The Identification of a Novel Nucleomodulin MbovP467 of Mycoplasmopsis bovis and Its Potential Contribution in Pathogenesis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Growth of Cells and Bacterial Strains
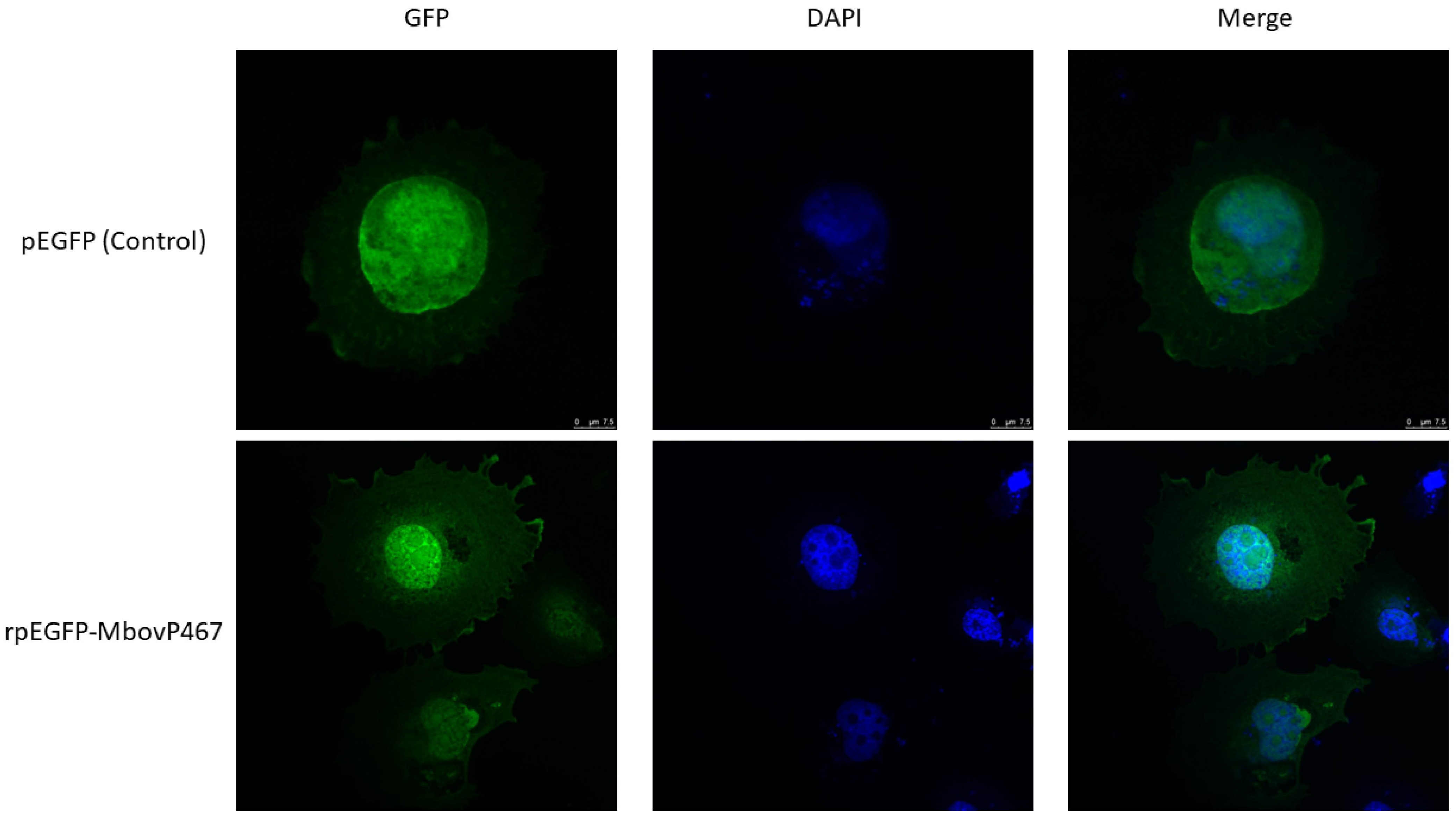
2.3. Confocal Microscopy
2.4. Gene Cloning, Expression of the Recombinant Proteins, and Polyclonal Antibody Production
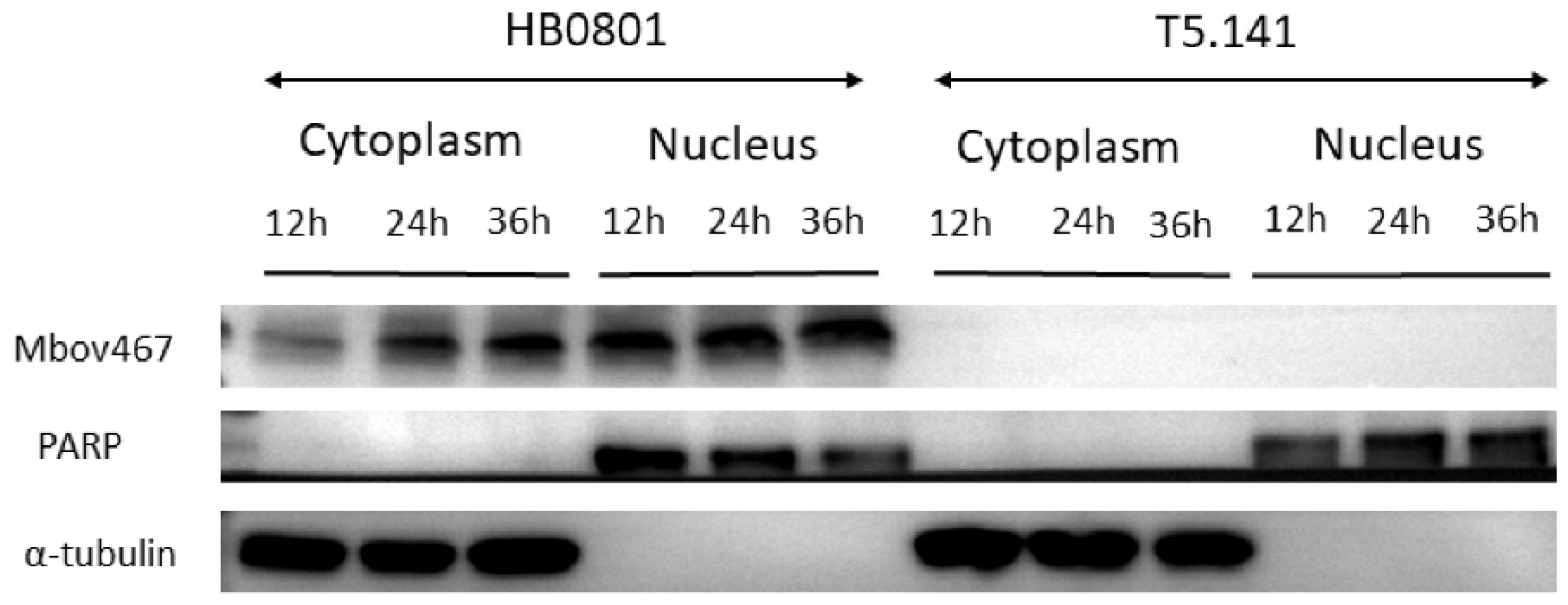
2.5. MbovP467 Location in BoMac Cells Detected with Western Blotting Assay
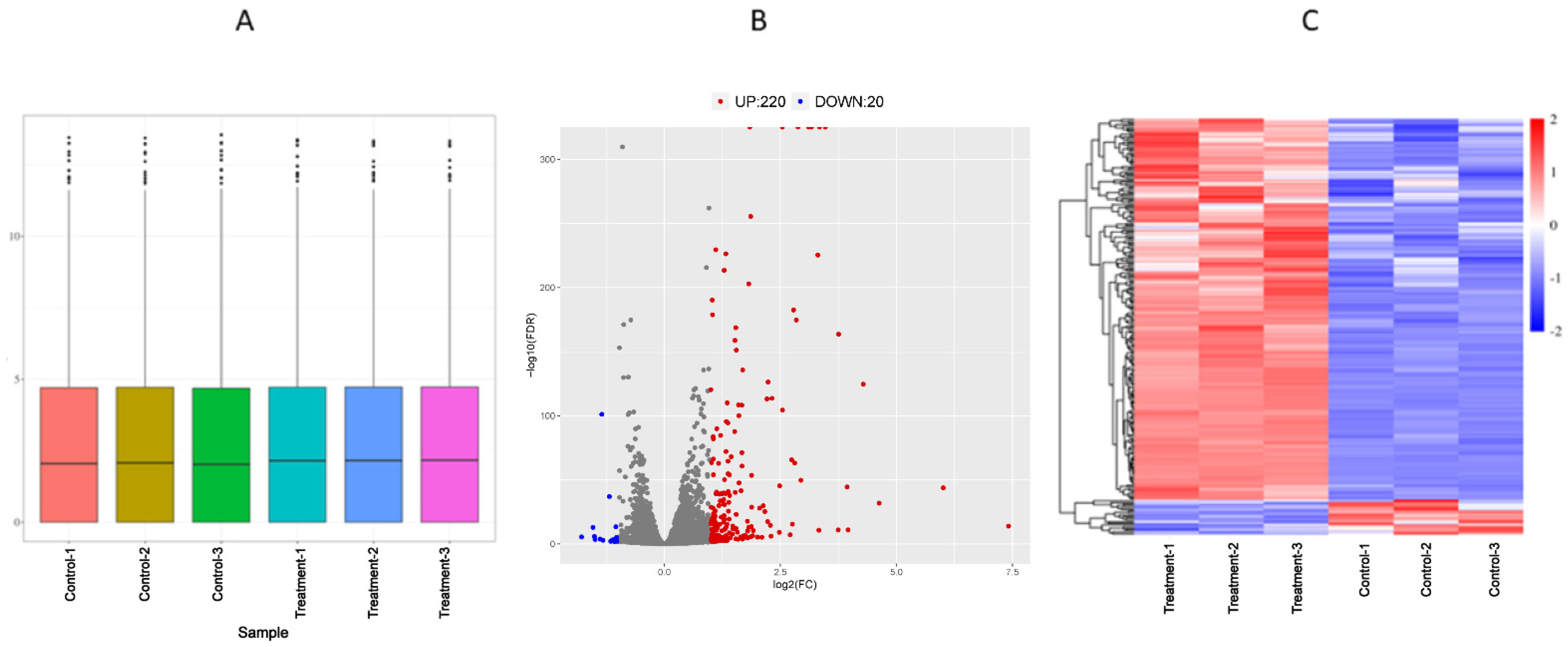
2.6. Transcriptomic Analysis
2.6.1. RNA Extraction, cDNA Library Preparation, and Sequencing
2.6.2. Data Processing
2.6.3. DEGs Detection and Functional Enrichment Analysis
2.6.4. Real-Time PCR for mRNA Expression Pro-Inflammatory Cytokines
2.7. Effects of rMbovP467 on the Promoter Region of TNF-α
2.8. Assays on BoMac Cell Viability
2.9. Assay on BoMac Cell Apoptosis
2.10. Statistical Analysis
3. Results
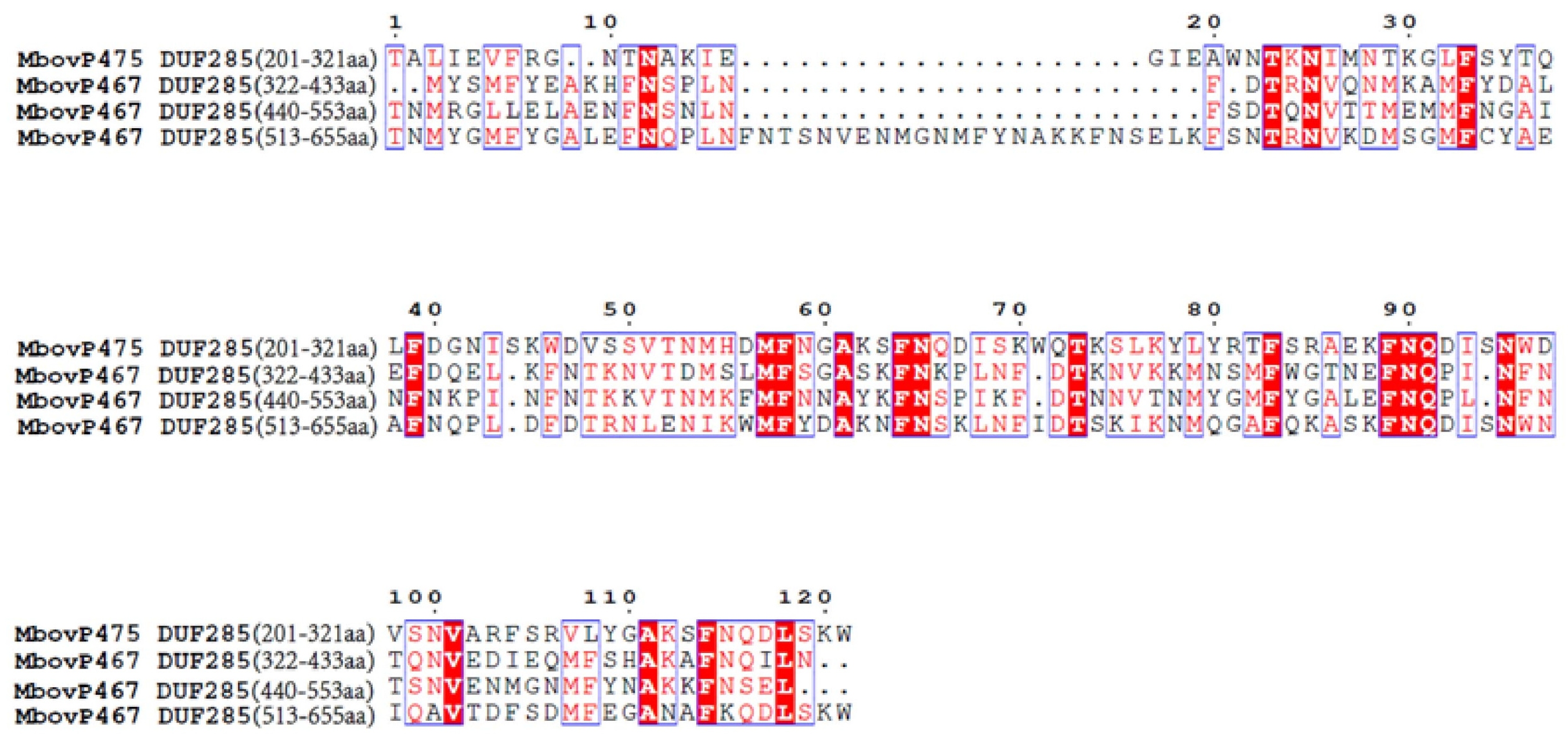
3.1. Nuclear Localization of MbovP467
3.2. Confirmation of Nuclear Localization of MbovP467 with Western Blotting
3.3. Analysis of Differentially Expressed Genes of Transcriptome
3.4. GO and KEGG Enrichment of Differential Expressed Genes
3.5. RNA-Seq Data Validation
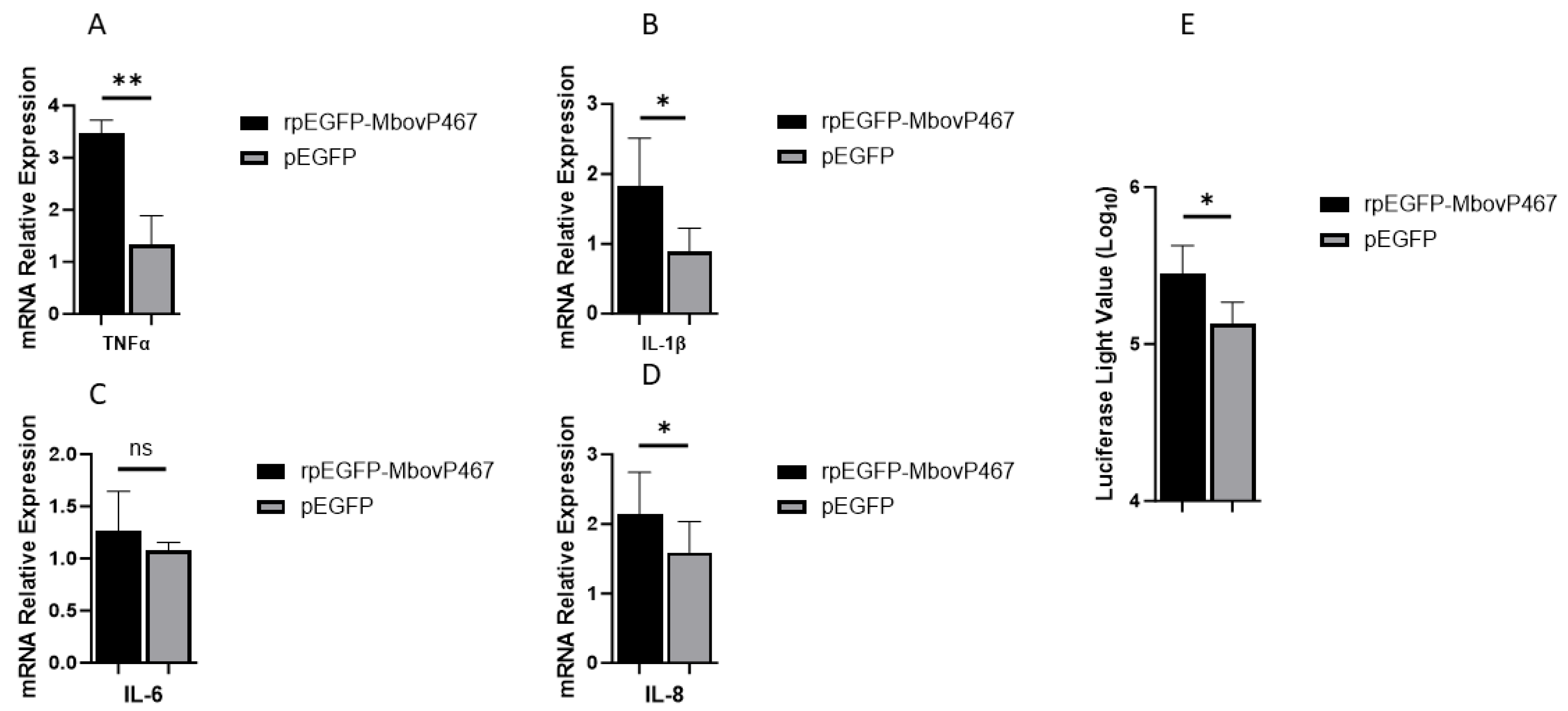
3.5.1. Effects of rMbovp467 on mRNA and Promoter Region of Pro-Inflammatory Cytokines
3.5.2. MbovP467 Reduces BoMac Cell Viability
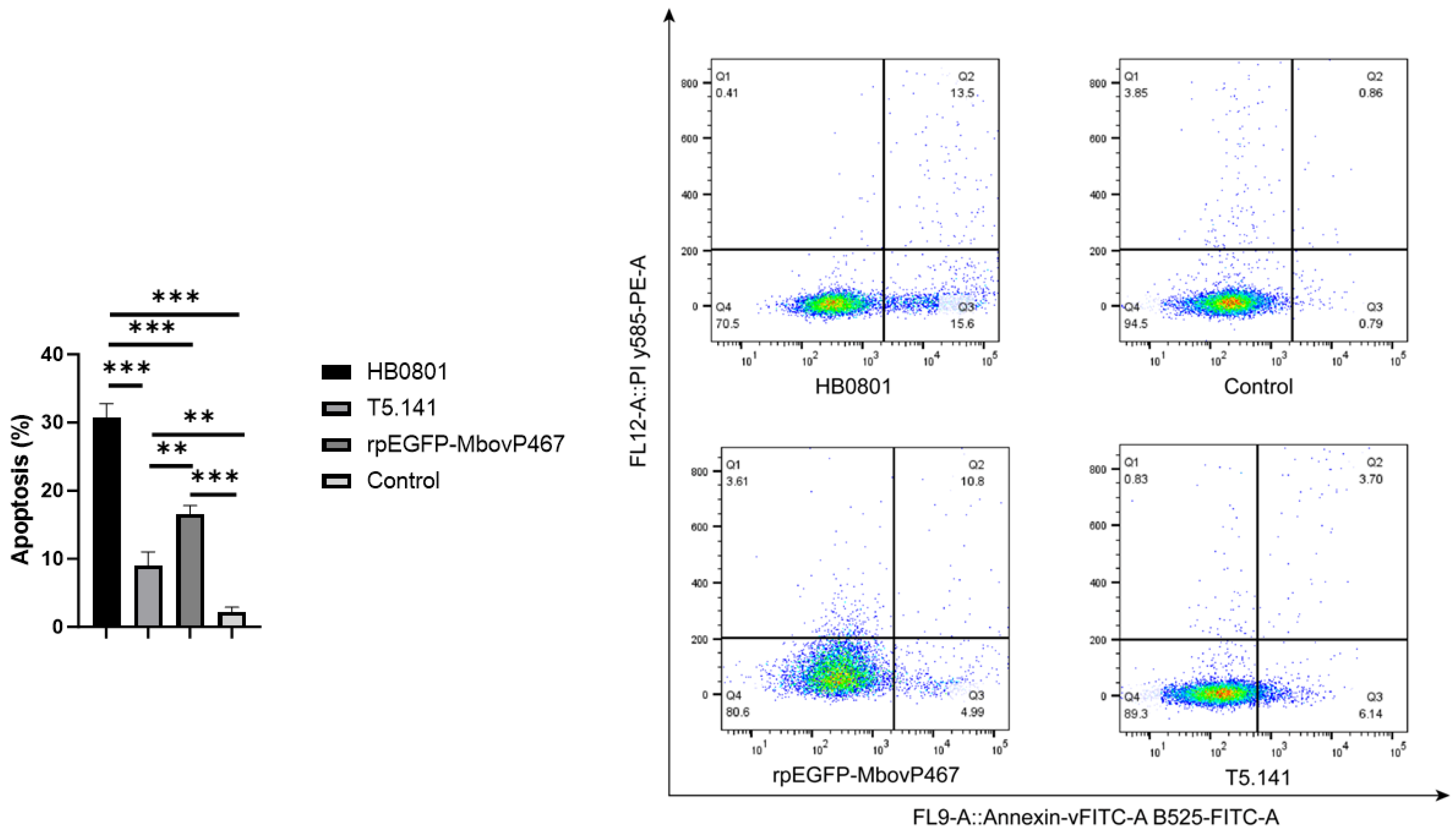
3.5.3. MbovP467 Induces Apoptosis in BoMac Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Razin, S. The minimal cellular genome of mycoplasma. Indian J. Biochem. Biophys. 1997, 34, 124–130. [Google Scholar] [PubMed]
- Arfi, Y.; Lartigue, C.; Sirand-Pugnet, P.; Blanchard, A. Beware of Mycoplasma Anti-immunoglobulin Strategies. mBio 2021, 12, e0197421. [Google Scholar] [CrossRef] [PubMed]
- Sachse, K.; Helbig, J.H.; Lysnyansky, I.; Grajetzki, C.; Müller, W.; Jacobs, E.; Yogev, D. Epitope mapping of immunogenic and adhesive structures in repetitive domains of Mycoplasma bovis variable surface lipoproteins. Infect. Immun. 2000, 68, 680–687. [Google Scholar] [CrossRef] [PubMed]
