Differentiation and Regulation of Bovine Th2 Cells In Vitro
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cattle
2.2. CD4+ T Cell Isolation
2.3. Differentiation of CD4+ T Cells
2.4. Flow Cytometry Analysis
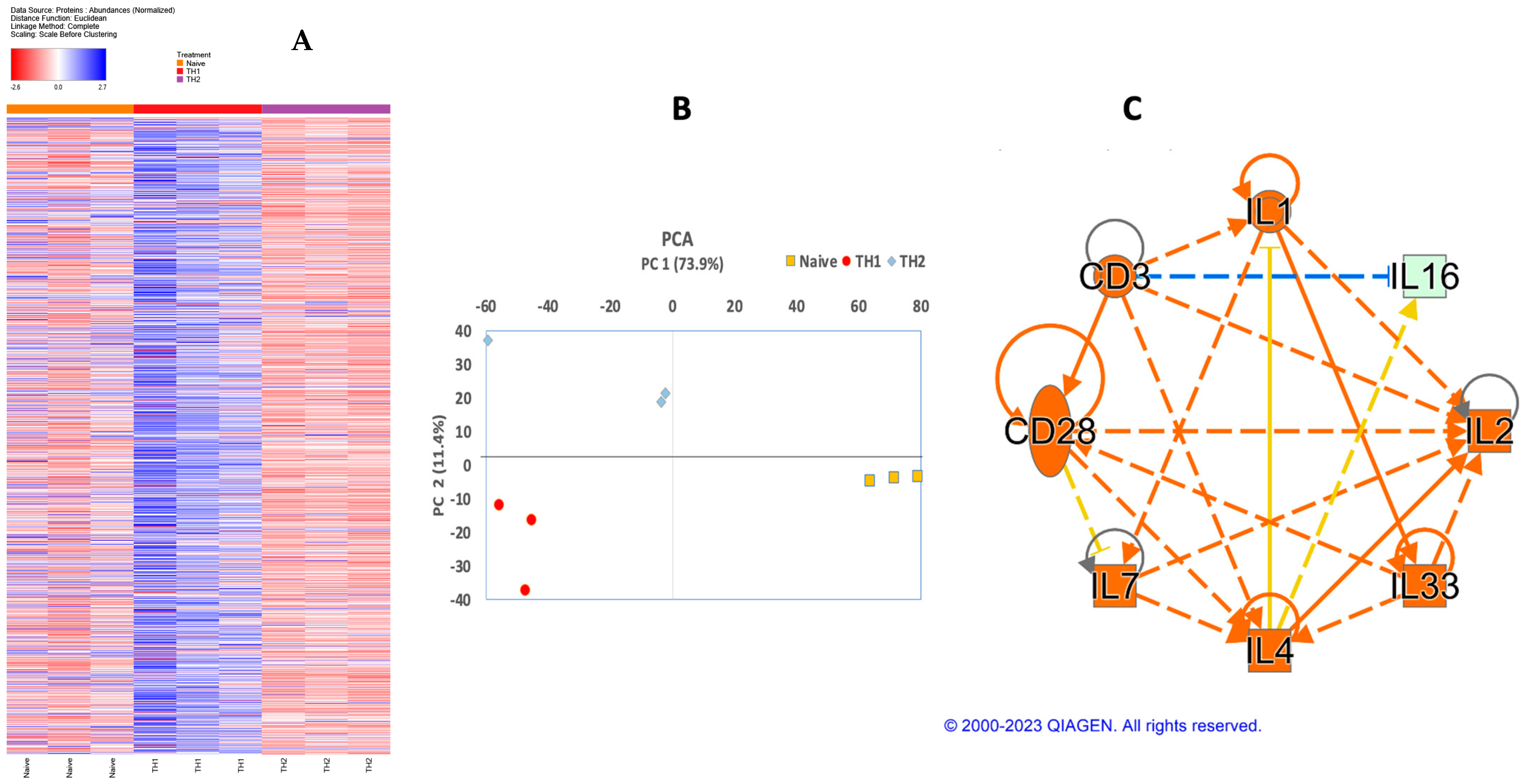
2.5. Proteomics
2.6. Quantitative PCR
2.7. Statistical Analysis
3. Results
3.1. Bovine Memory CD4+ T Cells Are a Mixture of IFNγ-Dominant Th0 Cells
3.2. In Vitro Programming under Th2 Leads to IFNγ-Dominant Th0 Mixture
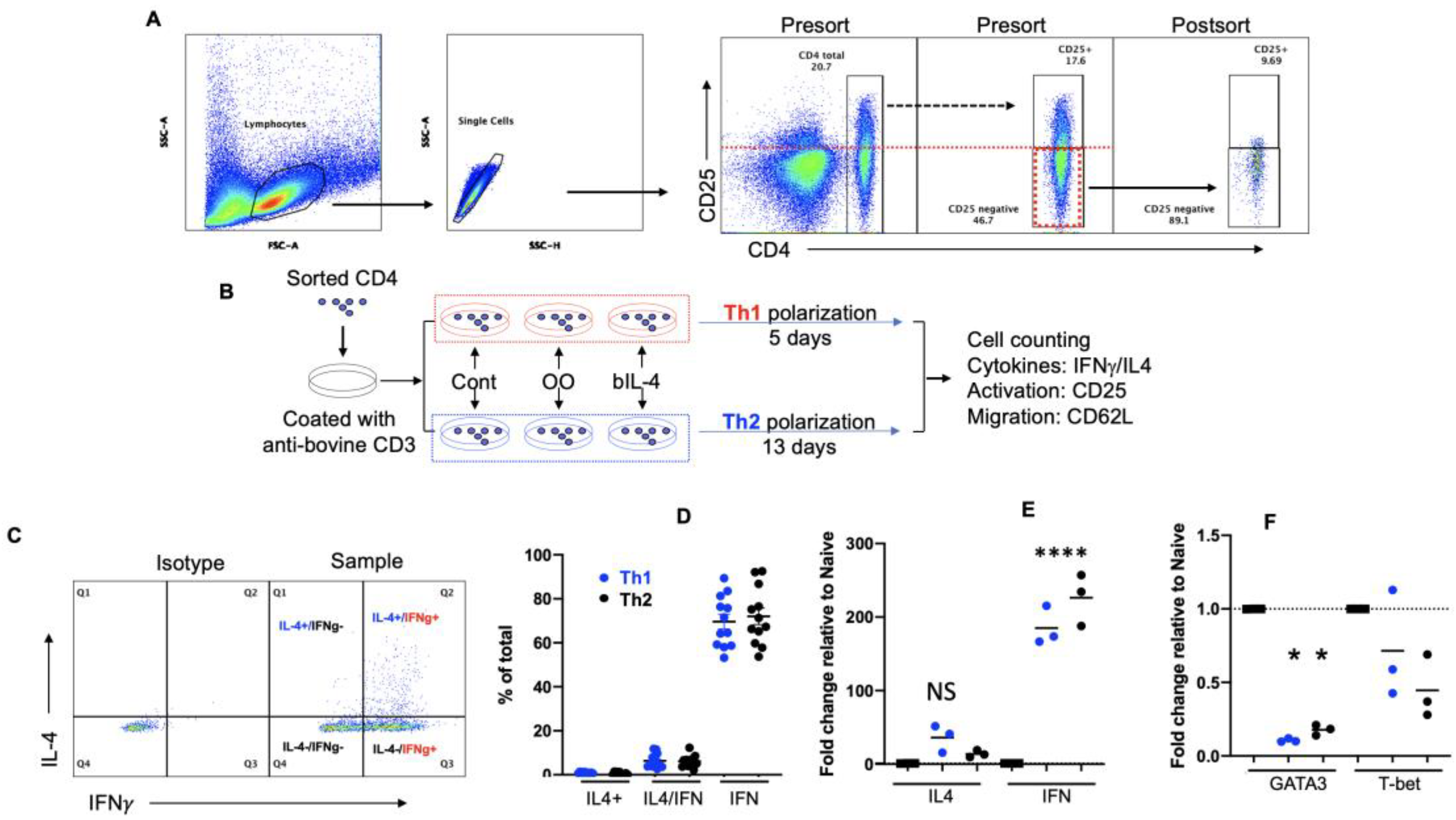
3.3. Validation of Th2 Differentiation
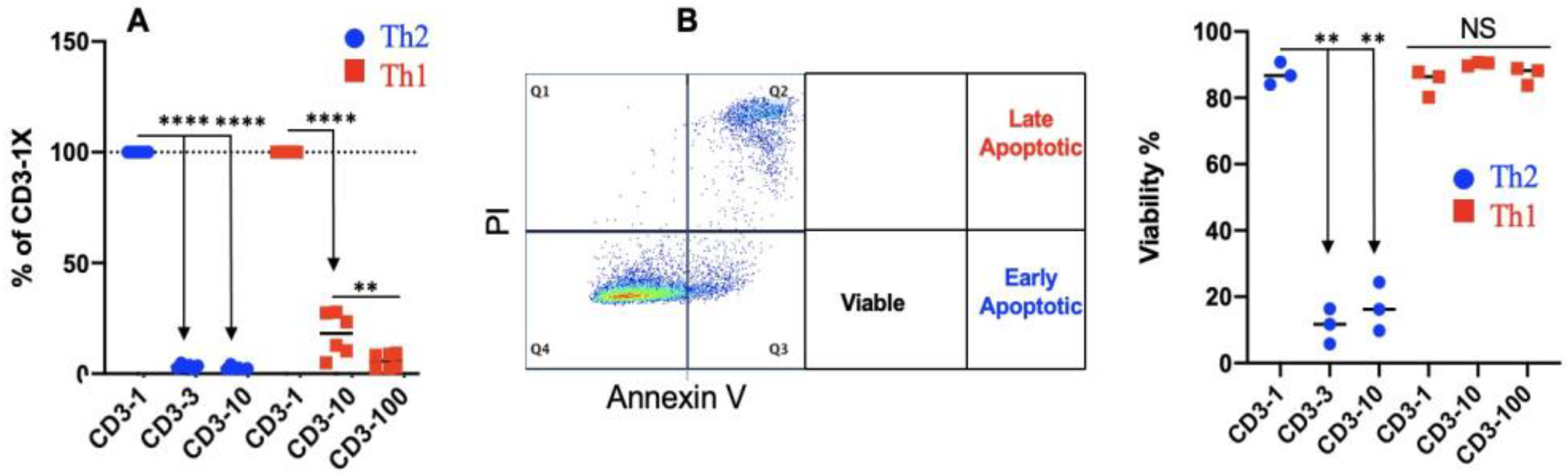
3.4. Th2 Differentiation Is Sensitive to TCR Stimulation Strength
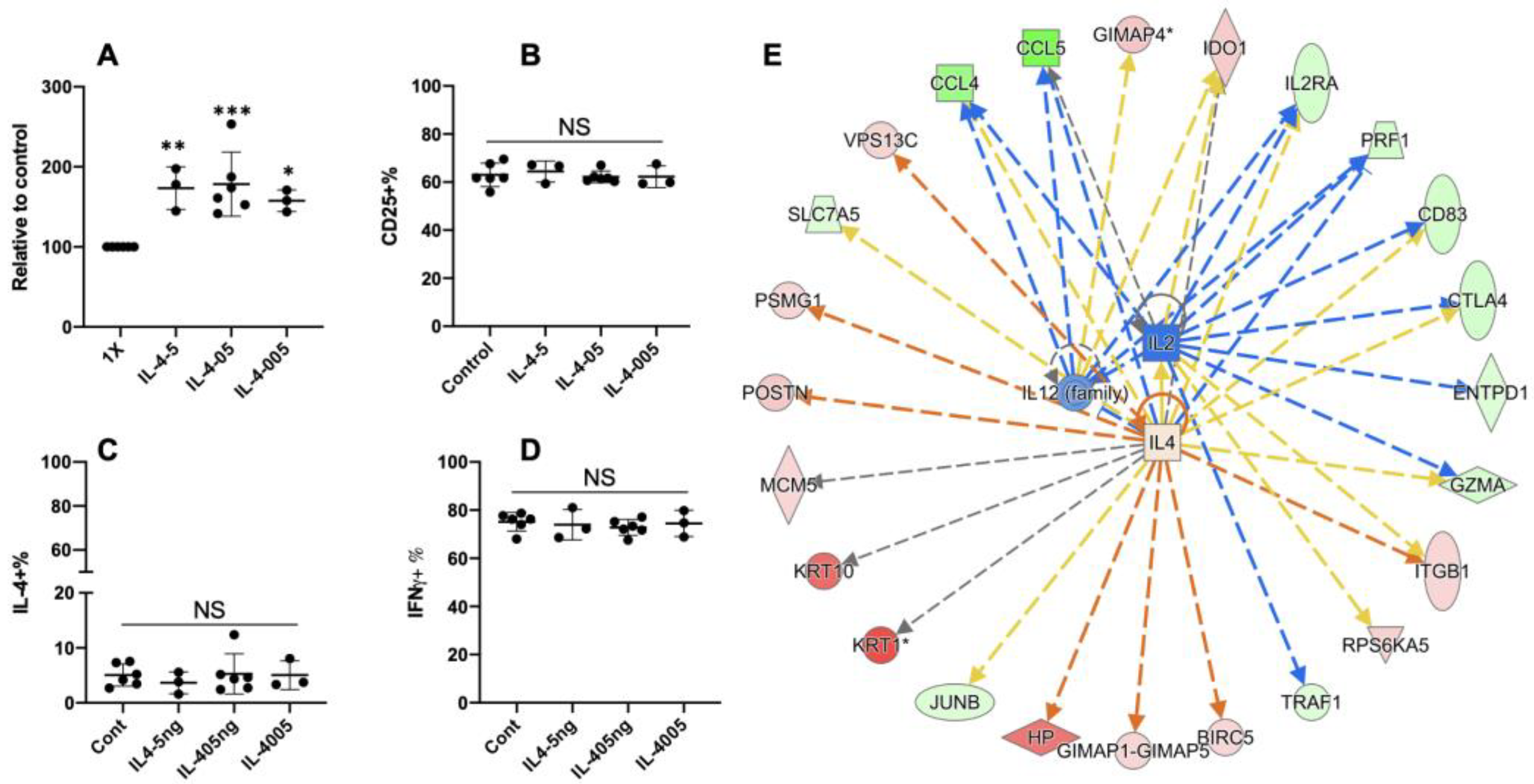
3.5. Extra Recombinant Bovine IL4(rbIL4) Leads to Enhanced Th2 Differentiation
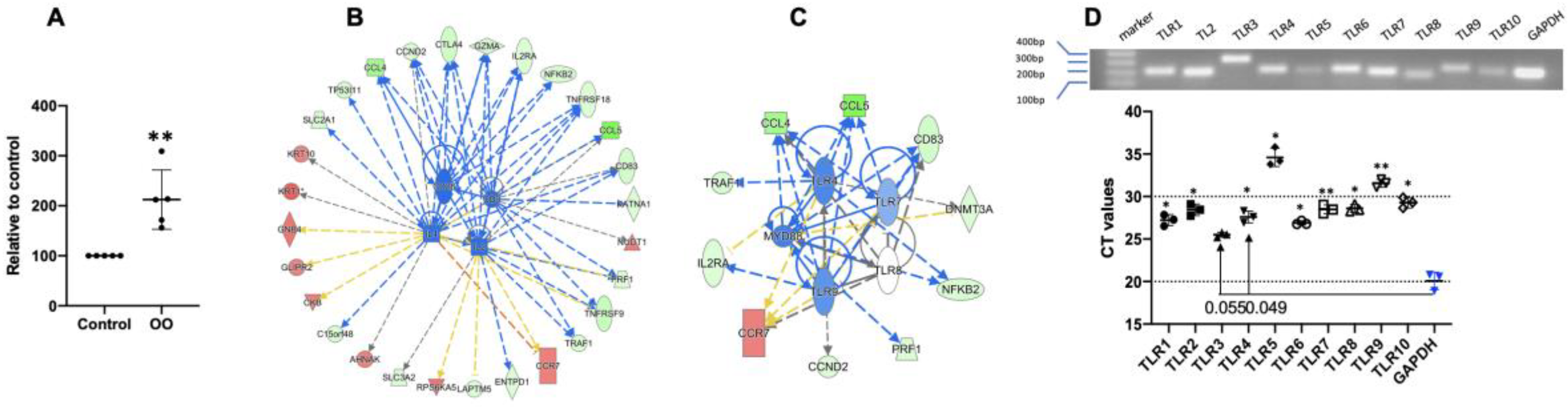
3.6. Ostertagia Ostertagi Extract (OO) Leads to the Inhibition of Th2 Differentiation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vlasova, A.N.; Saif, L.J. Bovine Immunology: Implications for Dairy Cattle. Front. Immunol. 2021, 12, 643206. [Google Scholar] [CrossRef] [PubMed]
- Claerebout, E.; Geldhof, P. Helminth vaccines in ruminants: From development to application. Vet. Clin. Food Anim. Pract. 2020, 36, 159–171. [Google Scholar] [CrossRef] [PubMed]
