocr. Rev. 2004, 25, 276–308. [Google Scholar] [CrossRef] [PubMed]
- Balbas, M.D.; Evans, M.J.; Hosfield, D.J.; Wongvipat, J.; Arora, V.K.; Watson, P.A.; Chen, Y.; Greene, G.L.; Shen, Y.; Sawyers, C.L. Overcoming Mutation-Based Resistance to Antiandrogens with Rational Drug Design. eLife 2013, 2, e00499. [Google Scholar] [CrossRef] [PubMed]
- Salami, J.; Alabi, S.; Willard, R.R.; Vitale, N.J.; Wang, J.; Dong, H.; Jin, M.; McDonnell, D.P.; Crew, A.P.; Neklesa, T.K.; et al. Androgen Receptor Degradation by the Proteolysis-Targeting Chimera ARCC-4 Outperforms Enzalutamide in Cellular Models of Prostate Cancer Drug Resistance. Commun. Biol. 2018, 1, 100. [Google Scholar] [CrossRef] [PubMed]
- Kregel, S.; Wang, C.; Han, X.; Xiao, L.; Fernandez-Salas, E.; Bawa, P.; McCollum, B.L.; Wilder-Romans, K.; Apel, I.J.; Cao, X.; et al. Androgen Receptor Degraders Overcome Common Resistance Mechanisms Developed during Prostate Cancer Treatment. Neoplasia 2020, 22, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.T.; Nagaya, N.; Desantis, J.; Madura, K.; Sabaawy, H.E.; Kim, W.-J.; Vaz, R.J.; Cruciani, G.; Kim, I.Y. Effects of MTX-23, a Novel PROTAC of Androgen Receptor Splice Variant-7 and Androgen Receptor, on CRPC Resistant to Second-Line Antiandrogen Therapy. Mol. Cancer Ther. 2021, 20, 490–499. [Google Scholar] [CrossRef] [PubMed]
- Shirasaki, R.; Matthews, G.M.; Gandolfi, S.; De Matos Simoes, R.; Buckley, D.L.; Raja Vora, J.; Sievers, Q.L.; Brüggenthies, J.B.; Dashevsky, O.; Poarch, H.; et al. Functional Genomics Identify Distinct and Overlapping Genes Mediating Resistance to Different Classes of Heterobifunctional Degraders of Oncoproteins. Cell Rep. 2021, 34, 108532. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Riley-Gillis, B.; Vijay, P.; Shen, Y. Acquired Resistance to BET-PROTACs (Proteolysis-Targeting Chimeras) Caused by Genomic Alterations in Core Components of E3 Ligase Complexes. Mol. Cancer Ther. 2019, 18, 1302–1311. [Google Scholar] [CrossRef] [PubMed]
- Ottis, P.; Palladino, C.; Thienger, P.; Britschgi, A.; Heichinger, C.; Berrera, M.; Julien-Laferriere, A.; Roudnicky, F.; Kam-Thong, T.; Bischoff, J.R.; et al. Cellular Resistance Mechanisms to Targeted Protein Degradation Converge Toward Impairment of the Engaged Ubiquitin Transfer Pathway. ACS Chem. Biol. 2019, 14, 2215–2223. [Google Scholar] [CrossRef] [PubMed]
- Békés, M.; Langley, D.R.; Crews, C.M. PROTAC Targeted Protein Degraders: The Past Is Prologue. Nat. Rev. Drug Discov. 2022, 21, 181–200. [Google Scholar] [CrossRef]
- Gechijian, L.N.; Buckley, D.L.; Lawlor, M.A.; Reyes, J.M.; Paulk, J.; Ott, C.J.; Winter, G.E.; Erb, M.A.; Scott, T.G.; Xu, M.; et al. Functional TRIM24 Degrader via Conjugation of Ineffectual Bromodomain and VHL Ligands. Nat. Chem. Biol. 2018, 14, 405–412. [Google Scholar] [CrossRef]
- Gironda-Martínez, A.; Donckele, E.J.; Samain, F.; Neri, D. DNA-Encoded Chemical Libraries: A Comprehensive Review with Succesful Stories and Future Challenges. ACS Pharmacol. Transl. Sci. 2021, 4, 1265–1279. [Google Scholar] [CrossRef]
- Satz, A.L.; Brunschweiger, A.; Flanagan, M.E.; Gloger, A.; Hansen, N.J.V.; Kuai, L.; Kunig, V.B.K.; Lu, X.; Madsen, D.; Marcaurelle, L.A.; et al. DNA-Encoded Chemical Libraries. Nat. Rev. Methods Primers 2022, 2, 3. [Google Scholar] [CrossRef]
- Prudent, R.; Annis, D.A.; Dandliker, P.J.; Ortholand, J.-Y.; Roche, D. Exploring New Targets and Chemical Space with Affinity Selection-Mass Spectrometry. Nat. Rev. Chem. 2020, 5, 62–71. [Google Scholar] [CrossRef]
- Disch, J.S.; Duffy, J.M.; Lee, E.C.Y.; Gikunju, D.; Chan, B.; Levin, B.; Monteiro, M.I.; Talcott, S.A.; Lau, A.C.; Zhou, F.; et al. Bispecific Estrogen Receptor α Degraders Incorporating Novel Binders Identified Using DNA-Encoded Chemical Library Screening. J. Med. Chem. 2021, 64, 5049–5066. [Google Scholar] [CrossRef] [PubMed]
- Chana, C.K.; Maisonneuve, P.; Posternak, G.; Grinberg, N.G.A.; Poirson, J.; Ona, S.M.; Ceccarelli, D.F.; Mader, P.; St-Cyr, D.J.; Pau, V.; et al. Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4. J. Med. Chem. 2022, 65, 12725–12746. [Google Scholar] [CrossRef] [PubMed]