- Nishi, K.; Gondaira, S.; Fujiki, J.; Katagata, M.; Sawada, C.; Eguchi, A.; Iwasaki, T.; Iwano, H.; Higuchi, H. Invasion of Mycoplasma bovis into bovine synovial cells utilizing the clathrin-dependent endocytosis pathway. Vet. Microbiol. 2021, 253, 108956. [Google Scholar] [CrossRef] [PubMed]
- Bürki, S.; Gaschen, V.; Stoffel, M.H.; Stojiljkovic, A.; Frey, J.; Kuehni-Boghenbor, K.; Pilo, P. Invasion and persistence of Mycoplasma bovis in embryonic calf turbinate cells. Vet. Res. 2015, 46, 53. [Google Scholar] [CrossRef] [PubMed]
- Askar, H.; Chen, S.; Hao, H.; Yan, X.; Ma, L.; Liu, Y.; Chu, Y. Immune Evasion of Mycoplasma bovis. Pathogens 2021, 10, 297. [Google Scholar] [CrossRef] [PubMed]
- Le, L.H.M.; Ying, L.; Ferrero, R.L. Nuclear trafficking of bacterial effector proteins. Cell. Microbiol. 2021, 23, e13320. [Google Scholar] [CrossRef] [PubMed]
- Bierne, H.; Cossart, P. When bacteria target the nucleus: The emerging family of nucleomodulins. Cell. Microbiol. 2012, 14, 622–633. [Google Scholar] [CrossRef] [PubMed]
- Nougayrède, J.P.; Taieb, F.; De Rycke, J.; Oswald, E. Cyclomodulins: Bacterial effectors that modulate the eukaryotic cell cycle. Trends Microbiol. 2005, 13, 103–110. [Google Scholar] [CrossRef]
- Bierne, H.; Pourpre, R. Bacterial Factors Targeting the Nucleus: The Growing Family of Nucleomodulins. Toxins 2020, 12, 220. [Google Scholar] [CrossRef]
- Hanford, H.E.; Von Dwingelo, J.; Abu Kwaik, Y. Bacterial nucleomodulins: A coevolutionary adaptation to the eukaryotic command center. PLoS Pathog. 2021, 17, e1009184. [Google Scholar] [CrossRef] [PubMed]
- Chernov, A.V.; Reyes, L.; Xu, Z.; Gonzalez, B.; Golovko, G.; Peterson, S.; Perucho, M.; Fofanov, Y.; Strongin, A.Y. Mycoplasma CG- and GATC-specific DNA methyltransferases selectively and efficiently methylate the host genome and alter the epigenetic landscape in human cells. Epigenetics 2015, 10, 303–318. [Google Scholar] [CrossRef] [PubMed]
