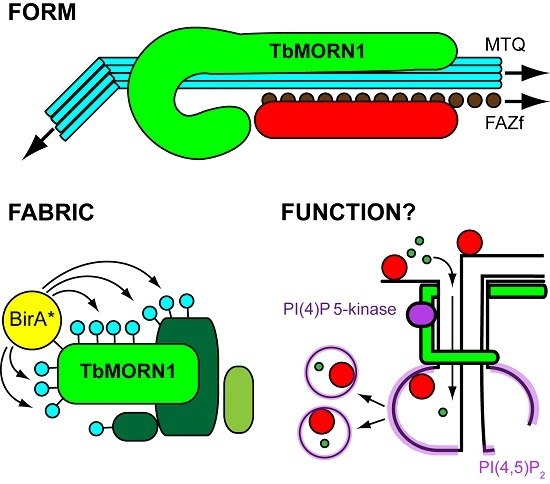
Form, Fabric, and Function of a Flagellum-Associated Cytoskeletal Structure
Abstract
:1. Introduction
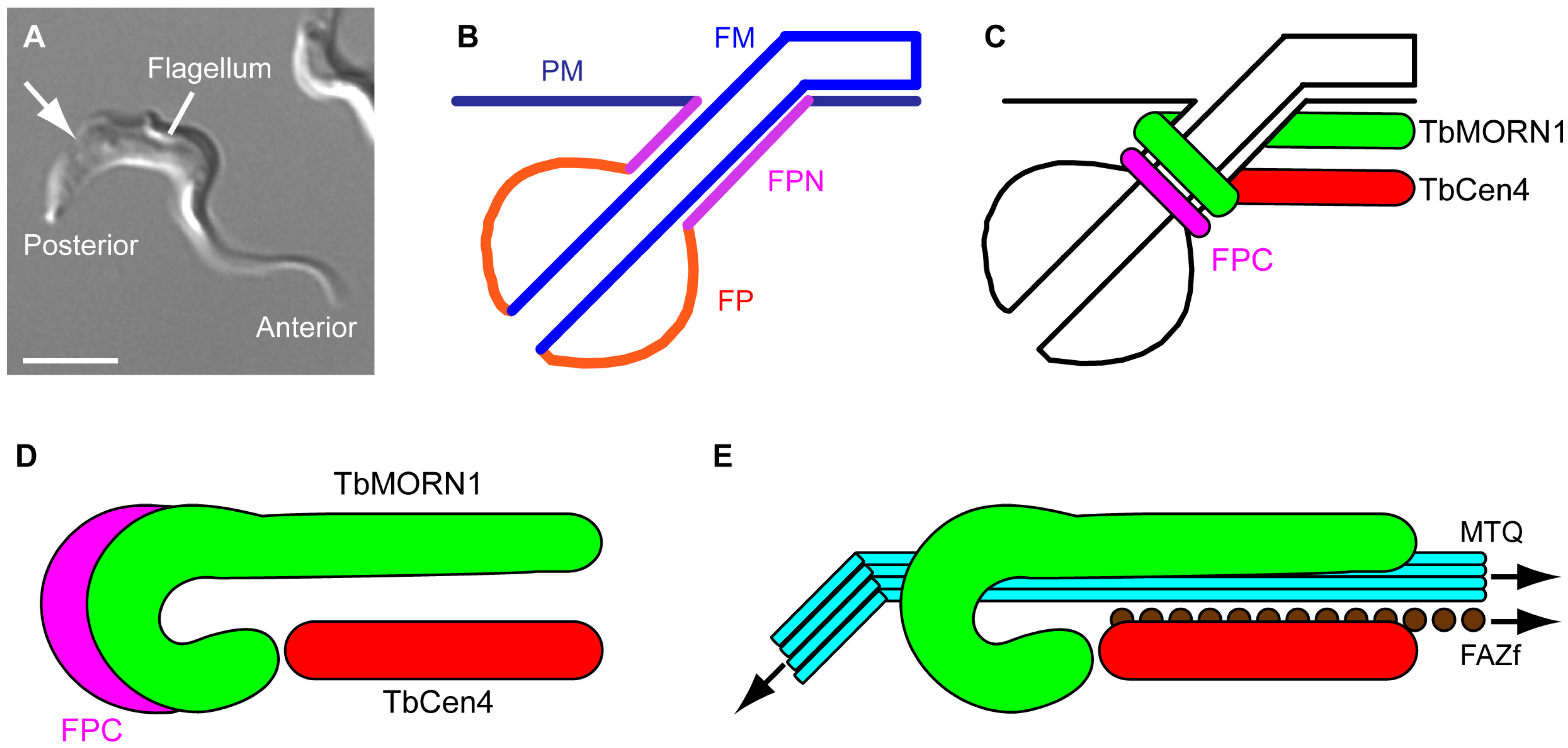
2. Form
2.1. The Shape of Things
2.2. Replication Mechanism
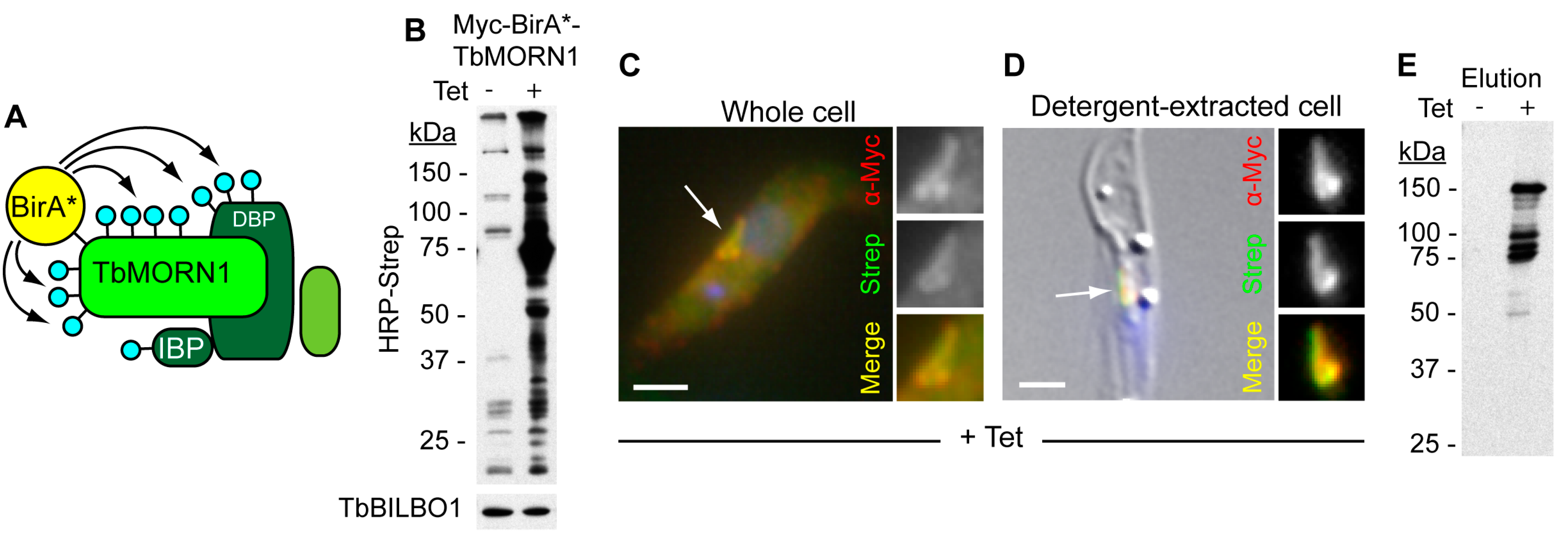
3. Fabric
Composition of the Hook Complex
| Gene Code, Name | Size (kDa) | Tritryp Presence | Domains (Including CC) | Repeat Motifs | References |
|---|---|---|---|---|---|
| Tb927.6.4670 (TbMORN1) | 41 | Tb, Tc, L | - | 15 × MORN | [34,36,38,51] |
| Tb927.4.3120 | 101 | Tb | - | - | [51] |
| Tb927.7.7000 | 175 | Tb, Tc | - | - | [51] |
| Tb927.8.3010 | 82 | Tb, Tc | 3 × CC | - | [51] |
| Tb927.10.850 | 168 | Tb, Tc | 7 × CC | 5 × WD40 | [42,51] |
| Tb927.10.3010 | 133 | Tb, Tc | 2 × CC | - | [42,51] |
| Tb927.10.8820 | 85 | Tb, Tc, L | - | - | [42,51] |
| Tb927.11.2440 (TBCCD1) | 59 | Tb, Tc, L | TBCC-like, 1 × CC | - | [58] |
| Tb927.11.8950 (TbLRRP1) | 79 | Tb, Tc, L | 1 × CC | 7 × LRR | [42,57,59,60] |
4. Function
4.1. History and Semantics
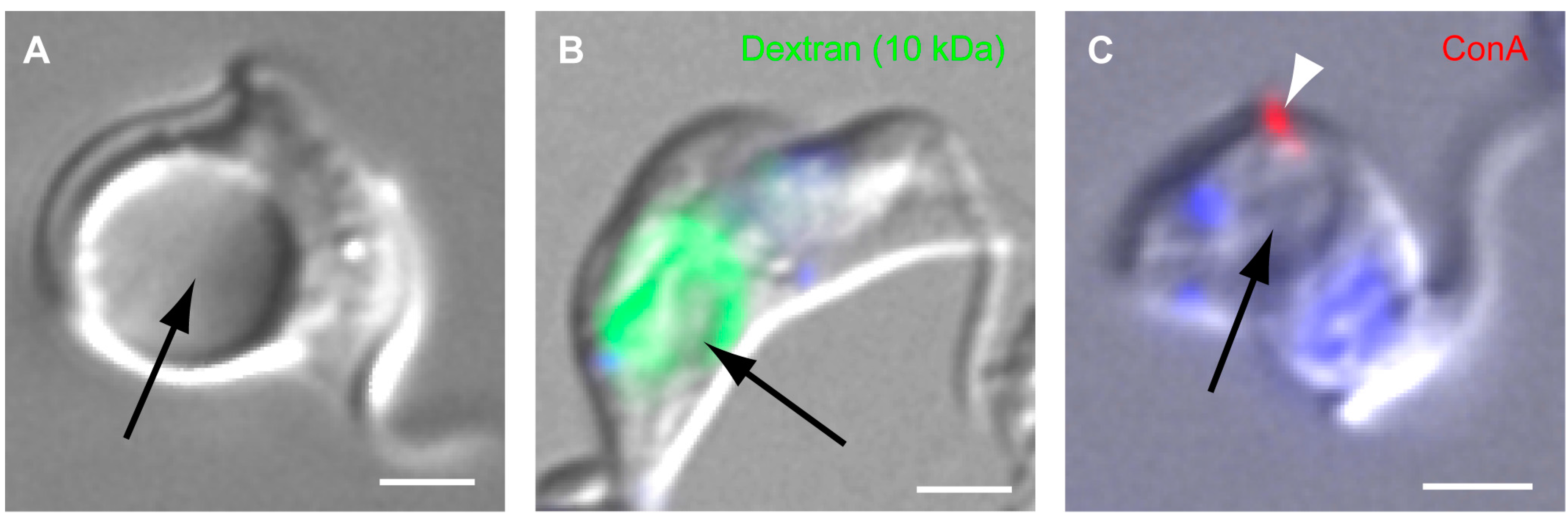
4.2. Function of TbMORN1
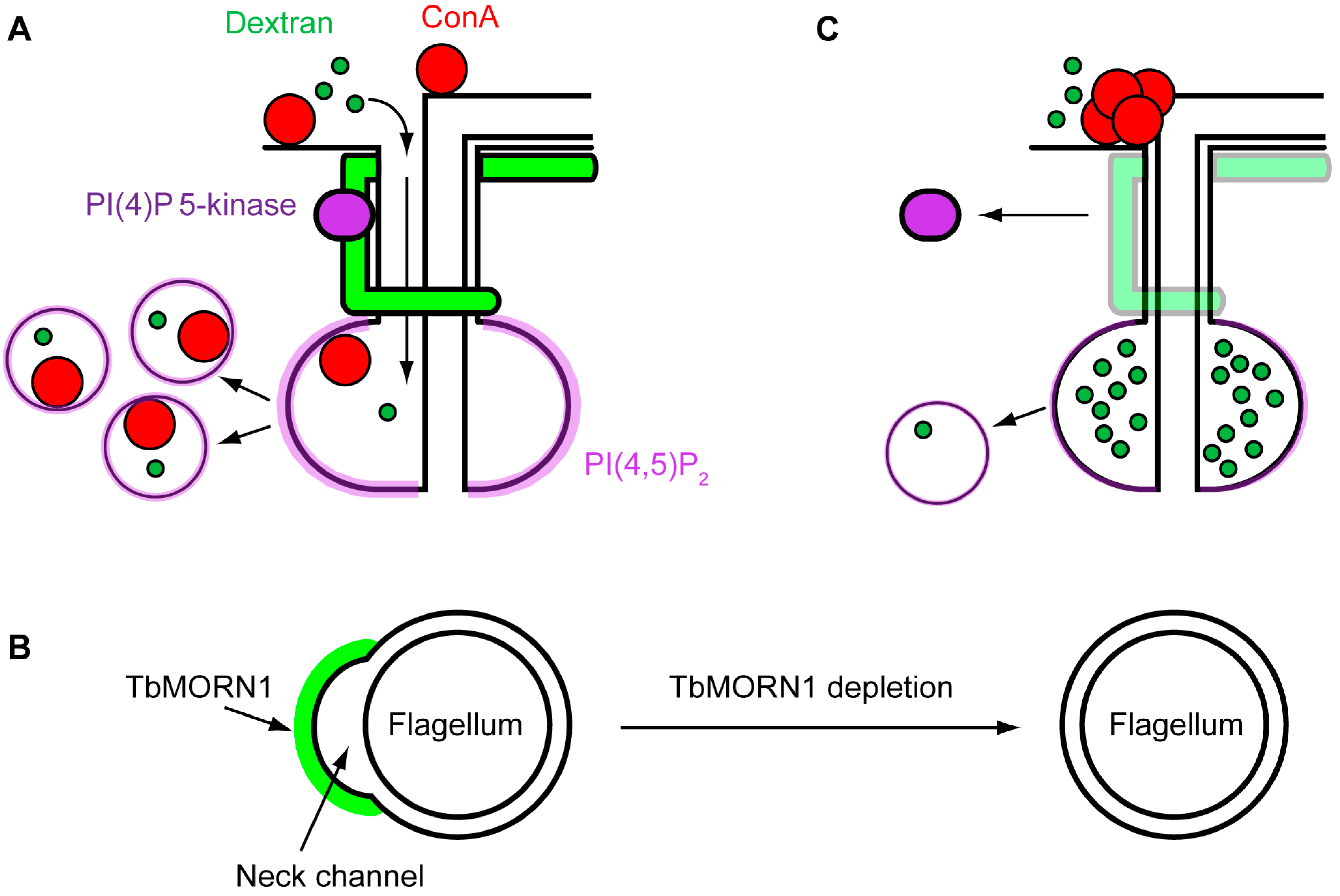
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Adl, S.M.; Simpson, A.G.; Lane, C.E.; Lukes, J.; Bass, D.; Bowser, S.S.; Brown, M.W.; Burki, F.; Dunthorn, M.; Hampl, V.; et al. The revised classification of eukaryotes. J. Eukaryot. Microbiol. 2012, 59, 429–493. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, C.S.; Yukich, J.; Goeree, R.; Tediosi, F. A literature review of economic evaluations for a neglected tropical disease: Human African trypanosomiasis (“sleeping sickness”). PLoS Negl. Trop. Dis. 2015, 9, e0003397. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Fiz-Palacios, O.; Fu, C.J.; Fehling, J.; Tsai, C.C.; Baldauf, S.L. An alternative root for the eukaryote tree of life. Curr. Biol. 2014, 24, 465–470. [Google Scholar] [CrossRef] [PubMed]