- Kebede, B.; Sori, T.; Kumssa, B. Review on current status of vaccines against parasitic diseases of animals. J. Vet. Sci. Technol. 2016, 7, 2. [Google Scholar] [CrossRef]
- Smith, W.; Zarlenga, D. Developments and hurdles in generating vaccines for controlling helminth parasites of grazing ruminants. Vet. Parasitol. 2006, 139, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Motran, C.C.; Silvane, L.; Chiapello, L.S.; Theumer, M.G.; Ambrosio, L.F.; Volpini, X.; Celias, D.P.; Cervi, L. Helminth infections: Recognition and modulation of the immune response by innate immune cells. Front. Immunol. 2018, 9, 664. [Google Scholar] [CrossRef] [PubMed]
- Gazzinelli-Guimaraes, P.H.; Nutman, T.B. Helminth parasites and immune regulation. F1000Research 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Anthony, R.M.; Urban, J.F., Jr.; Alem, F.; Hamed, H.A.; Rozo, C.T.; Boucher, J.-L.; Van Rooijen, N.; Gause, W.C. Memory TH2 cells induce alternatively activated macrophages to mediate protection against nematode parasites. Nat. Med. 2006, 12, 955–960. [Google Scholar] [CrossRef]
- Kandel, A.; Masello, M.; Xiao, Z. CD4+ T Cell Responses to Pathogens in Cattle In Bovine Science; IntechOpen: London, UK, 2021; Available online: https://www.intechopen.com/chapters/78918 (accessed on 20 April 2023).