- Sugio, A.; Kingdom, H.N.; MacLean, A.M.; Grieve, V.M.; Hogenhout, S.A. Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc. Natl. Acad. Sci. USA 2011, 108, E1254–E1263. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Lu, D.; Wang, S.; Zhang, H.; Zhu, X.; Hao, Z.; Dawood, A.; Chen, Y.; Schieck, E.; Hu, C.; et al. Novel mycoplasma nucleomodulin MbovP475 decreased cell viability by regulating expression of CRYAB and MCF2L2. Virulence 2022, 13, 1590–1613. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Zhu, X.; Zhang, H.; Chen, Y.; Schieck, E.; Hu, C.; Chen, H.; Guo, A. Novel Secreted Protein of Mycoplasma bovis MbovP280 Induces Macrophage Apoptosis Through CRYAB. Front. Immunol. 2021, 12, 619362. [Google Scholar] [CrossRef]
- Zubair, M.; Khan, F.A.; Menghwar, H.; Faisal, M.; Ashraf, M.; Rasheed, M.A.; Marawan, M.A.; Dawood, A.; Chen, Y.; Chen, H.; et al. Progresses on bacterial secretomes enlighten research on Mycoplasma secretome. Microb. Pathog. 2020, 144, 104160. [Google Scholar] [CrossRef] [PubMed]
- Voros, A.; DeLongchamp, J.; Saleh, M. The secretome of Mycoplasma capricolum subsp. capricolum in neutral and acidic media. J. Proteom. Bioinform. 2015, 8, 155–163. [Google Scholar]
- Qi, J.; Guo, A.; Cui, P.; Chen, Y.; Mustafa, R.; Ba, X.; Hu, C.; Bai, Z.; Chen, X.; Shi, L.; et al. Comparative geno-plasticity analysis of Mycoplasma bovis HB0801 (Chinese isolate). PLoS ONE 2012, 7, e38239. [Google Scholar] [CrossRef] [PubMed]
- Thorns, C.J. Identification of the products of the ‘film spots’ reaction of Mycoplasma bovis and Mycoplasma bovigenitalium. J. Biol. Stand. 1978, 6, 127–132. [Google Scholar] [CrossRef]
- Shan, B.; Liu, Y.; Yang, C.; Wang, L.; Li, Y.; Sun, D. De novo assembly and annotation of the whole transcriptome of Muraenesox cinereus. Mar. Genom. 2022, 61, 100910. [Google Scholar] [CrossRef]
- Bräutigam, A.; Mullick, T.; Schliesky, S.; Weber, A.P. Critical assessment of assembly strategies for non-model species mRNA-Seq data and application of next-generation sequencing to the comparison of C3 and C4 species. J. Exp. Bot. 2011, 62, 3093–3102. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, X.; Chu, Y.; Li, J.; Wang, W.; Hu, X.; Zhou, F.; Zhang, H.; Zhou, L.; Kuai, R.; et al. Analysis of transcript-wide profile regulated by microsatellite instability of colorectal cancer. Ann. Transl. Med. 2022, 10, 169. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Fan, H.; Pei, X.; Hua, X.; Xu, T.; He, Q. mRNA-Seq and miRNA-Seq Analyses Provide Insights into the Mechanism of Pinellia ternata Bulbil Initiation Induced by Phytohormones. Genes 2023, 14, 1727. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Masoudi-Nejad, A.; Goto, S.; Endo, T.R.; Kanehisa, M. KEGG bioinformatics resource for plant genomics research. Methods Mol. Biol. 2007, 406, 437–458. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.A.; Khan, Z. Bacterial nucleomodulins and cancer: An unresolved enigma. Transl. Oncol. 2021, 14, 100922. [Google Scholar] [CrossRef]
- Ma, Z.; Li, R.; Hu, R.; Deng, X.; Xu, Y.; Zheng, W.; Yi, J.; Wang, Y.; Chen, C. Brucella abortus BspJ Is a Nucleomodulin That Inhibits Macrophage Apoptosis and Promotes Intracellular Survival of Brucella. Front. Microbiol. 2020, 11, 599205. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; Choe, M.H.; Asaithambi, K.; Song, J.Y.; Lee, Y.S.; Lee, J.C.; Seo, J.H.; Kang, H.L.; Lee, K.H.; Lee, W.K.; et al. Helicobacter pylori HP0425 Targets the Nucleus with DNase I-Like Activity. Helicobacter 2016, 21, 218–225. [Google Scholar] [CrossRef]
- Shallberg, L.A.; Hunter, C.A. Long live the king: Toxoplasma gondii nucleomodulin inhibits necroptotic cell death. Cell Host Microbe 2021, 29, 1165–1166. [Google Scholar] [CrossRef]
- Ma, Z.; Yu, S.; Cheng, K.; Miao, Y.; Xu, Y.; Hu, R.; Zheng, W.; Yi, J.; Zhang, H.; Li, R.; et al. Nucleomodulin BspJ as an effector promotes the colonization of Brucella abortus in the host. J. Vet. Sci. 2022, 23, e8. [Google Scholar] [CrossRef] [PubMed]
- Adcox, H.E.; Hatke, A.L.; Andersen, S.E.; Gupta, S.; Otto, N.B.; Weber, M.M.; Marconi, R.T.; Carlyon, J.A. Orientia tsutsugamushi Nucleomodulin Ank13 Exploits the RaDAR Nuclear Import Pathway to Modulate Host Cell Transcription. mBio 2021, 12, e0181621. [Google Scholar] [CrossRef]