- Derelle, R.; Torruella, G.; Klimes, V.; Brinkmann, H.; Kim, E.; Vlcek, C.; Lang, B.F.; Elias, M. Bacterial proteins pinpoint a single eukaryotic root. Proc. Natl. Acad. Sci. USA 2015, 112, E693–E699. [Google Scholar] [CrossRef] [PubMed]
- Rodgers, J. Trypanosomiasis and the brain. Parasitology 2010, 137, 1995–2006. [Google Scholar] [CrossRef] [PubMed]
- Lundkvist, G.B.; Sellix, M.T.; Nygard, M.; Davis, E.; Straume, M.; Kristensson, K.; Block, G.D. Clock gene expression during chronic inflammation induced by infection with Trypanosoma brucei brucei in rats. J. Biol. Rhythm. 2010, 25, 92–102. [Google Scholar] [CrossRef] [PubMed]
- Vickerman, K. Developmental cycles and biology of pathogenic trypanosomes. Br. Med. Bull. 1985, 41, 105–114. [Google Scholar] [PubMed]
- Rotureau, B.; van Den Abbeele, J. Through the dark continent: African trypanosome development in the tsetse fly. Front. Cell. Infect. Microbiol. 2013, 3, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, R.; Gluenz, E.; Peacock, L.; Gibson, W.; Gull, K.; Carrington, M. The heart of darkness: Growth and form of Trypanosoma brucei in the tsetse fly. Trends Parasitol. 2009, 25, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, R.J.; Gluenz, E.; Gull, K. The limits on trypanosomatid morphological diversity. PLoS ONE 2013, 8, e79581. [Google Scholar] [CrossRef] [PubMed]