- Brown, W.; Rice-Ficht, A.C.; Estes, D.M. Bovine type 1 and type 2 responses. Vet. Immunol. Immunopathol. 1998, 63, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Boggiatto, P.M.; Schaut, R.G.; Olsen, S.C. Enhancing the Detection of Brucella-Specific CD4(+) T Cell Responses in Cattle via in vitro Antigenic Expansion and Restimulation. Front. Immunol. 2020, 11, 1944. [Google Scholar] [CrossRef] [PubMed]
- Carr, B.V.; Lefevre, E.A.; Windsor, M.A.; Inghese, C.; Gubbins, S.; Prentice, H.; Juleff, N.D.; Charleston, B. CD4+ T-cell responses to foot-and-mouth disease virus in vaccinated cattle. J. Gen. Virol. 2013, 94, 97–107. [Google Scholar] [CrossRef]
- Maggioli, M.F.; Palmer, M.V.; Thacker, T.C.; Vordermeier, H.M.; Waters, W.R. Characterization of effector and memory T cell subsets in the immune response to bovine tuberculosis in cattle. PLoS ONE 2015, 10, e0122571. [Google Scholar] [CrossRef] [PubMed]
- Blunt, L.; Hogarth, P.J.; Kaveh, D.A.; Webb, P.; Villarreal-Ramos, B.; Vordermeier, H.M. Phenotypic characterization of bovine memory cells responding to mycobacteria in IFNgamma enzyme linked immunospot assays. Vaccine 2015, 33, 7276–7282. [Google Scholar] [CrossRef] [PubMed]
- Bassey, E.; Collins, M.T. Study of T-lymphocyte subsets of healthy and Mycobacterium avium subsp. paratuberculosis-infected cattle. Infect. Immun. 1997, 65, 4869–4872. [Google Scholar] [CrossRef]
- Charleston, B.; Brackenbury, L.S.; Carr, B.V.; Fray, M.D.; Hope, J.C.; Howard, C.J.; Morrison, W.I. Alpha/beta and gamma interferons are induced by infection with noncytopathic bovine viral diarrhea virus in vivo. J. Virol. 2002, 76, 923–927. [Google Scholar] [CrossRef] [PubMed]
- Kanobana, K.; Koets, A.; Bakker, N.; Ploeger, H.; Vervelde, L. T-cell mediated immune responses in calves primary-infected or re-infected with Cooperia oncophora: Similar effector cells but different timing. Int. J. Parasitol. 2003, 33, 1503–1514. [Google Scholar] [CrossRef] [PubMed]
- Van Meulder, F.; Ratman, D.; Van Coppernolle, S.; Borloo, J.; Li, R.; Chiers, K.; Van Den Broeck, W.; De Bosscher, K.; Claerebout, E.; Geldhof, P. Analysis of the protective immune response following intramuscular vaccination of calves against the intestinal parasite Cooperia oncophora. Int. J. Parasitol. 2015, 45, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Parmentier, H.; Ploeger, H.; Nieuwland, M.; Souren, P.; Van Pinxteren, L.; Rietveld, F.; Reilingh, G.D.V.; Kloosterman, A. Low molecular weight Cooperia oncophora antigens: Characterization and humoral immune responses in calves mono-infected with 100 000 infective larvae. Vet. Parasitol. 1995, 59, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Strube, C.; Haake, C.; Sager, H.; Weber, S.S.; Kaminsky, R.; Buschbaum, S.; Joekel, D.; Schicht, S.; Kremmer, E.; Korrell, J. Vaccination with recombinant paramyosin against the bovine lungworm Dictyocaulus viviparus considerably reduces worm burden and larvae shedding. Parasites Vectors 2015, 8, 119. [Google Scholar] [CrossRef] [PubMed]
- Estes, D.M.; Brown, W.C. Type 1 and type 2 responses in regulation of Ig isotype expression in cattle. Vet. Immunol. Immunopathol. 2002, 90, 1–10. [Google Scholar] [CrossRef]
- Kaplan, M.H.; Schindler, U.; Smiley, S.T.; Grusby, M.J. Stat6 is required for mediating responses to IL-4 and for the development of Th2 cells. Immunity 1996, 4, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Tanaka, T.; Shi, W.; Matsumoto, M.; Minami, M.; Kashiwamura, S.-i.; Nakanishi, K.; Yoshida, N.; Kishimoto, T.; Akira, S. Essential role of Stat6 in IL-4 signalling. Nature 1996, 380, 627–630. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Takemoto, N.; Kurata, H.; Kamogawa, Y.; Miyatake, S.; O’Garra, A.; Arai, N. GATA-3 induces T helper cell type 2 (Th2) cytokine expression and chromatin remodeling in committed Th1 cells. J. Exp. Med. 2000, 192, 105–116. [Google Scholar] [CrossRef]
- Zheng, W.-p.; Flavell, R.A. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 1997, 89, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T.R.; Cherwinski, H.; Bond, M.W.; Giedlin, M.A.; Coffman, R.L. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J. Immunol. 1986, 136, 2348–2357. [Google Scholar] [CrossRef] [PubMed]
- Romagnani, S. Human TH1 and TH2 subsets: Doubt no more. Immunol. Today 1991, 12, 256–257. [Google Scholar] [CrossRef] [PubMed]
- Romagnani, S. Human TH1 and TH2 subsets: Regulation of differentiation and role in protection and immunopathology. Int. Arch. Allergy Immunol. 1992, 98, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Romagnani, S. T-cell subsets (Th1 versus Th2). Ann. Allergy Asthma Immunol. 2000, 85, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Mena, A.; Ioannou, X.P.; Van Kessel, A.; Little-Van Den, S.V.D.; Popowych, Y.; Babiuk, L.A.; Godson, D.L. Th1/Th2 biasing effects of vaccination in cattle as determined by real-time PCR. J. Immunol. Methods 2002, 263, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, S.; Buddle, B.; Hewinson, R.; Vordermeier, H. Bovine tuberculosis: Immune responses in the peripheral blood and at the site of active disease. Immunology 2000, 99, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Mohrs, M.; Shinkai, K.; Mohrs, K.; Locksley, R.M. Analysis of type 2 immunity in vivo with a bicistronic IL-4 reporter. Immunity 2001, 15, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Prout, M.; Ramshaw, H.; Lopez, A.F.; LeGros, G.; Min, B. Cutting edge: Basophils are transiently recruited into the draining lymph nodes during helminth infection via IL-3, but infection-induced Th2 immunity can develop without basophil lymph node recruitment or IL-3. J. Immunol. 2010, 184, 1143–1147. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, Q.; Pesce, J.; Whitmire, J.; Ekkens, M.J.; Foster, A.; VanNoy, J.; Sharpe, A.H.; Urban, J.F.; Gause, W.C. Nippostrongylus brasiliensis can induce B7-independent antigen-specific development of IL-4-producing T cells from naive CD4 T cells in vivo. J. Immunol. 2002, 169, 6959–6968. [Google Scholar] [CrossRef] [PubMed]
- Holland, M.J.; Harcus, Y.M.; Riches, P.L.; Maizels, R.M. Proteins secreted by the parasitic nematode Nippostrongylus brasiliensis act as adjuvants for Th2 responses. Eur. J. Immunol. 2000, 30, 1977–1987. [Google Scholar] [CrossRef] [PubMed]
- Osborne, J.; Devaney, E. The L3 of Brugia induces a Th2-polarized response following activation of an IL-4-producing CD4-CD8-alphabeta T cell population. Int. Immunol. 1998, 10, 1583–1590. [Google Scholar] [CrossRef] [PubMed]
- Kandel, A.; Li, L.; Hada, A.; Xiao, Z. Differential Expression of CD45RO and CD45RA in Bovine T Cells. Cells 2022, 11, 1844. [Google Scholar] [CrossRef] [PubMed]
- Tuo, W.; Li, L.; Lv, Y.; Carrillo, J.; Brown, D.; Davis, W.C.; Song, J.; Zarlenga, D.; Xiao, Z. Abomasal mucosal immune responses of cattle with limited or continuous exposure to pasture-borne gastrointestinal nematode parasite infection. Vet. Parasitol. 2016, 229, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Paul, W.E. Peripheral CD4+ T−cell differentiation regulated by networks of cytokines and transcription factors. Immunol. Rev. 2010, 238, 247–262. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.; Xie, H.; Dougan, S.K.; Ploegh, H.; van Oudenaarden, A. Stochastic cytokine expression induces mixed T helper cell states. PLoS Biol. 2013, 11, e1001618. [Google Scholar] [CrossRef] [PubMed]