- Yazdi, A.K.; Perveen, S.; Song, X.; Dong, A.; Szewczyk, M.M.; Calabrese, M.F.; Casimiro-Garcia, A.; Chakrapani, S.; Dowling, M.S.; Ficici, E.; et al. Chemical Tools for the Gid4 Subunit of the Human E3 Ligase C-Terminal to LisH (CTLH) Degradation Complex. bioRxiv 2023. [Google Scholar] [CrossRef] [PubMed]
- Schneekloth, A.R.; Pucheault, M.; Tae, H.S.; Crews, C.M. Targeted Intracellular Protein Degradation Induced by a Small Molecule: En Route to Chemical Proteomics. Bioorganic Med. Chem. Lett. 2008, 18, 5904–5908. [Google Scholar] [CrossRef] [PubMed]
- Buckley, D.L.; Raina, K.; Darricarrere, N.; Hines, J.; Gustafson, J.L.; Smith, I.E.; Miah, A.H.; Harling, J.D.; Crews, C.M. HaloPROTACS: Use of Small Molecule PROTACs to Induce Degradation of HaloTag Fusion Proteins. ACS Chem. Biol. 2015, 10, 1831–1837. [Google Scholar] [CrossRef] [PubMed]
- Nabet, B.; Roberts, J.M.; Buckley, D.L.; Paulk, J.; Dastjerdi, S.; Yang, A.; Leggett, A.L.; Erb, M.A.; Lawlor, M.A.; Souza, A.; et al. The DTAG System for Immediate and Target-Specific Protein Degradation. Nat. Chem. Biol. 2018, 14, 431–441. [Google Scholar] [CrossRef]
- Bensimon, A.; Pizzagalli, M.D.; Kartnig, F.; Dvorak, V.; Essletzbichler, P.; Winter, G.E.; Superti-Furga, G. Targeted Degradation of SLC Transporters Reveals Amenability of Multi-Pass Transmembrane Proteins to Ligand-Induced Proteolysis. Cell Chem. Biol. 2020, 27, 728–739.e9. [Google Scholar] [CrossRef]
- Ege, N.; Bouguenina, H.; Tatari, M.; Chopra, R. Phenotypic Screening with Target Identification and Validation in the Discovery and Development of E3 Ligase Modulators. Cell Chem. Biol. 2021, 28, 283–299. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; He, M.; Wang, L.; He, Y.; Rao, Y. Chemistries of Bifunctional PROTAC Degraders. Chem. Soc. Rev. 2022, 51, 7066–7114. [Google Scholar] [CrossRef]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Žídek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A.; et al. Highly Accurate Protein Structure Prediction for the Human Proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D.; et al. Accurate Prediction of Protein Structures and Interactions Using a Three-Track Neural Network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef]
- Drummond, M.L.; Williams, C.I. In Silico Modeling of PROTAC-Mediated Ternary Complexes: Validation and Application. J. Chem. Inf. Model. 2019, 59, 1634–1644. [Google Scholar] [CrossRef] [PubMed]
- Drummond, M.L.; Henry, A.; Li, H.; Williams, C.I. Improved Accuracy for Modeling PROTAC-Mediated Ternary Complex Formation and Targeted Protein Degradation via New In Silico Methodologies. J. Chem. Inf. Model. 2020, 60, 5234–5254. [Google Scholar] [CrossRef]
- Zaidman, D.; Prilusky, J.; London, N. PRosettaC: Rosetta Based Modeling of PROTAC Mediated Ternary Complexes. J. Chem. Inf. Model. 2020, 60, 4894–4903. [Google Scholar] [CrossRef] [PubMed]
- Bai, N.; Miller, S.A.; Andrianov, G.V.; Yates, M.; Kirubakaran, P.; Karanicolas, J. Rationalizing PROTAC-Mediated Ternary Complex Formation Using Rosetta. J. Chem. Inf. Model. 2021, 61, 1368–1382. [Google Scholar] [CrossRef]
- Weng, G.; Li, D.; Kang, Y.; Hou, T. Integrative Modeling of PROTAC-Mediated Ternary Complexes. J. Med. Chem. 2021, 64, 16271–16281. [Google Scholar] [CrossRef]
- Troup, R.I.; Fallan, C.; Baud, M.G.J. Current Strategies for the Design of PROTAC Linkers: A Critical Review. Explor. Target. Anti-Tumor Ther. 2020, 1, 273–312. [Google Scholar] [CrossRef]
- Sun, X.; Gao, H.; Yang, Y.; He, M.; Wu, Y.; Song, Y.; Tong, Y.; Rao, Y. PROTACs: Great Opportunities for Academia and Industry. Signal Transduct. Target. Ther. 2019, 4, 64. [Google Scholar] [CrossRef] [PubMed]
- Burslem, G.M.; Crews, C.M. Small-Molecule Modulation of Protein Homeostasis. Chem. Rev. 2017, 117, 11269–11301. [Google Scholar] [CrossRef] [PubMed]
- Maple, H.J.; Clayden, N.; Baron, A.; Stacey, C.; Felix, R. Developing Degraders: Principles and Perspectives on Design and Chemical Space. Med. Chem. Commun. 2019, 10, 1755–1764. [Google Scholar] [CrossRef] [PubMed]