- Gull, K. The cytoskeleton of trypanosomatid parasites. Annu. Rev. Microbiol. 1999, 53, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D.R.; Sherwin, T.; Ploubidou, A.; Byard, E.H.; Gull, K. Microtubule polarity and dynamics in the control of organelle positioning, segregation, and cytokinesis in the trypanosome cell cycle. J. Cell Biol. 1995, 128, 1163–1172. [Google Scholar] [CrossRef] [PubMed]
- Engstler, M.; Thilo, L.; Weise, F.; Grunfelder, C.G.; Schwarz, H.; Boshart, M.; Overath, P. Kinetics of endocytosis and recycling of the GPI-anchored variant surface glycoprotein in Trypanosoma brucei. J. Cell Sci. 2004, 117, 1105–1115. [Google Scholar] [CrossRef] [PubMed]
- Grunfelder, C.G.; Engstler, M.; Weise, F.; Schwarz, H.; Stierhof, Y.D.; Morgan, G.W.; Field, M.C.; Overath, P. Endocytosis of a glycosylphosphatidylinositol-anchored protein via clathrin-coated vesicles, sorting by default in endosomes, and exocytosis via RAB11-positive carriers. Mol. Biol. Cell 2003, 14, 2029–2040. [Google Scholar] [CrossRef] [PubMed]
- Lacomble, S.; Vaughan, S.; Gadelha, C.; Morphew, M.K.; Shaw, M.K.; McIntosh, J.R.; Gull, K. Basal body movements orchestrate membrane organelle division and cell morphogenesis in Trypanosoma brucei. J. Cell Sci. 2010, 123, 2884–2891. [Google Scholar] [CrossRef] [PubMed]
- Heddergott, N.; Kruger, T.; Babu, S.B.; Wei, A.; Stellamanns, E.; Uppaluri, S.; Pfohl, T.; Stark, H.; Engstler, M. Trypanosome motion represents an adaptation to the crowded environment of the vertebrate bloodstream. PLoS Pathog. 2012, 8, e1003023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Branche, C.; Kohl, L.; Toutirais, G.; Buisson, J.; Cosson, J.; Bastin, P. Conserved and specific functions of axoneme components in trypanosome motility. J. Cell Sci. 2006, 119, 3443–3455. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, S.; Kohl, L.; Ngai, I.; Wheeler, R.J.; Gull, K. A repetitive protein essential for the flagellum attachment zone filament structure and function in Trypanosoma brucei. Protist 2008, 159, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Sunter, J.D.; Varga, V.; Dean, S.; Gull, K. A dynamic coordination of flagellum and cytoplasmic cytoskeleton assembly specifies cell morphogenesis in trypanosomes. J. Cell Sci. 2015, 128, 1580–1594. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Hu, H.; He, C.Y.; Li, Z. Assembly and maintenance of the flagellum attachment zone filament in Trypanosoma brucei. J. Cell Sci. 2015, 128, 2361–2372. [Google Scholar] [CrossRef] [PubMed]
- Vickerman, K. On the surface coat and flagellar adhesion in trypanosomes. J. Cell Sci. 1969, 5, 163–193. [Google Scholar] [PubMed]
- Hoog, J.L.; Bouchet-Marquis, C.; McIntosh, J.R.; Hoenger, A.; Gull, K. Cryo-electron tomography and 3-D analysis of the intact flagellum in Trypanosoma brucei. J. Struct. Biol. 2012, 178, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.Y.; Wang, C.; Yuan, Y.A.; He, C.Y. An intracellular membrane junction consisting of flagellum adhesion glycoproteins links flagellum biogenesis to cell morphogenesis in Trypanosoma brucei. J. Cell Sci. 2013, 126, 520–531. [Google Scholar] [CrossRef] [PubMed]
- Lacomble, S.; Vaughan, S.; Gadelha, C.; Morphew, M.K.; Shaw, M.K.; McIntosh, J.R.; Gull, K. Three-dimensional cellular architecture of the flagellar pocket and associated cytoskeleton in trypanosomes revealed by electron microscope tomography. J. Cell Sci. 2009, 122, 1081–1090. [Google Scholar] [CrossRef] [PubMed]