- Mestas, J.; Hughes, C.C. Of mice and not men: Differences between mouse and human immunology. J. Immunol. 2004, 172, 2731–2738. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, A.; Omer, S.B. Why and How Vaccines Work. Cell 2020, 183, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Lau, C.M.; Sun, J.C. The widening spectrum of immunological memory. Curr. Opin. Immunol. 2018, 54, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Kandel, A.; Li, L. Synergistic Activation of Bovine CD4+ T Cells by Neutrophils and IL-12. Pathogens 2021, 10, 694. [Google Scholar] [CrossRef] [PubMed]
- Maura, D.; Elmekki, N.; Goddard, C.A. The ammonia oxidizing bacterium Nitrosomonas eutropha blocks T helper 2 cell polarization via the anti-inflammatory cytokine IL-10. Sci. Rep. 2021, 11, 14162. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.A.; Patel, P.; Viney, J.M.; Phillips, R.M.; Solari, R.; Pease, J.E. A degradatory fate for CCR4 suggests a primary role in Th2 inflammation. J. Leukoc. Biol. 2020, 107, 455–466. [Google Scholar] [CrossRef] [PubMed]
- Mandala, W.; Harawa, V.; Munyenyembe, A.; Soko, M.; Longwe, H. Optimization of stimulation and staining conditions for intracellular cytokine staining (ICS) for determination of cytokine-producing T cells and monocytes. Curr. Res. Immunol. 2021, 2, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Ai, W.; Li, H.; Song, N.; Li, L.; Chen, H. Optimal Method to Stimulate Cytokine Production and Its Use in Immunotoxicity Assessment. Int. J. Environ. Res. Public. Health 2013, 10, 3834–3842. [Google Scholar] [CrossRef] [PubMed]
- Elnaggar, M.M.; Knowles, D.P.; Davis, W.C.; Fry, L.M. Flow Cytometric Analysis of the Cytotoxic T-Cell Recall Response to Theileria parva in Cattle Following Vaccination by the Infection and Treatment Method. Vet. Sci. 2021, 8, 114. [Google Scholar] [CrossRef] [PubMed]
- Panagioti, E.; Klenerman, P.; Lee, L.N.; Van Der Burg, S.H.; Arens, R. Features of effective T cell-inducing vaccines against chronic viral infections. Front. Immunol. 2018, 9, 276. [Google Scholar] [CrossRef] [PubMed]
- Flaxman, A.; Ewer, K.J. Methods for Measuring T-Cell Memory to Vaccination: From Mouse to Man. Vaccines 2018, 6, 43. [Google Scholar] [CrossRef] [PubMed]
- Mescher, M.F.; Curtsinger, J.M.; Agarwal, P.; Casey, K.A.; Gerner, M.; Hammerbeck, C.D.; Popescu, F.; Xiao, Z. Signals required for programming effector and memory development by CD8+ T cells. Immunol. Rev. 2006, 211, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Harty, J.T.; Badovinac, V.P. Shaping and reshaping CD8+ T-cell memory. Nat. Rev. Immunol. 2008, 8, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Pepper, M.; Jenkins, M.K. Origins of CD4(+) effector and central memory T cells. Nat. Immunol. 2011, 12, 467–471. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T.R.; Coffman, R.L. TH1 and TH2 cells: Different patterns of lymphokine secretion lead to different functional properties. Annu. Rev. Immunol. 1989, 7, 145–173. [Google Scholar] [CrossRef] [PubMed]
- Pulendran, B. Modulating TH1/TH2 responses with microbes, dendritic cells, and pathogen recognition receptors. Immunol. Res. 2004, 29, 187–196. [Google Scholar] [CrossRef]
- Liu, Y.J. Thymic stromal lymphopoietin: Master switch for allergic inflammation. J. Exp. Med. 2006, 203, 269–273. [Google Scholar] [CrossRef]
- Ochiai, S.; Jagot, F.; Kyle, R.L.; Hyde, E.; White, R.F.; Prout, M.; Schmidt, A.J.; Yamane, H.; Lamiable, O.; Le Gros, G.; et al. Thymic stromal lymphopoietin drives the development of IL-13(+) Th2 cells. Proc. Natl. Acad. Sci. USA 2018, 115, 1033–1038. [Google Scholar] [CrossRef] [PubMed]
- Elhmouzi-Younes, J.; Storset, A.K.; Boysen, P.; Laurent, F.; Drouet, F. Bovine neonate natural killer cells are fully functional and highly responsive to interleukin-15 and to NKp46 receptor stimulation. Vet. Res. 2009, 40, 54. [Google Scholar] [CrossRef] [PubMed]
- Ethier, M.F.; Madison, J.M. IL-4 inhibits calcium transients in bovine trachealis cells by a ryanodine receptor-dependent mechanism. Faseb J. 2006, 20, 154–156. [Google Scholar] [CrossRef]
- Goff, W.L.; Storset, A.K.; Johnson, W.C.; Brown, W.C. Bovine splenic NK cells synthesize IFN-gamma in response to IL-12-containing supernatants from Babesia bovis-exposed monocyte cultures. Parasite Immunol. 2006, 28, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Akbar, A.; Terry, L.; Timms, A.; Beverley, P.; Janossy, G. Unidirectional phenotypic changes within the T200 complex during activation of T cells. J. Immunol. 1988, 140, 2171. [Google Scholar] [CrossRef]
- Sanders, M.E.; Makgoba, M.W.; Shaw, S. Human naive and memory T cells: Reinterpretation of helper-inducer and suppressor-inducer subsets. Immunol. Today 1988, 9, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Anmol, K.; Akanksha, H.; Zhengguo, X. Are CD45RO+ and CD45RA-genuine markers for bovine memory T cells? Anim. Dis. 2022, 2, 1–9. [Google Scholar] [CrossRef]
- Sekiya, T.; Yoshimura, A. In Vitro Th Differentiation Protocol. Methods Mol. Biol. 2016, 1344, 183–191. [Google Scholar] [PubMed]
- Li, L.; Si, H.; Wu, S.W.; Mendez, J.O.; Zarlenga, D.; Tuo, W.; Xiao, Z. Characterization of IL-10-producing neutrophils in cattle infected with Ostertagia ostertagi. Sci. Rep. 2019, 9, 20292. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.; Scozzi, D.; Toth, K.A.; Ponti, D.; Kreisel, D.; Menna, C.; De Falco, E.; D’Andrilli, A.; Rendina, E.A.; Calogero, A.; et al. Naive CD4(+) T Cells Carrying a TLR2 Agonist Overcome TGF-beta-Mediated Tumor Immune Evasion. J. Immunol. 2018, 200, 847–856. [Google Scholar] [CrossRef]
- van Beek, J.J.P.; Florez-Grau, G.; Gorris, M.A.J.; Mathan, T.S.M.; Schreibelt, G.; Bol, K.F.; Textor, J.; de Vries, I.J.M. Human pDCs Are Superior to cDC2s in Attracting Cytolytic Lymphocytes in Melanoma Patients Receiving DC Vaccination. Cell Rep. 2020, 30, 1027–1038.e4. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Yamane, H.; Cote-Sierra, J.; Guo, L.; Paul, W.E. GATA-3 promotes Th2 responses through three different mechanisms: Induction of Th2 cytokine production, selective growth of Th2 cells and inhibition of Th1 cell-specific factors. Cell Res. 2006, 16, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Min, B.; Hu-Li, J.; Watson, C.J.; Grinberg, A.; Wang, Q.; Killeen, N.; Urban, J.F.; Guo, L.; Paul, W.E. Conditional deletion of Gata3 shows its essential function in TH 1-TH 2 responses. Nat. Immunol. 2004, 5, 1157–1165. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Jankovic, D.; Oler, A.J.; Wei, G.; Sharma, S.; Hu, G.; Guo, L.; Yagi, R.; Yamane, H.; Punkosdy, G. The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity 2012, 37, 660–673. [Google Scholar] [CrossRef] [PubMed]
- Rincón, M.; Dérijard, B.; Chow, C.W.; Davis, R.; Flavell, R. Reprogramming the signalling requirement for AP-1 (activator protein-1) activation during differentiation of precursor CD4+ T-cells into effector Th1 and Th2 cells. Genes. Funct. 1997, 1, 51–68. [Google Scholar] [CrossRef] [PubMed]