- Borsari, C.; Trader, D.J.; Tait, A.; Costi, M.P. Designing Chimeric Molecules for Drug Discovery by Leveraging Chemical Biology. J. Med. Chem. 2020, 63, 1908–1928. [Google Scholar] [CrossRef] [PubMed]
- Hendrick, C.E.; Jorgensen, J.R.; Chaudhry, C.; Strambeanu, I.I.; Brazeau, J.-F.; Schiffer, J.; Shi, Z.; Venable, J.D.; Wolkenberg, S.E. Direct-to-Biology Accelerates PROTAC Synthesis and the Evaluation of Linker Effects on Permeability and Degradation. ACS Med. Chem. Lett. 2022, 13, 1182–1190. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.; Merritt, A.; Skerratt, S.; Swarbrick, M.E. Recent Trends in Medicinal Chemistry and Enabling Technologies. Highlights from the Society for Medicines Research Conference. London—December 8, 2022. Drugs Future 2023, 48, 211–219. [Google Scholar] [CrossRef]
- Plesniak, M.P.; Taylor, E.K.; Eisele, F.; Kourra, C.M.B.K.; Michaelides, I.N.; Oram, A.; Wernevik, J.; Valencia, Z.S.; Rowbottom, H.; Mann, N.; et al. Rapid PROTAC Discovery Platform: Nanomole-Scale Array Synthesis and Direct Screening of Reaction Mixtures. ACS Med. Chem. Lett. 2023, 14, 1882–1890. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Girona, A.; Mendy, D.; Ito, T.; Miller, K.; Gandhi, A.K.; Kang, J.; Karasawa, S.; Carmel, G.; Jackson, P.; Abbasian, M.; et al. Cereblon Is a Direct Protein Target for Immunomodulatory and Antiproliferative Activities of Lenalidomide and Pomalidomide. Leukemia 2012, 26, 2326–2335. [Google Scholar] [CrossRef] [PubMed]
- Owa, T.; Yoshino, H.; Okauchi, T.; Yoshimatsu, K.; Ozawa, Y.; Sugi, N.H.; Nagasu, T.; Koyanagi, N.; Kitoh, K. Discovery of Novel Antitumor Sulfonamides Targeting G1 Phase of the Cell Cycle. J. Med. Chem. 1999, 42, 3789–3799. [Google Scholar] [CrossRef]
- Han, T.; Goralski, M.; Gaskill, N.; Capota, E.; Kim, J.; Ting, T.C.; Xie, Y.; Williams, N.S.; Nijhawan, D. Anticancer Sulfonamides Target Splicing by Inducing RBM39 Degradation via Recruitment to DCAF15. Science 2017, 356, eaal3755. [Google Scholar] [CrossRef]
- Bussiere, D.E.; Xie, L.; Srinivas, H.; Shu, W.; Burke, A.; Be, C.; Zhao, J.; Godbole, A.; King, D.; Karki, R.G.; et al. Structural Basis of Indisulam-Mediated RBM39 Recruitment to DCAF15 E3 Ligase Complex. Nat. Chem. Biol. 2020, 16, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Kozicka, Z.; Suchyta, D.J.; Focht, V.; Kempf, G.; Petzold, G.; Jentzsch, M.; Zou, C.; Di Genua, C.; Donovan, K.A.; Coomar, S.; et al. Design Principles for Cyclin K Molecular Glue Degraders. Nat. Chem. Biol. 2024, 20, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Bray, M.-A.; Singh, S.; Han, H.; Davis, C.T.; Borgeson, B.; Hartland, C.; Kost-Alimova, M.; Gustafsdottir, S.M.; Gibson, C.C.; Carpenter, A.E. Cell Painting, a High-Content Image-Based Assay for Morphological Profiling Using Multiplexed Fluorescent Dyes. Nat. Protoc. 2016, 11, 1757–1774. [Google Scholar] [CrossRef] [PubMed]
- Ng, A.; Offensperger, F.; Cisneros, J.A.; Scholes, N.S.; Malik, M.; Villanti, L.; Rukavina, A.; Ferrada, E.; Hannich, J.T.; Koren, A.; et al. Discovery of Molecular Glue Degraders via Isogenic Morphological Profiling. ACS Chem. Biol. 2023, 18, 2464–2473. [Google Scholar] [CrossRef] [PubMed]
- Bonazzi, S.; d’Hennezel, E.; Beckwith, R.E.J.; Xu, L.; Fazal, A.; Magracheva, A.; Ramesh, R.; Cernijenko, A.; Antonakos, B.; Bhang, H.C.; et al. Discovery and Characterization of a Selective IKZF2 Glue Degrader for Cancer Immunotherapy. Cell Chem. Biol. 2023, 30, 235–247.e12. [Google Scholar] [CrossRef] [PubMed]
- Tutter, A.; Buckley, D.; Golosov, A.A.; Ma, X.; Shu, W.; McKay, D.J.J.; Darsigny, V.; Dovala, D.; Beckwith, R.; Solomon, J.; et al. A Small Molecule VHL Molecular Glue Degrader for Cysteine Dioxygenase 1. bioRxiv 2024. [Google Scholar] [CrossRef]
- Simonetta, K.R.; Taygerly, J.; Boyle, K.; Basham, S.E.; Padovani, C.; Lou, Y.; Cummins, T.J.; Yung, S.L.; Von Soly, S.K.; Kayser, F.; et al. Prospective Discovery of Small Molecule Enhancers of an E3 Ligase-Substrate Interaction. Nat. Commun. 2019, 10, 1402. [Google Scholar] [CrossRef] [PubMed]