- Lacomble, S.; Vaughan, S.; Deghelt, M.; Moreira-Leite, F.F.; Gull, K. A Trypanosoma brucei protein required for maintenance of the flagellum attachment zone and flagellar pocket ER domains. Protist 2012, 163, 602–615. [Google Scholar] [CrossRef] [PubMed]
- Henley, G.L.; Lee, C.M.; Takeuchi, A. Electron microscopy observations on Trypanosoma brucei: Freeze-cleaving and thin-sectioning study of the apical part of the flagellar pocket. Z. Parasitenkd. 1978, 55, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Gadelha, C.; Rothery, S.; Morphew, M.; McIntosh, J.R.; Severs, N.J.; Gull, K. Membrane domains and flagellar pocket boundaries are influenced by the cytoskeleton in African trypanosomes. Proc. Natl. Acad. Sci. USA 2009, 106, 17425–17430. [Google Scholar] [CrossRef] [PubMed]
- Bonhivers, M.; Nowacki, S.; Landrein, N.; Robinson, D.R. Biogenesis of the trypanosome endo-exocytotic organelle is cytoskeleton mediated. PLoS Biol. 2008, 6, e105. [Google Scholar] [CrossRef] [PubMed]
- Florimond, C.; Sahin, A.; Vidilaseris, K.; Dong, G.; Landrein, N.; Dacheux, D.; Albisetti, A.; Byard, E.H.; Bonhivers, M.; Robinson, D.R. BILBO1 Is a Scaffold Protein of the Flagellar Pocket Collar in the Pathogen Trypanosoma brucei. PLoS Pathog. 2015, 11, e1004654. [Google Scholar] [CrossRef] [PubMed]
- Vidilaseris, K.; Morriswood, B.; Kontaxis, G.; Dong, G. Structure of the TbBILBO1 protein N-terminal domain from Trypanosoma brucei reveals an essential requirement for a conserved surface patch. J. Biol. Chem. 2014, 289, 3724–3735. [Google Scholar] [CrossRef] [PubMed]