- Canonne, J.; Rivas, S. Bacterial effectors target the plant cell nucleus to subvert host transcription. Plant Signal. Behav. 2012, 7, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Seibel, N.M.; Eljouni, J.; Nalaskowski, M.M.; Hampe, W. Nuclear localization of enhanced green fluorescent protein homomultimers. Anal. Biochem. 2007, 368, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Yang, Y.; Wang, L.; Chen, Y.; Han, X.; Sun, L.; Chen, H.; Chen, Q. Effect of Bifidobacterium on osteoclasts: TNF-α/NF-κB inflammatory signal pathway-mediated mechanism. Front. Endocrinol. 2023, 14, 1109296. [Google Scholar] [CrossRef]
- Carswell, E.A.; Old, L.J.; Kassel, R.L.; Green, S.; Fiore, N.; Williamson, B. An endotoxin-induced serum factor that causes necrosis of tumors. Proc. Natl. Acad. Sci. USA 1975, 72, 3666–3670. [Google Scholar] [CrossRef]
- Baud, V.; Karin, M. Signal transduction by tumor necrosis factor and its relatives. Trends Cell Biol. 2001, 11, 372–377. [Google Scholar] [CrossRef]
- Pomerantz, J.L.; Baltimore, D. Two pathways to NF-kappaB. Mol. Cell 2002, 10, 693–695. [Google Scholar] [CrossRef]
- Hayden, M.S.; Ghosh, S. Signaling to NF-kappaB. Genes Dev. 2004, 18, 2195–2224. [Google Scholar] [CrossRef]
- Upadhayay, S.; Gupta, R.; Singh, S.; Mundkar, M.; Singh, G.; Kumar, P. Involvement of the G-Protein-Coupled Estrogen Receptor-1 (GPER) Signaling Pathway in Neurodegenerative Disorders: A Review. Cell. Mol. Neurobiol. 2023, 43, 1833–1847. [Google Scholar] [CrossRef]
- New, D.C.; Wu, K.; Kwok, A.W.; Wong, Y.H. G protein-coupled receptor-induced Akt activity in cellular proliferation and apoptosis. FEBS J. 2007, 274, 6025–6036. [Google Scholar] [CrossRef]
- Revankar, C.M.; Vines, C.M.; Cimino, D.F.; Prossnitz, E.R. Arrestins block G protein-coupled receptor-mediated apoptosis. J. Biol. Chem. 2004, 279, 24578–24584. [Google Scholar] [CrossRef]
- Pawlak, M.; Lefebvre, P.; Staels, B. General molecular biology and architecture of nuclear receptors. Curr. Top. Med. Chem. 2012, 12, 486–504. [Google Scholar] [CrossRef]
- Qureshi, S.A.; Jackson, S.P. Sequence-specific DNA binding by the S. shibatae TFIIB homolog, TFB, and its effect on promoter strength. Mol. Cell 1998, 1, 389–400. [Google Scholar] [CrossRef] [PubMed]
- García-Rodríguez, F.M.; Schrammeijer, B.; Hooykaas, P.J. The Agrobacterium VirE3 effector protein: A potential plant transcriptional activator. Nucleic Acids Res. 2006, 34, 6496–6504. [Google Scholar] [CrossRef] [PubMed]