- Buckley, D.L.; Van Molle, I.; Gareiss, P.C.; Tae, H.S.; Michel, J.; Noblin, D.J.; Jorgensen, W.L.; Ciulli, A.; Crews, C.M. Targeting the von Hippel–Lindau E3 Ubiquitin Ligase Using Small Molecules To Disrupt the VHL/HIF-1α Interaction. J. Am. Chem. Soc. 2012, 134, 4465–4468. [Google Scholar] [CrossRef] [PubMed]
- Zoppi, V.; Hughes, S.J.; Maniaci, C.; Testa, A.; Gmaschitz, T.; Wieshofer, C.; Koegl, M.; Riching, K.M.; Daniels, D.L.; Spallarossa, A.; et al. Iterative Design and Optimization of Initially Inactive Proteolysis Targeting Chimeras (PROTACs) Identify VZ185 as a Potent, Fast, and Selective von Hippel–Lindau (VHL) Based Dual Degrader Probe of BRD9 and BRD7. J. Med. Chem. 2019, 62, 699–726. [Google Scholar] [CrossRef]
- Smith, B.E.; Wang, S.L.; Jaime-Figueroa, S.; Harbin, A.; Wang, J.; Hamman, B.D.; Crews, C.M. Differential PROTAC Substrate Specificity Dictated by Orientation of Recruited E3 Ligase. Nat. Commun. 2019, 10, 131. [Google Scholar] [CrossRef]
- Daniels, D.L.; Riching, K.M.; Urh, M. Monitoring and Deciphering Protein Degradation Pathways inside Cells. Drug Discov. Today Technol. 2019, 31, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Riching, K.M.; Mahan, S.; Corona, C.R.; McDougall, M.; Vasta, J.D.; Robers, M.B.; Urh, M.; Daniels, D.L. Quantitative Live-Cell Kinetic Degradation and Mechanistic Profiling of PROTAC Mode of Action. ACS Chem. Biol. 2018, 13, 2758–2770. [Google Scholar] [CrossRef] [PubMed]
- Hyer, M.L.; Milhollen, M.A.; Ciavarri, J.; Fleming, P.; Traore, T.; Sappal, D.; Huck, J.; Shi, J.; Gavin, J.; Brownell, J.; et al. A Small-Molecule Inhibitor of the Ubiquitin Activating Enzyme for Cancer Treatment. Nat. Med. 2018, 24, 186–193. [Google Scholar] [CrossRef]
- Soucy, T.A.; Smith, P.G.; Milhollen, M.A.; Berger, A.J.; Gavin, J.M.; Adhikari, S.; Brownell, J.E.; Burke, K.E.; Cardin, D.P.; Critchley, S.; et al. An Inhibitor of NEDD8-Activating Enzyme as a New Approach to Treat Cancer. Nature 2009, 458, 732–736. [Google Scholar] [CrossRef] [PubMed]
- Degorce, F. HTRF: A Technology Tailored for Drug Discovery—A Review of Theoretical Aspects and Recent Applications. Curr. Chem. Genom. 2009, 3, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Beaudet, L.; Rodriguez-Suarez, R.; Venne, M.-H.; Caron, M.; Bédard, J.; Brechler, V.; Parent, S.; Bielefeld-Sévigny, M. AlphaLISA Immunoassays: The No-Wash Alternative to ELISAs for Research and Drug Discovery. Nat. Methods 2008, 5, an8–an9. [Google Scholar] [CrossRef]
- Vetma, V.; Casarez-Perez, L.; Eliaš, J.; Stingu, A.; Kombara, A.; Gmaschitz, T.; Braun, N.; Ciftci, T.; Dahmann, G.; Diers, E.; et al. Confounding Factors in Targeted Degradation of Short-Lived Proteins. bioRxiv 2024. [Google Scholar] [CrossRef]
- Hjerpe, R.; Aillet, F.; Lopitz-Otsoa, F.; Lang, V.; England, P.; Rodriguez, M.S. Efficient Protection and Isolation of Ubiquitylated Proteins Using Tandem Ubiquitin-binding Entities. EMBO Rep. 2009, 10, 1250–1258. [Google Scholar] [CrossRef]
- Serna, S.; Xolalpa, W.; Lang, V.; Aillet, F.; England, P.; Reichardt, N.; Rodriguez, M.S. Efficient Monitoring of Protein Ubiquitylation Levels Using TUBE S-based Microarrays. FEBS Lett. 2016, 590, 2748–2756. [Google Scholar] [CrossRef]
- Huang, H.-T.; Dobrovolsky, D.; Paulk, J.; Yang, G.; Weisberg, E.L.; Doctor, Z.M.; Buckley, D.L.; Cho, J.-H.; Ko, E.; Jang, J.; et al. A Chemoproteomic Approach to Query the Degradable Kinome Using a Multi-Kinase Degrader. Cell Chem. Biol. 2018, 25, 88–99.e6. [Google Scholar] [CrossRef]
- Zeng, S.; Ye, Y.; Xia, H.; Min, J.; Xu, J.; Wang, Z.; Pan, Y.; Zhou, X.; Huang, W. Current Advances and Development Strategies of Orally Bioavailable PROTACs. Eur. J. Med. Chem. 2023, 261, 115793. [Google Scholar] [CrossRef] [PubMed]
- Schott, A.F.; Hurvitz, S.; Ma, C.; Hamilton, E.; Nanda, R.; Zahrah, G.; Hunter, N.; Tan, A.R.; Telli, M.; Mesias, J.A.; et al. Abstract GS3-03: GS3-03 ARV-471, a PROTAC® Estrogen Receptor (ER) Degrader in Advanced ER-Positive/Human Epidermal Growth Factor Receptor 2 (HER2)-Negative Breast Cancer: Phase 2 Expansion (VERITAC) of a Phase 1/2 Study. Cancer Res. 2023, 83, GS3-03. [Google Scholar] [CrossRef]