- Hartenstein, B.; Teurich, S.; Hess, J.; Schenkel, J.; Schorpp-Kistner, M.; Angel, P. Th2 cell-specific cytokine expression and allergen-induced airway inflammation depend on JunB. EMBO J. 2002, 21, 6321–6329. [Google Scholar] [CrossRef] [PubMed]
- Fang, D.; Elly, C.; Gao, B.; Fang, N.; Altman, Y.; Joazeiro, C.; Hunter, T.; Copeland, N.; Jenkins, N.; Liu, Y.-C. Dysregulation of T lymphocyte function in itchy mice: A role for Itch in TH2 differentiation. Nat. Immunol. 2002, 3, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Tournier, C.; Davis, R.J.; Flavell, R.A. Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. Embo J. 1999, 18, 420–432. [Google Scholar] [CrossRef] [PubMed]
- Seumois, G.; Chavez, L.; Gerasimova, A.; Lienhard, M.; Omran, N.; Kalinke, L.; Vedanayagam, M.; Ganesan, A.P.V.; Chawla, A.; Djukanović, R. Epigenomic analysis of primary human T cells reveals enhancers associated with TH2 memory cell differentiation and asthma susceptibility. Nat. Immunol. 2014, 15, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Lund, R.J.; Löytömöki, M.; Naumanen, T.; Dixon, C.; Chen, Z.; Ahlfors, H.; Tuomela, S.; Tahvanainen, J.; Scheinin, J.; Henttinen, T. Genome-wide identification of novel genes involved in early Th1 and Th2 cell differentiation. J. Immunol. 2007, 178, 3648–3660. [Google Scholar] [CrossRef] [PubMed]
- Voice, J.; Donnelly, S.; Dorsam, G.; Dolganov, G.; Paul, S.; Goetzl, E.J. c-Maf and JunB mediation of Th2 differentiation induced by the type 2 G protein-coupled receptor (VPAC2) for vasoactive intestinal peptide. J. Immunol. 2004, 172, 7289–7296. [Google Scholar] [CrossRef]
- Ho, I.-C.; Lo, D.; Glimcher, L.H. c-maf promotes T helper cell type 2 (Th2) and attenuates Th1 differentiation by both interleukin 4–dependent and–independent mechanisms. J. Exp. Med. 1998, 188, 1859–1866. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.R. Transcriptional regulation of T helper type 2 differentiation. Immunology 2014, 141, 498–505. [Google Scholar] [CrossRef]
- Kozuka, T.; Sugita, M.; Shetzline, S.; Gewirtz, A.M.; Nakata, Y. c-Myb and GATA-3 cooperatively regulate IL-13 expression via conserved GATA-3 response element and recruit mixed lineage leukemia (MLL) for histone modification of the IL-13 locus. J. Immunol. 2011, 187, 5974–5982. [Google Scholar] [CrossRef]
- Khare, S.P.; Shetty, A.; Biradar, R.; Patta, I.; Chen, Z.J.; Sathe, A.V.; Reddy, P.C.; Lahesmaa, R.; Galande, S. NF-κB signaling and IL-4 signaling regulate SATB1 expression via alternative promoter usage during Th2 differentiation. Front. Immunol. 2019, 10, 667. [Google Scholar] [CrossRef] [PubMed]
- Santarlasci, V.; Cosmi, L.; Maggi, L.; Liotta, F.; Annunziato, F. IL-1 and T Helper Immune Responses. Front. Immunol. 2013, 4, 182. [Google Scholar] [CrossRef] [PubMed]
- Caucheteux, S.M.; Hu-Li, J.; Guo, L.; Bhattacharyya, N.; Crank, M.; Collins, M.T.; Paul, W.E. IL-1β enhances inflammatory TH2 differentiation. J. Allergy Clin. Immunol. 2016, 138, 898–901.e4. [Google Scholar] [CrossRef] [PubMed]
- Cayrol, C.; Girard, J.P. Interleukin-33 (IL-33): A nuclear cytokine from the IL-1 family. Immunol. Rev. 2018, 281, 154–168. [Google Scholar] [CrossRef]
- Leal-Silva, T.; Vieira-Santos, F.; Oliveira, F.M.S.; Padrão, L.L.S.; Kraemer, L.; da Paixão Matias, P.H.; de Almeida Lopes, C.; Loiola Ruas, A.C.; de Azevedo, I.C.; Nogueira, D.S.; et al. Detrimental role of IL-33/ST2 pathway sustaining a chronic eosinophil-dependent Th2 inflammatory response, tissue damage and parasite burden during Toxocara canis infection in mice. PLoS Negl. Trop. Dis. 2021, 15, e0009639. [Google Scholar] [CrossRef] [PubMed]
- Yeon, S.M.; Halim, L.; Chandele, A.; Perry, C.J.; Kim, S.H.; Kim, S.U.; Byun, Y.; Yuk, S.H.; Kaech, S.M.; Jung, Y.W. IL-7 plays a critical role for the homeostasis of allergen-specific memory CD4 T cells in the lung and airways. Sci. Rep. 2017, 7, 11155. [Google Scholar] [CrossRef] [PubMed]
- Bošnjak, B.; Kazemi, S.; Altenburger, L.M.; Mokrović, G.; Epstein, M.M. Th2-T(RMs) Maintain Life-Long Allergic Memory in Experimental Asthma in Mice. Front. Immunol. 2019. 10, 840.
- Tubo, N.J.; Jenkins, M.K. TCR signal quantity and quality in CD4+ T cell differentiation. Trends Immunol. 2014, 35, 591–596. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, N.D.; Feng, C.G. Regulation of T helper cell fate by TCR signal strength. Front. Immunol. 2020, 11, 624. [Google Scholar] [CrossRef] [PubMed]
- Guy, C.S.; Vignali, K.M.; Temirov, J.; Bettini, M.L.; Overacre, A.E.; Smeltzer, M.; Zhang, H.; Huppa, J.B.; Tsai, Y.-H.; Lobry, C. Distinct TCR signaling pathways drive proliferation and cytokine production in T cells. Nat. Immunol. 2013, 14, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Brogdon, J.L.; Leitenberg, D.; Bottomly, K. The potency of TCR signaling differentially regulates NFATc/p activity and early IL-4 transcription in naive CD4+ T cells. J. Immunol. 2002, 168, 3825–3832. [Google Scholar] [CrossRef] [PubMed]
- Cook, K.D.; Miller, J. TCR-dependent translational control of GATA-3 enhances Th2 differentiation. J. Immunol. 2010, 185, 3209–3216. [Google Scholar] [CrossRef] [PubMed]
- Meningher, T.; Barsheshet, Y.; Ofir-Birin, Y.; Gold, D.; Brant, B.; Dekel, E.; Sidi, Y.; Schwartz, E.; Regev-Rudzki, N.; Avni, O.; et al. Schistosomal extracellular vesicle-enclosed miRNAs modulate host T helper cell differentiation. EMBO Rep. 2020, 21, e47882. [Google Scholar] [CrossRef] [PubMed]
- Mendez, J.; Sun, D.; Tuo, W.; Xiao, Z. Bovine neutrophils form extracellular traps in response to the gastrointestinal parasite Ostertagia ostertagi. Sci. Rep. 2018, 8, 17598. [Google Scholar] [CrossRef]
- Bakshi, M.; Hebert, D.; Gulbronson, C.; Bauchan, G.; Tuo, W.; Zarlenga, D. Ostertagia ostertagi Mediates Early Host Immune Responses via Macrophage and Toll-Like Receptor Pathways. Infect. Immun. 2021, 89, e00017–e00021. [Google Scholar] [CrossRef] [PubMed]
- Seya, T.; Funami, K.; Taniguchi, M.; Matsumoto, M. Antibodies against human Toll-like receptors (TLRs): TLR distribution and localization in human dendritic cells. J. Endotoxin Res. 2005, 11, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Kumar, H.; Kawai, T.; Akira, S. Toll-like receptors and innate immunity. Biochem. Biophys. Res. Commun. 2009, 388, 621–625. [Google Scholar] [CrossRef] [PubMed]
- Mojtabavi, N.; Dekan, G.; Stingl, G.; Epstein, M.M. Long-lived Th2 memory in experimental allergic asthma. J. Immunol. 2002, 169, 4788–4796. [Google Scholar] [CrossRef] [PubMed]
- Steinfelder, S.; Rausch, S.; Michael, D.; Kühl, A.A.; Hartmann, S. Intestinal helminth infection induces highly functional resident memory CD4+ T cells in mice. Eur. J. Immunol. 2017, 47, 353–363. [Google Scholar] [CrossRef]
- Lederer, J.A.; Perez, V.L.; DesRoches, L.; Kim, S.M.; Abbas, A.K.; Lichtman, A.H. Cytokine transcriptional events during helper T cell subset differentiation. J. Exp. Med. 1996, 184, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Grogan, J.L.; Mohrs, M.; Harmon, B.; Lacy, D.A.; Sedat, J.W.; Locksley, R.M. Early transcription and silencing of cytokine genes underlie polarization of T helper cell subsets. Immunity 2001, 14, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Peine, M.; Rausch, S.; Helmstetter, C.; Fröhlich, A.; Hegazy, A.N.; Kühl, A.A.; Grevelding, C.G.; Höfer, T.; Hartmann, S.; Löhning, M. Stable T-bet+ GATA-3+ Th1/Th2 hybrid cells arise in vivo, can develop directly from naive precursors, and limit immunopathologic inflammation. PLoS Biol. 2013, 11, e1001633. [Google Scholar] [CrossRef] [PubMed]