- Vidilaseris, K.; Shimanovskaya, E.; Esson, H.J.; Morriswood, B.; Dong, G. Assembly mechanism of Trypanosoma brucei BILBO1, a multidomain cytoskeletal protein. J. Biol. Chem. 2014, 289, 23870–23881. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Franklin, J.B.; Yelinek, J.T.; Ebersberger, I.; Warren, G.; He, C.Y. Centrin4 coordinates cell and nuclear division in T. brucei. J. Cell Sci. 2008, 121, 3062–3070. [Google Scholar] [CrossRef] [PubMed]
- Selvapandiyan, A.; Kumar, P.; Morris, J.C.; Salisbury, J.L.; Wang, C.C.; Nakhasi, H.L. Centrin1 is required for organelle segregation and cytokinesis in Trypanosoma brucei. Mol. Biol. Cell 2007, 18, 3290–3301. [Google Scholar] [CrossRef] [PubMed]
- Morriswood, B.; He, C.Y.; Sealey-Cardona, M.; Yelinek, J.; Pypaert, M.; Warren, G. The bilobe structure of Trypanosoma brucei contains a MORN-repeat protein. Mol. Biochem. Parasitol. 2009, 167, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Takeshima, H.; Komazaki, S.; Nishi, M.; Iino, M.; Kangawa, K. Junctophilins: A novel family of junctional membrane complex proteins. Mol. Cell 2000, 6, 11–22. [Google Scholar] [CrossRef]
- Esson, H.J.; Morriswood, B.; Yavuz, S.; Vidilaseris, K.; Dong, G.; Warren, G. Morphology of the trypanosome bilobe, a novel cytoskeletal structure. Eukaryot. Cell 2012, 11, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D.; Beattie, P.; Sherwin, T.; Gull, K. Microtubules, tubulin, and microtubule-associated proteins of trypanosomes. Methods Enzymol. 1991, 196, 285–299. [Google Scholar] [PubMed]
- Morriswood, B.; Schmidt, K. A MORN-repeat protein facilitates protein entry into the flagellar pocket of Trypanosoma brucei. Eukaryot. Cell 2015, 14, 1081–1093. [Google Scholar] [CrossRef] [PubMed]