- Weinthal, D.M.; Barash, I.; Tzfira, T.; Gaba, V.; Teper, D.; Sessa, G.; Manulis-Sasson, S. Characterization of nuclear localization signals in the type III effectors HsvG and HsvB of the gall-forming bacterium Pantoea agglomerans. Microbiology 2011, 157, 1500–1508. [Google Scholar] [CrossRef]
- Lebreton, A.; Job, V.; Ragon, M.; Le Monnier, A.; Dessen, A.; Cossart, P.; Bierne, H. Structural basis for the inhibition of the chromatin repressor BAHD1 by the bacterial nucleomodulin LntA. mBio 2014, 5, e00775-13. [Google Scholar] [CrossRef]
- Kwon, Y.C.; Kim, S.; Lee, Y.S.; Lee, J.C.; Cho, M.J.; Lee, W.K.; Kang, H.L.; Song, J.Y.; Baik, S.C.; Ro, H.S. Novel nuclear targeting coiled-coil protein of Helicobacter pylori showing Ca2+-independent, Mg2+-dependent DNase I activity. J. Microbiol. 2016, 54, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Haraga, A.; Miller, S.I. A Salmonella enterica serovar typhimurium translocated leucine-rich repeat effector protein inhibits NF-kappa B-dependent gene expression. Infect. Immun. 2003, 71, 4052–4058. [Google Scholar] [CrossRef]
- Nishi, K.; Okada, J.; Iwasaki, T.; Gondaira, S.; Higuchi, H. Characteristics of Mycoplasma bovis, Mycoplasma arginini, and Mycoplasma californicum on immunological response of bovine synovial cells. Vet. Immunol. Immunopathol. 2023, 260, 110608. [Google Scholar] [CrossRef]
- Suwanruengsri, M.; Uemura, R.; Kanda, T.; Fuke, N.; Nueangphuet, P.; Pornthummawat, A.; Yasuda, M.; Hirai, T.; Yamaguchi, R. Production of granulomas in Mycoplasma bovis infection associated with meningitis-meningoencephalitis, endocarditis, and pneumonia in cattle. J. Vet. Diagn. Investig. Off. Publ. Am. Assoc. Vet. Lab. Diagn. Inc. 2022, 34, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Wang, G.; Lv, T.; Song, X.; Wang, T.; Xie, G.; Cao, Y.; Zhang, N.; Cao, R. Endometrial inflammation and abnormal expression of extracellular matrix proteins induced by Mycoplasma bovis in dairy cows. Theriogenology 2014, 81, 669–674. [Google Scholar] [CrossRef] [PubMed]