- Jenner, R.G.; Townsend, M.J.; Jackson, I.; Sun, K.; Bouwman, R.D.; Young, R.A.; Glimcher, L.H.; Lord, G.M. The transcription factors T-bet and GATA-3 control alternative pathways of T-cell differentiation through a shared set of target genes. Proc. Natl. Acad. Sci. 2009, 106, 17876–17881. [Google Scholar] [CrossRef] [PubMed]
- Cousins, D.J.; Lee, T.H.; Staynov, D.Z. Cytokine coexpression during human Th1/Th2 cell differentiation: Direct evidence for coordinated expression of Th2 cytokines. J. Immunol. 2002, 169, 2498–2506. [Google Scholar] [CrossRef] [PubMed]
- Messi, M.; Giacchetto, I.; Nagata, K.; Lanzavecchia, A.; Natoli, G.; Sallusto, F. Memory and flexibility of cytokine gene expression as separable properties of human TH1 and TH2 lymphocytes. Nat. Immunol. 2003, 4, 78–86. [Google Scholar] [CrossRef]
- Ferber, I.A.; Lee, H.-J.; Zonin, F.; Heath, V.; Mui, A.; Arai, N.; O’Garra, A. GATA-3 significantly downregulates IFN-γ production from developing Th1 cells in addition to inducing IL-4 and IL-5 levels. Clin. Immunol. 1999, 91, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Yagi, R.; Junttila, I.S.; Wei, G.; Urban Jr, J.F.; Zhao, K.; Paul, W.E.; Zhu, J. The transcription factor GATA3 actively represses RUNX3 protein-regulated production of interferon-γ. Immunity 2010, 32, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Yagi, R.; Zhu, J.; Paul, W.E. An updated view on transcription factor GATA3-mediated regulation of Th1 and Th2 cell differentiation. Int. Immunol. 2011, 23, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Collier, S.P.; Henderson, M.A.; Tossberg, J.T.; Aune, T.M. Regulation of the Th1 genomic locus from Ifng through Tmevpg1 by T-bet. J. Immunol. 2014, 193, 3959–3965. [Google Scholar] [CrossRef] [PubMed]
- Usui, T.; Nishikomori, R.; Kitani, A.; Strober, W. GATA-3 suppresses Th1 development by downregulation of Stat4 and not through effects on IL-12Rβ2 chain or T-bet. Immunity 2003, 18, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Paul, W.E. What determines Th2 differentiation, in vitro and in vivo? Immunol. Cell Biol. 2010, 88, 236–239. [Google Scholar] [CrossRef] [PubMed]
- Taylor-Robinson, A.; Phillips, R. Expression of the IL-1 receptor discriminates Th2 from Th1 cloned CD4+ T cells specific for Plasmodium chabaudi. Immunology 1994, 81, 216. [Google Scholar] [PubMed]
- Greenbaum, L.A.; Horowitz, J.; Woods, A.; Pasqualini, T.; Reich, E.; Bottomly, K. Autocrine growth of CD4+ T cells. Differential effects of IL-1 on helper and inflammatory T cells. J. Immunol. 1988, 140, 1555–1560. [Google Scholar] [CrossRef]
- Lichtman, A.H.; Chin, J.; Schmidt, J.A.; Abbas, A.K. Role of interleukin 1 in the activation of T lymphocytes. Proc. Natl. Acad. Sci. USA 1988, 85, 9699–9703. [Google Scholar] [CrossRef] [PubMed]
- Helmby, H.; Grencis, R.K. Interleukin 1 plays a major role in the development of Th2-mediated immunity. Eur. J. Immunol. 2004, 34, 3674–3681. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Guo, L.; Min, B.; Watson, C.J.; Hu-Li, J.; Young, H.A.; Tsichlis, P.N.; Paul, W.E. Growth factor independent-1 induced by IL-4 regulates Th2 cell proliferation. Immunity 2002, 16, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Cote-Sierra, J.; Foucras, G.; Guo, L.; Chiodetti, L.; Young, H.A.; Hu-Li, J.; Zhu, J.; Paul, W.E. Interleukin 2 plays a central role in Th2 differentiation. Proc. Natl. Acad. Sci. USA 2004, 101, 3880–3885. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Jankovic, D.; Grinberg, A.; Guo, L.; Paul, W.E. Gfi-1 plays an important role in IL-2-mediated Th2 cell expansion. Proc. Natl. Acad. Sci. USA 2006, 103, 18214–18219. [Google Scholar] [CrossRef] [PubMed]
- Yamane, H.; Zhu, J.; Paul, W.E. Independent roles for IL-2 and GATA-3 in stimulating naive CD4+ T cells to generate a Th2-inducing cytokine environment. J. Exp. Med. 2005, 202, 793–804. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Huang, Y.; Chen, X.; Hu-Li, J.; Urban, J.F., Jr.; Paul, W.E. Innate immunological function of TH2 cells in vivo. Nat. Immunol. 2015, 16, c1051–c1059. [Google Scholar] [CrossRef]
- Wurster, A.L.; Rodgers, V.L.; Satoskar, A.R.; Whitters, M.J.; Young, D.A.; Collins, M.; Grusby, M.J. Interleukin 21 is a T helper (Th) cell 2 cytokine that specifically inhibits the differentiation of naive Th cells into interferon gamma-producing Th1 cells. J. Exp. Med. 2002, 196, 969–977. [Google Scholar] [CrossRef]
- Fröhlich, A.; Marsland, B.J.; Sonderegger, I.; Kurrer, M.; Hodge, M.R.; Harris, N.L.; Kopf, M. IL-21 receptor signaling is integral to the development of Th2 effector responses in vivo. Blood 2007, 109, 2023–2031. [Google Scholar] [CrossRef] [PubMed]
- Davydov, I.V.; Krammer, P.H.; Li-Weber, M. Nuclear factor-IL6 activates the human IL-4 promoter in T cells. J. Immunol. 1995, 155, 5273–5279. [Google Scholar] [CrossRef]
- Park, J.; Chung, S.W.; Kim, S.H.; Kim, T.S. Up-regulation of interleukin-4 production via NF-AT/AP-1 activation in T cells by biochanin A, a phytoestrogen and its metabolites. Toxicol. Appl. Pharmacol. 2006, 212, 188–199. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Y.; Gu, W.; Sun, B. TH1/TH2 cell differentiation and molecular signals. Adv. Exp. Med. Biol. 2014, 841, E1–E2. [Google Scholar] [PubMed]
- Liao, W.; Schones, D.E.; Oh, J.; Cui, Y.; Cui, K.; Roh, T.-Y.; Zhao, K.; Leonard, W.J. Priming for T helper type 2 differentiation by interleukin 2–mediated induction of interleukin 4 receptor α-chain expression. Nat. Immunol. 2008, 9, 1288–1296. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Cote-Sierra, J.; Guo, L.; Paul, W.E. Stat5 activation plays a critical role in Th2 differentiation. Immunity 2003, 19, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Heussler, V.T.; Eichhora, M.; Dobbelaere, D.A. Cloning of a full-length cDNA encoding bovine interleukin 4 by the polymerase chain reaction. Gene 1992, 114, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Prashanth, T.; Reddy, G.R.; Suryanaryana, V.V.; Dechamma, H.J. Cloning and expression of Bos indicus interleukin-4 in mammalian cells. Indian. J. Exp. Biol. 2013, 51, 352–356. [Google Scholar] [PubMed]
- Olsen, S.; Stevens, M. Effects of recombinant human cytokines on mitogen-induced bovine peripheral blood mononuclear cell proliferation. Cytokine 1993, 5, 498–505. [Google Scholar] [CrossRef] [PubMed]
- Tuo, W.; Estes, D.M.; Brown, W.C. Comparative effects of interleukin-12 and interleukin-4 on cytokine responses by antigen-stimulated memory CD4+ T cells of cattle: IL-12 enhances IFN-gamma production, whereas IL-4 has marginal effects on cytokine expression. J. Interferon Cytokine Res. 1999, 19, 741–749. [Google Scholar] [CrossRef] [PubMed]
- Parronchi, P.; De Carli, M.; Manetti, R.; Simonelli, C.; Sampognaro, S.; Piccinni, M.P.; Macchia, D.; Maggi, E.; Del Prete, G.; Romagnani, S. IL-4 and IFN (alpha and gamma) exert opposite regulatory effects on the development of cytolytic potential by Th1 or Th2 human T cell clones. J. Immunol. 1992, 149, 2977–2983. [Google Scholar] [CrossRef] [PubMed]