- Gao, X.; Burris Iii, H.A.; Vuky, J.; Dreicer, R.; Sartor, A.O.; Sternberg, C.N.; Percent, I.J.; Hussain, M.H.A.; Rezazadeh Kalebasty, A.; Shen, J.; et al. Phase 1/2 Study of ARV-110, an Androgen Receptor (AR) PROTAC Degrader, in Metastatic Castration-Resistant Prostate Cancer (MCRPC). JCO 2022, 40, 17. [Google Scholar] [CrossRef]
- Bartlett, D.W.; Gilbert, A.M. Translational PK–PD for Targeted Protein Degradation. Chem. Soc. Rev. 2022, 51, 3477–3486. [Google Scholar] [CrossRef]
- Lee, D.K.; Chang, C. Expression and Degradation of Androgen Receptor: Mechanism and Clinical Implication. J. Clin. Endocrinol. Metab. 2003, 88, 4043–4054. [Google Scholar] [CrossRef]
- Lung, D.K.; Warrick, J.W.; Hematti, P.; Callander, N.S.; Mark, C.J.; Miyamoto, S.; Alarid, E.T. Bone Marrow Stromal Cells Transcriptionally Repress ESR1 but Cannot Overcome Constitutive ESR1 Mutant Activity. Endocrinology 2019, 160, 2427–2440. [Google Scholar] [CrossRef] [PubMed]
- Chirnomas, D.; Hornberger, K.R.; Crews, C.M. Protein Degraders Enter the Clinic—A New Approach to Cancer Therapy. Nat. Rev. Clin. Oncol. 2023, 20, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Muller, F.L.; Aquilanti, E.A.; DePinho, R.A. Collateral Lethality: A New Therapeutic Strategy in Oncology. Trends Cancer 2015, 1, 161–173. [Google Scholar] [CrossRef]
- Hoffman, G.R.; Rahal, R.; Buxton, F.; Xiang, K.; McAllister, G.; Frias, E.; Bagdasarian, L.; Huber, J.; Lindeman, A.; Chen, D.; et al. Functional Epigenetics Approach Identifies BRM/SMARCA2 as a Critical Synthetic Lethal Target in BRG1-Deficient Cancers. Proc. Natl. Acad. Sci. USA 2014, 111, 3128–3133. [Google Scholar] [CrossRef]
- Oike, T.; Ogiwara, H.; Tominaga, Y.; Ito, K.; Ando, O.; Tsuta, K.; Mizukami, T.; Shimada, Y.; Isomura, H.; Komachi, M.; et al. A Synthetic Lethality–Based Strategy to Treat Cancers Harboring a Genetic Deficiency in the Chromatin Remodeling Factor BRG1. Cancer Res. 2013, 73, 5508–5518. [Google Scholar] [CrossRef]
- Ehrenhöfer-Wölfer, K.; Puchner, T.; Schwarz, C.; Rippka, J.; Blaha-Ostermann, S.; Strobl, U.; Hörmann, A.; Bader, G.; Kornigg, S.; Zahn, S.; et al. SMARCA2-Deficiency Confers Sensitivity to Targeted Inhibition of SMARCA4 in Esophageal Squamous Cell Carcinoma Cell Lines. Sci. Rep. 2019, 9, 11661. [Google Scholar] [CrossRef] [PubMed]
- Cantley, J.; Ye, X.; Rousseau, E.; Januario, T.; Hamman, B.D.; Rose, C.M.; Cheung, T.K.; Hinkle, T.; Soto, L.; Quinn, C.; et al. Selective PROTAC-Mediated Degradation of SMARCA2 Is Efficacious in SMARCA4 Mutant Cancers. Nat. Commun. 2022, 13, 6814. [Google Scholar] [CrossRef] [PubMed]
- Hulse, M.; Agarwal, A.; Wang, M.; Carter, J.; Sivakumar, M.; Vidal, B.; Brown, J.; Moore, A.; Grego, A.; Bhagwat, N.; et al. Abstract 3263: Preclinical Characterization of PRT3789, a Potent and Selective SMARCA2 Targeted Degrader. Cancer Res. 2022, 82, 3263. [Google Scholar] [CrossRef]
- Miah, A.H.; Smith, I.E.D.; Rackham, M.; Mares, A.; Thawani, A.R.; Nagilla, R.; Haile, P.A.; Votta, B.J.; Gordon, L.J.; Watt, G.; et al. Optimization of a Series of RIPK2 PROTACs. J. Med. Chem. 2021, 64, 12978–13003. [Google Scholar] [CrossRef] [PubMed]
- Kato, J.; Korenaga, S.; Iwakura, M. Discovery of a Potent and Subtype-Selective TYK2 Degrader Based on an Allosteric TYK2 Inhibitor. Bioorg. Med. Chem. Lett. 2023, 79, 129083. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Ma, Z.; Yang, Z.; He, Z.; Bao, J.; Peng, X.; Liu, Y.; Chen, T.; Cai, S.; Chen, J.; et al. Discovery of Ibrutinib-Based BTK PROTACs with in Vivo Anti-Inflammatory Efficacy by Inhibiting NF-κB Activation. Eur. J. Med. Chem. 2023, 259, 115664. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.-R.; Yang, W.-G.; Hou, X.-H.; Shen, D.-D.; Zhang, S.-N.; Li, Y.; Qiao, Y.-Y.; Wang, S.-Q.; Yuan, S.; Liu, H.-M. The Recent Advance of Interleukin-1 Receptor Associated Kinase 4 Inhibitors for the Treatment of Inflammation and Related Diseases. Eur. J. Med. Chem. 2023, 258, 115606. [Google Scholar] [CrossRef] [PubMed]