- Baquero, M.; Vulikh, K.; Wong, C.; Domony, M.; Burrows, D.; Marom, D.; Perez-Casal, J.; Cai, H.Y.; Caswell, J.L. Effects of inflammatory stimuli on responses of macrophages to Mycoplasma bovis infection. Vet. Microbiol. 2021, 262, 109235. [Google Scholar] [CrossRef] [PubMed]
- Jose, L.; Ramachandran, R.; Bhagavat, R.; Gomez, R.L.; Chandran, A.; Raghunandanan, S.; Omkumar, R.V.; Chandra, N.; Mundayoor, S.; Kumar, R.A. Hypothetical protein Rv3423.1 of Mycobacterium tuberculosis is a histone acetyltransferase. FEBS J. 2016, 283, 265–281. [Google Scholar] [CrossRef]
- Pennini, M.E.; Perrinet, S.; Dautry-Varsat, A.; Subtil, A. Histone methylation by NUE, a novel nuclear effector of the intracellular pathogen Chlamydia trachomatis. PLoS Pathog. 2010, 6, e1000995. [Google Scholar] [CrossRef] [PubMed]
- Murata, M.; Azuma, Y.; Miura, K.; Rahman, M.A.; Matsutani, M.; Aoyama, M.; Suzuki, H.; Sugi, K.; Shirai, M. Chlamydial SET domain protein functions as a histone methyltransferase. Microbiology 2007, 153, 585–592. [Google Scholar] [CrossRef]
- Arbibe, L.; Kim, D.W.; Batsche, E.; Pedron, T.; Mateescu, B.; Muchardt, C.; Parsot, C.; Sansonetti, P.J. An injected bacterial effector targets chromatin access for transcription factor NF-kappaB to alter transcription of host genes involved in immune responses. Nat. Immunol. 2007, 8, 47–56. [Google Scholar] [CrossRef]




| Samples | Raw Reads | Raw Bases | Clean Reads | Clean Bases | Clean Ratio | Q20 | Q30 | GC |
|---|---|---|---|---|---|---|---|---|
| Control 1 | 59,992,918 | 8,998,937,700 | 59,543,994 | 8.92 × 109 | 99.25% | 98.04% | 94.35% | 48.33% |
| Control 2 | 42,413,822 | 6,362,073,300 | 42,136,802 | 6.31 × 109 | 99.35% | 98.26% | 94.83% | 48.37% |
| Control 3 | 58,935,792 | 8,840,368,800 | 58,511,866 | 8.76 × 109 | 99.28% | 98.08% | 94.42% | 48.35% |
| Treatment 1 | 83,245,064 | 12,486,759,600 | 82,730,332 | 1.24 × 1010 | 99.38% | 98.35% | 95.10% | 48.48% |
| Treatment 2 | 70,364,650 | 10,554,697,500 | 69,876,584 | 1.05 × 1010 | 99.31% | 98.15% | 94.61% | 48.46% |
| Treatment 3 | 69,921,538 | 10,488,230,700 | 69,479,750 | 1.04 × 1010 | 99.37% | 98.27% | 94.92% | 48.65% |
| Samples | Total Reads | Total Mapped Reads | Map Rate | Unique | Paired | Single | Self and Mate | Map Diff CHR |
|---|---|---|---|---|---|---|---|---|