- Yarovinsky, T.O.; Butler, N.S.; Monick, M.M.; Hunninghake, G.W. Early exposure to IL-4 stabilizes IL-4 mRNA in CD4+ T cells via RNA-binding protein HuR. J. Immunol. 2006, 177, 4426–4435. [Google Scholar] [CrossRef] [PubMed]
- Renz, H.; Domenico, J.; Gelfand, E.W. IL-4-dependent up-regulation of IL-4 receptor expression in murine T and B cells. J. Immunol. 1991, 146, 3049–3055. [Google Scholar] [CrossRef] [PubMed]
- Imrie, H.; Williams, D.J. Stimulation of bovine monocyte-derived macrophages with lipopolysaccharide, interferon-ɣ, Interleukin-4 or Interleukin-13 does not induce detectable changes in nitric oxide or arginase activity. BMC Vet. Res. 2019, 15, 45. [Google Scholar] [CrossRef] [PubMed]
- Nuntaprasert, A.; Mori, Y.; Muneta, Y.; Yoshihara, K.; Tsukiyama-Kohara, K.; Kai, C. The effect of recombinant swine interleukin-4 on swine immune cells and on pro-inflammatory cytokine productions in pigs. Comp. Immunol. Microbiol. Infect. Dis. 2005, 28, 83–101. [Google Scholar] [CrossRef] [PubMed]
- Ebner, S.; Hofer, S.; Nguyen, V.A.; Fürhapter, C.; Herold, M.; Fritsch, P.; Heufler, C.; Romani, N. A novel role for IL-3: Human monocytes cultured in the presence of IL-3 and IL-4 differentiate into dendritic cells that produce less IL-12 and shift Th cell responses toward a Th2 cytokine pattern. J. Immunol. 2002, 168, 6199–6207. [Google Scholar] [CrossRef] [PubMed]
- Selliah, N.; Finkel, T.H. Cutting edge: JAK3 activation and rescue of T cells from HIV gp120-induced unresponsiveness. J. Immunol. 1998, 160, 5697–5701. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Guo, L.; Gogate, S.; Karim, Z.; Hanifi, A.; Leung, D.Y.; Gorska, M.M.; Alam, R. IL-2 and IL-4 Stimulate MEK1 Expression and Contribute to T Cell Resistance against Suppression by TGF-β and IL-10 in Asthma. J. Immunol. 2010, 185, 5704–5713. [Google Scholar] [CrossRef] [PubMed]
- Ohkubo, Y.; Arima, M.; Arguni, E.; Okada, S.; Yamashita, K.; Asari, S.; Obata, S.; Sakamoto, A.; Hatano, M.; Ebara, M.; et al. A role for c-fos/activator protein 1 in B lymphocyte terminal differentiation. J. Immunol. 2005, 174, 7703–7710. [Google Scholar] [CrossRef]
- Zhang, L.F.; Okuma, K.; Tanaka, R.; Kodama, A.; Kondo, K.; Ansari, A.A.; Tanaka, Y. Generation of mature dendritic cells with unique phenotype and function by in vitro short-term culture of human monocytes in the presence of interleukin-4 and interferon-beta. Exp. Biol. Med. 2008, 233, 721–731. [Google Scholar] [CrossRef] [PubMed]
- Dauer, M.; Obermaier, B.; Herten, J.; Haerle, C.; Pohl, K.; Rothenfusser, S.; Schnurr, M.; Endres, S.; Eigler, A. Mature dendritic cells derived from human monocytes within 48 hours: A novel strategy for dendritic cell differentiation from blood precursors. J. Immunol. 2003, 170, 4069–4076. [Google Scholar] [CrossRef] [PubMed]
- Liedtke, K.; Alter, C.; Günther, A.; Hövelmeyer, N.; Klopfleisch, R.; Naumann, R.; Wunderlich, F.T.; Buer, J.; Westendorf, A.M.; Hansen, W. Endogenous CD83 Expression in CD4(+) Conventional T Cells Controls Inflammatory Immune Responses. J. Immunol. 2020, 204, 3217–3226. [Google Scholar] [CrossRef] [PubMed]
- Hock, B.; Fernyhough, L.; Gough, S.; Steinkasserer, A.; Cox, A.; McKenzie, J. Release and clinical significance of soluble CD83 in chronic lymphocytic leukemia. Leuk. Res. 2009, 33, 1089–1095. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Rani, L.; Mhaske, S.T.; Pote, S.T.; Behera, S.; Mishra, G.C.; Wani, M.R. IL-3 receptor expression on activated human Th cells is regulated by IL-4, and IL-3 synergizes with IL-4 to enhance Th2 cell differentiation. J. Immunol. 2020, 204, 819–831. [Google Scholar] [CrossRef]
- Maizels, R.M.; McSorley, H.J. Regulation of the host immune system by helminth parasites. J. Allergy Clin. Immunol. 2016, 138, 666–675. [Google Scholar] [CrossRef] [PubMed]
- Maizels, R.M.; Smits, H.H.; McSorley, H.J. Modulation of host immunity by helminths: The expanding repertoire of parasite effector molecules. Immunity 2018, 49, 801–818. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.D.; van der Werf, N.; Maizels, R.M. T cells in helminth infection: The regulators and the regulated. Trends Immunol. 2012, 33, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Bouchery, T.; Kyle, R.; Ronchese, F.; Le Gros, G. The differentiation of CD4+ T-helper cell subsets in the context of helminth parasite infection. Front. Immunol. 2014, 5, 487. [Google Scholar] [CrossRef] [PubMed]
- Wiggin, C.; Gibbs, H. Studies of the immunomodulatory effects of low-level infection with Ostertagia ostertagi in calves. Am. J. Vet. Res. 1989, 50, 1764–1770. [Google Scholar] [PubMed]
- Wiggin, C.; Gibbs, H. Adverse immune reactions and the pathogenesis of Ostertagia ostertagi infections in calves. Am. J. Vet. Res. 1990, 51, 825–832. [Google Scholar] [CrossRef]
- De Marez, T.; Cox, E.; Claerebout, E.; Vercruysse, J.; Goddeeris, B. Induction and suppression of lymphocyte proliferation by antigen extracts of Ostertagia ostertagi. Vet. Immunol. Immunopathol. 1997, 57, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Hagberg, M.; Lundén, A.; Höglund, J.; Morrison, D.A.; Waller, K.P.; Wattrang, E. Characterization of bovine lymphocytes stimulated in vitro by Dictyocaulus viviparus homogenate. Parasite Immunol. 2008, 30, 342–353. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.; Howie, S.; Odling, K.; Glass, E. Theileria annulata induces abberrant T cell activation in vitro and in vivo. Clin. Exp. Immunol. 1995, 99, 203–210. [Google Scholar] [CrossRef] [PubMed]
- McNeilly, T.N.; Nisbet, A.J. Immune modulation by helminth parasites of ruminants: Implications for vaccine development and host immune competence. Parasite 2014, 21, 51. [Google Scholar] [CrossRef] [PubMed]
- Noland, G.S.; Chowdhury, D.R.; Urban, J.F., Jr.; Zavala, F.; Kumar, N. Helminth infection impairs the immunogenicity of a Plasmodium falciparum DNA vaccine, but not irradiated sporozoites, in mice. Vaccine 2010, 28, 2917–2923. [Google Scholar] [CrossRef]
- van der Zande, H.J.; Zawistowska-Deniziak, A.; Guigas, B. Immune regulation of metabolic homeostasis by helminths and their molecules. Trends Parasitol. 2019, 35, 795–808. [Google Scholar] [CrossRef] [PubMed]
- White, R.R.; Artavanis-Tsakonas, K. How helminths use excretory secretory fractions to modulate dendritic cells. Virulence 2012, 3, 668–677. [Google Scholar] [CrossRef]
- Nono, J.K.; Ndlovu, H.; Abdel Aziz, N.; Mpotje, T.; Hlaka, L.; Brombacher, F. Interleukin-4 receptor alpha is still required after Th2 polarization for the maintenance and the recall of protective immunity to Nematode infection. PLoS Negl. Trop. Dis. 2017, 11, e0005675. [Google Scholar] [CrossRef] [PubMed]
- Bian, K.; Zhong, M.; Harari, Y.; Lai, M.; Weisbrodt, N.; Murad, F. Helminth regulation of host IL-4Rα/Stat6 signaling: Mechanism underlying NOS-2 inhibition by Trichinella spiralis. Proc. Natl. Acad. Sci. USA 2005, 102, 3936–3941. [Google Scholar] [CrossRef] [PubMed]