- Gribkoff, V.K.; Kaczmarek, L.K. The Need for New Approaches in CNS Drug Discovery: Why Drugs Have Failed, and What Can Be Done to Improve Outcomes. Neuropharmacology 2017, 120, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Tashima, T. Proteolysis-Targeting Chimera (PROTAC) Delivery into the Brain across the Blood-Brain Barrier. Antibodies 2023, 12, 43. [Google Scholar] [CrossRef]
- Tashima, T. Delivery of Intravenously Administered Antibodies Targeting Alzheimer’s Disease-Relevant Tau Species into the Brain Based on Receptor-Mediated Transcytosis. Pharmaceutics 2022, 14, 411. [Google Scholar] [CrossRef]
- Potjewyd, F.M.; Axtman, A.D. Exploration of Aberrant E3 Ligases Implicated in Alzheimer’s Disease and Development of Chemical Tools to Modulate Their Function. Front. Cell. Neurosci. 2021, 15, 768655. [Google Scholar] [CrossRef] [PubMed]
- Thomas, B.A.I.; Lewis, H.L.; Jones, D.H.; Ward, S.E. Central Nervous System Targeted Protein Degraders. Biomolecules 2023, 13, 1164. [Google Scholar] [CrossRef] [PubMed]
- Chu, T.-T.; Gao, N.; Li, Q.-Q.; Chen, P.-G.; Yang, X.-F.; Chen, Y.-X.; Zhao, Y.-F.; Li, Y.-M. Specific Knockdown of Endogenous Tau Protein by Peptide-Directed Ubiquitin-Proteasome Degradation. Cell Chem. Biol. 2016, 23, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Liu, T.; Jiao, Q.; Ji, J.; Tao, M.; Liu, Y.; You, Q.; Jiang, Z. Discovery of a Keap1-Dependent Peptide PROTAC to Knockdown Tau by Ubiquitination-Proteasome Degradation Pathway. Eur. J. Med. Chem. 2018, 146, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.C.; Ferguson, F.M.; Cai, Q.; Donovan, K.A.; Nandi, G.; Patnaik, D.; Zhang, T.; Huang, H.-T.; Lucente, D.E.; Dickerson, B.C.; et al. Targeted Degradation of Aberrant Tau in Frontotemporal Dementia Patient-Derived Neuronal Cell Models. eLife 2019, 8, e45457. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.C.; Nandi, G.; Donovan, K.A.; Cai, Q.; Berry, B.C.; Nowak, R.P.; Fischer, E.S.; Gray, N.S.; Ferguson, F.M.; Haggarty, S.J. Discovery and Optimization of Tau Targeted Protein Degraders Enabled by Patient Induced Pluripotent Stem Cells-Derived Neuronal Models of Tauopathy. Front. Cell. Neurosci. 2022, 16, 801179. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhou, Q.; Jiang, T.; Li, S.; Ye, J.; Zheng, J.; Wang, X.; Liu, Y.; Deng, M.; Ke, D.; et al. A Novel Small-Molecule PROTAC Selectively Promotes Tau Clearance to Improve Cognitive Functions in Alzheimer-like Models. Theranostics 2021, 11, 5279–5295. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; Gu, L.; Zhang, H.; Min, J.; Wang, Z.; Ma, Z.; Zhang, C.; Zeng, S.; Pan, Y.; Yan, D.; et al. Design, Synthesis, and Bioactivity of Novel Bifunctional Small Molecules for Alzheimer’s Disease. ACS Omega 2022, 7, 26308–26315. [Google Scholar] [CrossRef] [PubMed]
- Rui, Q.; Ni, H.; Li, D.; Gao, R.; Chen, G. The Role of LRRK2 in Neurodegeneration of Parkinson Disease. Curr. Neuropharmacol. 2018, 16, 1348–1357. [Google Scholar] [CrossRef]
- Sparks, S.M.; Araujo, E.; Berlin, M.; Zhang, W.; Wang, J. Selective Modulators of Mutant Lrrk2 Proteolysis and Associated Methods of Use. U.S. Patent No. 11858940B2, 2 January 2024. [Google Scholar]
- Araujo, E.; Sparks, S.M.; Berlin, M.; Zhang, W.; Wang, J. Indazole Based Compounds and Associated Methods of Use. U.S. Patent No. 20210315896A1, 14 October 2021. [Google Scholar]
- Liu, X.; Kalogeropulou, A.F.; Domingos, S.; Makukhin, N.; Nirujogi, R.S.; Singh, F.; Shpiro, N.; Saalfrank, A.; Sammler, E.; Ganley, I.G.; et al. Discovery of XL01126: A Potent, Fast, Cooperative, Selective, Orally Bioavailable, and Blood–Brain Barrier Penetrant PROTAC Degrader of Leucine-Rich Repeat Kinase 2. J. Am. Chem. Soc. 2022, 144, 16930–16952. [Google Scholar] [CrossRef]
- Qu, J.; Ren, X.; Xue, F.; He, Y.; Zhang, R.; Zheng, Y.; Huang, H.; Wang, W.; Zhang, J. Specific Knockdown of α-Synuclein by Peptide-Directed Proteasome Degradation Rescued Its Associated Neurotoxicity. Cell Chem. Biol. 2020, 27, 751–762.e4. [Google Scholar] [CrossRef] [PubMed]