| Control 1 | 59,543,994 | 57,467,676 | 96.51% | 55,323,506 | 53,793,422 | 1,616,810 | 55,850,866 | 391,760 |
| Control 2 | 42,136,802 | 40,710,940 | 96.62% | 39,379,997 | 38,294,926 | 1,134,004 | 39,576,936 | 321,496 |
| Control 3 | 58,511,866 | 56,444,119 | 96.47% | 54,499,140 | 52,903,316 | 1,671,653 | 54,772,466 | 467,834 |
| Treatment 1 | 82,730,332 | 79,971,255 | 96.66% | 77,250,388 | 75,151,040 | 2,199,405 | 77,771,850 | 616,782 |
| Treatment 2 | 69,876,584 | 67,520,868 | 96.63% | 65,269,363 | 63,463,956 | 1,887,772 | 65,633,096 | 533,124 |
| Treatment 3 | 69,479,750 | 67,058,501 | 96.52% | 64,775,746 | 62,935,550 | 1,923,865 | 65,134,636 | 566,662 |
| Genes | Forward Sequence | Reverse Sequence |
|---|---|---|
| Caspase 3 | ACCTCTTCTGCCCTGACTTC | TGTAACTACTGAAGCCCGCA |
| Caspase 2 | TTTCTCCCTGCTCACCTCAG | TCTGCCTTCATACTGTGCCA |
| Caspase 8 | TTCCTTTGCTGCCTCGAGTA | GTTGGGAGTGGGGAGATTCA |
| Caspase 9 | CTCCTCTCCTTTTGCCCTCA | TGGACCATAAAGCAGGCTGA |
| IL-1β | TCCGACGATTTCTGTGTTGA | ACGAGGAGGCCAAGAAAAGA |
| IL-6 | GGTCAAGATGCCAAGTCAGC | TGGGATCTTCACTGAATGCT |
| IL-8 | ACTGTGTGGGTCTGGTGTAG | AGGCGAGGGTTGCAAGATTA |
| TNF-α | TTGTTCCTCACCCACACCAT | CCAAAGTAGACCTGCCCAGA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raheem, A.; Lu, D.; Khalid, A.K.; Zhao, G.; Fu, Y.; Chen, Y.; Chen, X.; Hu, C.; Chen, J.; Chen, H.; et al. The Identification of a Novel Nucleomodulin MbovP467 of Mycoplasmopsis bovis and Its Potential Contribution in Pathogenesis. Cells 2024, 13, 604. https://doi.org/10.3390/cells13070604
Raheem A, Lu D, Khalid AK, Zhao G, Fu Y, Chen Y, Chen X, Hu C, Chen J, Chen H, et al. The Identification of a Novel Nucleomodulin MbovP467 of Mycoplasmopsis bovis and Its Potential Contribution in Pathogenesis. Cells. 2024; 13(7):604. https://doi.org/10.3390/cells13070604
Chicago/Turabian StyleRaheem, Abdul, Doukun Lu, Abdul Karim Khalid, Gang Zhao, Yingjie Fu, Yingyu Chen, Xi Chen, Changmin Hu, Jianguo Chen, Huanchun Chen, and et al. 2024. "The Identification of a Novel Nucleomodulin MbovP467 of Mycoplasmopsis bovis and Its Potential Contribution in Pathogenesis" Cells 13, no. 7: 604. https://doi.org/10.3390/cells13070604