- Babu, S.; Blauvelt, C.P.; Kumaraswami, V.; Nutman, T.B. Cutting edge: Diminished T cell TLR expression and function modulates the immune response in human filarial infection. J. Immunol. 2006, 176, 3885–3889. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Zhao, Y.; Feng, Y.; Jin, C.; Yang, Q.; Qiu, H.; Xie, H.; Xie, S.; Zhou, Y.; Huang, J. Expression of TLR2, TLR3, TLR4, and TLR7 on pulmonary lymphocytes of Schistosoma japonicum-infected C57BL/6 mice. Innate Immun. 2019, 25, 224–234. [Google Scholar] [CrossRef]
- Pineda, M.A.; McGrath, M.A.; Smith, P.C.; Al-Riyami, L.; Rzepecka, J.; Gracie, J.A.; Harnett, W.; Harnett, M.M. The parasitic helminth product ES-62 suppresses pathogenesis in collagen-induced arthritis by targeting the interleukin-17–producing cellular network at multiple sites. Arthritis Rheum. 2012, 64, 3168–3178. [Google Scholar] [CrossRef] [PubMed]
- Schenten, D.; Nish, S.A.; Yu, S.; Yan, X.; Lee, H.K.; Brodsky, I.; Pasman, L.; Yordy, B.; Wunderlich, F.T.; Brüning, J.C.; et al. Signaling through the adaptor molecule MyD88 in CD4+ T cells is required to overcome suppression by regulatory T cells. Immunity 2014, 40, 78–90. [Google Scholar] [CrossRef] [PubMed]
- Nouri, Y.; Weinkove, R.; Perret, R. T-cell intrinsic Toll-like receptor signaling: Implications for cancer immunotherapy and CAR T-cells. J. Immunother. Cancer 2021, 9, e003065. [Google Scholar] [CrossRef] [PubMed]
- Cottalorda, A.; Verschelde, C.; Marçais, A.; Tomkowiak, M.; Musette, P.; Uematsu, S.; Akira, S.; Marvel, J.; Bonnefoy-Berard, N. TLR2 engagement on CD8 T cells lowers the thresholdfor optimal antigen-induced T cell activation. Eur. J. Immunol. 2006, 36, 1684–1693. [Google Scholar] [CrossRef] [PubMed]
- Komai-Koma, M.; Jones, L.; Ogg, G.S.; Xu, D.; Liew, F.Y. TLR2 is expressed on activated T cells as a costimulatory receptor. Proc. Natl. Acad. Sci. USA 2004, 101, 3029–3034. [Google Scholar] [CrossRef] [PubMed]
- Gelman, A.E.; Zhang, J.; Choi, Y.; Turka, L.A. Toll-like receptor ligands directly promote activated CD4+ T cell survival. J. Immunol. 2004, 172, 6065–6073. [Google Scholar] [CrossRef] [PubMed]
- Mansson, A.; Adner, M.; Cardell, L.O. Toll-like receptors in cellular subsets of human tonsil T cells: Altered expression during recurrent tonsillitis. Respir. Res. 2006, 7, 36. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.H.; Taylor, D.K.; Turka, L.A. The contribution of direct TLR signaling to T cell responses. Immunol. Res. 2009, 45, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Kelso, A.; Troutt, A.B.; Maraskovsky, E.; Gough, N.M.; Morris, L.; Pech, M.H.; Thomson, J.A. Heterogeneity in lymphokine profiles of CD4+ and CD8+ T cells and clones activated in vivo and in vitro. Immunol. Rev. 1991, 123, 85–114. [Google Scholar] [CrossRef] [PubMed]
- Paliard, X.; de Waal Malefijt, R.; Yssel, H.; Blanchard, D.; Chretien, I.; Abrams, J.; De Vries, J.; Spits, H. Simultaneous production of IL-2, IL-4, and IFN-gamma by activated human CD4+ and CD8+ T cell clones. J. Immunol. 1988, 141, 849–855. [Google Scholar] [CrossRef] [PubMed]
- Miner, K.T.; Croft, M. Generation, persistence, and modulation of Th0 effector cells: Role of autocrine IL-4 and IFN-γ. J. Immunol. 1998, 160, 5280–5287. [Google Scholar] [CrossRef]
- Bock, C.N.; Babu, S.; Breloer, M.; Rajamanickam, A.; Boothra, Y.; Brunn, M.-L.; Kühl, A.A.; Merle, R.; Löhning, M.; Hartmann, S. Th2/1 hybrid cells occurring in murine and human strongyloidiasis share effector functions of Th1 cells. Front. Cell. Infect. Microbiol. 2017, 7, 261. [Google Scholar] [CrossRef]
- Burt, P.; Peine, M.; Peine, C.; Borek, Z.; Serve, S.; Flossdorf, M.; Hegazy, A.N.; Hoefer, T.; Loehning, M.; Thurley, K. Dissecting the dynamic transcriptional landscape of early T helper cell differentiation into Th1, Th2, and Th1/2 hybrid cells. Front. Immunol. 2022, 13, 4407. [Google Scholar] [CrossRef] [PubMed]
- Norimine, J.; Suarez, C.E.; McElwain, T.F.; Florin-Christensen, M.; Brown, W.C. Immunodominant epitopes in Babesia bovis rhoptry-associated protein 1 that elicit memory CD4(+)-T-lymphocyte responses in B-bovis-immune individuals are located in the amino-terminal domain. Infect. Immun. 2002, 70, 2039–2048. [Google Scholar] [CrossRef] [PubMed]
- Brown, W.C.; Davis, W.C.; Dobbelaere, D.A.; Rice-Ficht, A.C. CD4+ T-cell clones obtained from cattle chronically infected with Fasciola hepatica and specific for adult worm antigen express both unrestricted and Th2 cytokine profiles. Infect. Immun. 1994, 62, 818–827. [Google Scholar] [CrossRef] [PubMed]
- Rosbottom, A.; Guy, C.; Gibney, E.; Smith, R.; Valarcher, J.; Taylor, G.; Williams, D.J. Peripheral immune responses in pregnant cattle following Neospora caninum infection. Parasite Immunol. 2007, 29, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Afkarian, M.; Sedy, J.R.; Yang, J.; Jacobson, N.G.; Cereb, N.; Yang, S.Y.; Murphy, T.L.; Murphy, K.M. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat. Immunol. 2002, 3, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Szabo, S.J.; Kim, S.T.; Costa, G.L.; Zhang, X.; Fathman, C.G.; Glimcher, L.H. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 2000, 100, 655–669. [Google Scholar] [CrossRef]
- Szabo, S.J.; Sullivan, B.M.; Stemmann, C.; Satoskar, A.R.; Sleckman, B.P.; Glimcher, L.H. Distinct effects of T-bet in TH1 lineage commitment and IFN-γ production in CD4 and CD8 T cells. Science 2002, 295, 338–342. [Google Scholar] [CrossRef]

| Specificity | Clone | Isotype | Colors | Source |
|---|---|---|---|---|
| bCD3 | MM1A | IgG1 | - | WSUMAC |
| bCD4 | CC8 | IgG2a | FITC | Bio-Rad |
| bCD25 | LCTB2A | IgG3 | PE | WSUMAC |
| bCD62L | BAQ92A | IgG1 | FITC | WSUMAC |
| bIFNγ | CC302 | IgG1 | PE | Bio-Rad |
| bIL4 | CC303 | IgG2a | - | Bio-Rad |
| Specificity | Secondary Antibodies | Source |
|---|---|---|
| IgG1 | Anti-mouse IgG1-APC | BioLegend |
| IgG2a | Anti-mouse IgG2a-APC | BioLegend |
| Upstream Regulator | Th1/Naïve | Th2/Naïve | ||||||
|---|---|---|---|---|---|---|---|---|
| Z-Score | Predicted Activation State | p-Value | Target Molecules in Database | Z-Score | Predicted Activation State | p-Value | Target Molecules in Database | |
| CEBPB | 3.466 | Activated | 2.54 × 10−12 | 36 | 2.859 | Activated | 3.16 × 10−15 | 41 |
| ETV5 | 2 | Activated | 0.0167 | 5 | 2.449 | Activated | 0.000158 | 8 |
| MYC | 4.449 | Activated | 4.51 × 10−6 | 40 | 3.667 | Activated | 7.34 × 10−11 | 52 |
| TBX21 | 2.449 | Activated | 9.78 × 10−4 | 6 | 2 | Activated | 0.0308 | 4 |
| FOXM1 | 2.47 | Activated | 2.3 × 10−6 | 12 | N/A | |||
| AP1 | N/A | 2.372 | Activated | 2.05 × 10−2 | 7 | |||
| E2F2 | N/A | 2 | Activated | 0.00593 | 5 | |||
| MAF | N/A | 2.219 | Activated | 8.91 × 10−5 | 7 | |||
| MYB | N/A | 2.157 | Activated | 0.00012 | 11 | |||
| NFKB1 | N/A | 2.578 | Activated | 6.56 × 10−5 | 15 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kandel, A.; Li, L.; Wang, Y.; Tuo, W.; Xiao, Z. Differentiation and Regulation of Bovine Th2 Cells In Vitro. Cells 2024, 13, 738. https://doi.org/10.3390/cells13090738
Kandel A, Li L, Wang Y, Tuo W, Xiao Z. Differentiation and Regulation of Bovine Th2 Cells In Vitro. Cells. 2024; 13(9):738. https://doi.org/10.3390/cells13090738
Chicago/Turabian StyleKandel, Anmol, Lei Li, Yan Wang, Wenbin Tuo, and Zhengguo Xiao. 2024. "Differentiation and Regulation of Bovine Th2 Cells In Vitro" Cells 13, no. 9: 738. https://doi.org/10.3390/cells13090738