- Andrew, P.C.; Dong, H.; Berlin, M.; Sparks, S.M. Proteolysis Targeting Chimeric (Protac) Compound with E3 Ubiquitin Ligase Binding Activity and Targeting Alpha-Synuclein Protein for Treating Neurodegenerative Diseases. WO Patent No. WO2020041331A1, 27 February 2020. [Google Scholar]
- Kargbo, R.B. PROTAC Compounds Targeting α-Synuclein Protein for Treating Neurogenerative Disorders: Alzheimer’s and Parkinson’s Diseases. ACS Med. Chem. Lett. 2020, 11, 1086–1087. [Google Scholar] [CrossRef] [PubMed]
- Tomoshige, S.; Nomura, S.; Ohgane, K.; Hashimoto, Y.; Ishikawa, M. Discovery of Small Molecules That Induce the Degradation of Huntingtin. Angew. Chem. Int. Ed. 2017, 56, 11530–11533. [Google Scholar] [CrossRef] [PubMed]
- Tomoshige, S.; Ishikawa, M. PROTACs and Other Chemical Protein Degradation Technologies for the Treatment of Neurodegenerative Disorders. Angew. Chem. Int. Ed. 2021, 60, 3346–3354. [Google Scholar] [CrossRef] [PubMed]
- Tseng, Y.-L.; Lu, P.-C.; Lee, C.-C.; He, R.-Y.; Huang, Y.-A.; Tseng, Y.-C.; Cheng, T.-J.R.; Huang, J.J.-T.; Fang, J.-M. Degradation of Neurodegenerative Disease-Associated TDP-43 Aggregates and Oligomers via a Proteolysis-Targeting Chimera. J. Biomed. Sci. 2023, 30, 27. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Ruan, Y.; Wang, D.; Fan, J.; Luo, N.; Chen, H.; Li, X.; Chen, W.; Wang, X. VHL-Recruiting PROTAC Attenuates Renal Fibrosis and Preserves Renal Function via Simultaneous Degradation of Smad3 and Stabilization of HIF-2α. Cell Biosci. 2022, 12, 203. [Google Scholar] [CrossRef] [PubMed]
- Hoi, D.M.; Junker, S.; Junk, L.; Schwechel, K.; Fischel, K.; Podlesainski, D.; Hawkins, P.M.E.; Van Geelen, L.; Kaschani, F.; Leodolter, J.; et al. Clp-Targeting BacPROTACs Impair Mycobacterial Proteostasis and Survival. Cell 2023, 186, 2176–2192.e22. [Google Scholar] [CrossRef] [PubMed]
- Morreale, F.E.; Kleine, S.; Leodolter, J.; Junker, S.; Hoi, D.M.; Ovchinnikov, S.; Okun, A.; Kley, J.; Kurzbauer, R.; Junk, L.; et al. BacPROTACs Mediate Targeted Protein Degradation in Bacteria. Cell 2022, 185, 2338–2353.e18. [Google Scholar] [CrossRef]
- Zhou, Q.-Q.; Xiao, H.-T.; Yang, F.; Wang, Y.-D.; Li, P.; Zheng, Z.-G. Advancing Targeted Protein Degradation for Metabolic Diseases Therapy. Pharmacol. Res. 2023, 188, 106627. [Google Scholar] [CrossRef]


| Compound | Company | Protein Target | Disease Setting | Dose Route | Current Phase | Clinical Trial ID |
|---|---|---|---|---|---|---|
| ARV-471 | Arvinas, New Haven, CT, USA | Estrogen Receptor | BC | Oral | Phase 3 | NCT05909397; NCT05654623 |
| ARV-110 | Arvinas, New Haven, CT, USA | Androgen Receptor | mCRPC | Oral | Phase 2 | NCT03888612 |
| ARV-766 | Arvinas, New Haven, CT, USA | Androgen Receptor | mCRPC | Oral | Phase 2 | NCT05067140 |
| RNK05047 | Ranok Therapeutics, Hangzhou, China | BRD4 | DLBCL | iv | Phase 1/2 | NCT05487170 |
| BGB-16673 | Beigene, Beijing, China | BTK | B-cell lymphomas | Oral | Phase 1/2 | NCT05294731; NCT05006716 |
| CFT1946 | C4 Therapeutics, Watertown, MA, USA | mtBRAF V600 | NSCLC, mCRC, melanoma | Oral | Phase 1/2 | NCT05668585 |
| AC176 | Accutar Biotech, Cranbury, NJ, USA | Androgen Receptor | mCRPC | Oral | Phase 1 | NCT05673109; NCT05241613 |
| ARD-LCC/CC-94676 | Celgene/BMS, Lawrenceville, NJ, USA | Androgen Receptor | mCRPC | Oral | Phase 1 | NCT04428788 |
| RO7656594 | Gemicure/Genentech, South San Francisco, CA, USA | Androgen Receptor | mCRPC | Oral | Phase 1 | NCT05800665 |
| HP518 | Hinova, Beijing, China | Androgen Receptor | mCRPC | Oral | Phase 1 | NCT05252364 |
| DT-2216 | Dialectic Therapeutics, Dallas, TX, USA | BCL-xL | Solid tumors/ Hematological tumors | iv | Phase 1 | NCT04886622 |
| CFT8634 | C4 Therapeutics, Watertown, MA, USA | BRD9 | Synovial Sarcoma | Oral | Phase 1 | NCT05355753 |
| FHD-609 | Foghorn Therapeutics, Cambridge, MA, USA | BRD9 | Synovial Sarcoma | iv | Phase 1 | NCT04965753 |
| AC676 | Accutar Biotech, Cranbury, NJ, USA | BTK (wt/C481S) | B-cell lymphomas | Oral | Phase 1 | NCT05780034 |
| NX-2127 | Nurix Therapeutics, San Francisco, CA, USA | BTK | B-cell lymphomas | Oral | Phase 1 | NCT04830137 |
| NX-5948 | Nurix Therapeutics, San Francisco, CA, USA | BTK | B-cell lymphomas | Oral | Phase 1 | NCT05131022 |
| AC682 | Accutar Biotech, Cranbury, NJ, USA | Estrogen Receptor | BC | Oral | Phase 1 | NCT05489679; NCT05080842 |
| AC699 | Accutar Biotech, Cranbury, NJ, USA | Estrogen Receptor | BC | Oral | Phase 1 | NCT05654532 |
| KT-253 | Kymera Therapeutics, Watertown, MA, USA | MDM2 | Various tumors | iv | Phase 1 | NCT05775406 |
| PRT3789 | Prelude Therapeutics, Wilmington, DE, USA | SMARCA2 | SMARCA4-del NSCLC | iv | Phase 1 | NCT05639751 |
| KT-333 | Kymera Therapeutics, Watertown, MA, USA | STAT3 | Various lymphoma, leukemia, solid tumors | iv | Phase 1 | NCT05225584 |
| Compound | Company | Protein Target | Disease Setting | Dose Route | Current Phase | Clinical Trial ID |
|---|---|---|---|---|---|---|
| Thalidomide (Thalomid) | Celgene/BMS, Lawrenceville, NJ, USA | IKZF1/3 | Multiple myeloma | Oral | Approved | |
| Lenalidomide (Revlimid) | Celgene/BMS, Lawrenceville, NJ, USA | IKZF1/3 | Multiple myeloma, Myleodysplastic syndrome | Oral | Approved | |
| Pomalidomide (Pomalyst) | Celgene/BMS, Lawrenceville, NJ, USA | IKZF1/3 | Multiple myeloma | Oral | Approved | |
| CC-122 | Celgene/BMS, Lawrenceville, NJ, USA | IKZF1/3 | Melanoma, Hepatocellular carcinoma, Chronic lymphocytic leukemia | Oral | Phase II | NCT03834623, NCT02859324, NCT02406742 |
| CC-92480 (Mezigdomide) | Celgene/BMS, Lawrenceville, NJ, USA | IKZF1/3 | Multiple myeloma | Oral | Phase II | NCT05372354 |
| CC-220 (Iberdomide) | Celgene/BMS, Lawrenceville, NJ, USA | IKZF1/3 | Multiple myeloma | Oral | Phase II | NCT05199311 |
| ICP-490 | InnoCare Pharma, Beijing, China | IKZF1/3 | Multiple myeloma | Oral | Phase II | NCT05719701 |
| CC-90009 | BMS/Celgene, Lawrenceville, NJ, USA | GSPT1 | AML | iv | Phase II | NCT04336982 NCT02848001 |
| CQS | City of Hope Medical Center, Duarte, CA, USA | RBM39 | Colorectal cancer, Small-cell lung cancer | iv | Phase II | NCT00005864; NCT00008372 |
| E7070 (Indisulam) | Eisai, Tokyo, Japan | RBM39 | BC, AML, Melanoma, Renal cell carcinoma, Colorectal cancer, | iv | Phase II | NCT00080197, NCT01692197; NCT00014625; NCT00059735; NCT00165867, NCT00165854 |
| E7820 | Eisai, Tokyo, Japan | RBM39 | AML, Colorectal | Oral | Phase II | NCT05024994; NCT00309179 |
| MRT-2359 | Monte Rosa Therapeutics, Boston, MA, USA | GSPT1 | Myc-driven tumors | Oral | Phase I/II | NCT05546268 |
| CC-99282 (Golcadomide) | BMS/Celgene, Lawrenceville, NJ, USA | IKZF1/3 | Non-Hodgkin and B-Cell Lymphomas | Oral | Phase I/II | NCT03930953; NCT04884035 |
| CFT-7455 | C4 Therapeutics, Watertown, MA, USA | IKZF1/3 | Multiple myeloma, Non-Hodgkin and B-Cell Lymphomas | Oral | Phase I/II | NCT04756726 |
| BTX-1188 | Biotheryx, San Diego, CA, USA | IKZF1/3 | Non-Hodgkin lymphoma, AML, Solid tumors | Oral | Phase I | NCT05144334 |
| GT-919 | Gluetacs Therapeutics, Shanghai, China | IKZF1/3 | Hematological tumors | Oral | Phase I | Undefined |
| GT-929 | Gluetacs Therapeutics, Shanghai, China | IKZF1/3 | Hematological tumors | Oral | Phase I | Undefined |
| SP-3164 | Salarius Pharmaceuticals, Houston TX, USA | IKZF1/3 | Non-Hodgkin lymphoma | Oral | Phase I | NCT05979857 |
| DKY-709 | Novartis Pharmaceuticals, Basel, Switzerland | IKZF2 | Advanced solid tumors | Oral | Phase I/Ib | NCT03891953 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bouvier, C.; Lawrence, R.; Cavallo, F.; Xolalpa, W.; Jordan, A.; Hjerpe, R.; Rodriguez, M.S. Breaking Bad Proteins—Discovery Approaches and the Road to Clinic for Degraders. Cells 2024, 13, 578. https://doi.org/10.3390/cells13070578
Bouvier C, Lawrence R, Cavallo F, Xolalpa W, Jordan A, Hjerpe R, Rodriguez MS. Breaking Bad Proteins—Discovery Approaches and the Road to Clinic for Degraders. Cells. 2024; 13(7):578. https://doi.org/10.3390/cells13070578
Chicago/Turabian StyleBouvier, Corentin, Rachel Lawrence, Francesca Cavallo, Wendy Xolalpa, Allan Jordan, Roland Hjerpe, and Manuel S. Rodriguez. 2024. "Breaking Bad Proteins—Discovery Approaches and the Road to Clinic for Degraders" Cells 13, no. 7: 578. https://doi.org/10.3390/cells13070578