- De Graffenried, C.L.; Ho, H.H.; Warren, G. Polo-like kinase is required for Golgi and bilobe biogenesis in Trypanosoma brucei. J. Cell Biol. 2008, 181, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K.N.; de Graffenried, C.L. Polo-like kinase is necessary for flagellum inheritance in Trypanosoma brucei. J. Cell Sci. 2012, 125, 3173–3184. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Nunez, A.; Ikeda, K.N.; Sauer, T.; de Graffenried, C.L. An analogue-sensitive approach identifies basal body rotation and flagellum attachment zone elongation as key functions of PLK in Trypanosoma brucei. Mol. Biol. Cell 2013, 24, 1321–1333. [Google Scholar] [CrossRef] [PubMed]
- McAllaster, M.R.; Ikeda, K.N.; Lozano-Nunez, A.; Anrather, D.; Unterwurzacher, V.; Gossenreiter, T.; Perry, J.A.; Crickley, R.; Mercadante, C.J.; Vaughan, S.; et al. Proteomic identification of novel cytoskeletal proteins associated with TbPLK, an essential regulator of cell morphogenesis in T. brucei. Mol. Biol. Cell 2015, 26, 3013–3029. [Google Scholar] [CrossRef] [PubMed]
- Gubbels, M.J.; Vaishnava, S.; Boot, N.; Dubremetz, J.F.; Striepen, B. A MORN-repeat protein is a dynamic component of the Toxoplasma gondii cell division apparatus. J. Cell Sci. 2006, 119, 2236–2245. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.; Johnson, J.; Florens, L.; Fraunholz, M.; Suravajjala, S.; DiLullo, C.; Yates, J.; Roos, D.S.; Murray, J.M. Cytoskeletal components of an invasion machine—The apical complex of Toxoplasma gondii. PLoS Pathog. 2006, 2, e13. [Google Scholar] [CrossRef] [PubMed]
- Hu, K. Organizational changes of the daughter basal complex during the parasite replication of Toxoplasma gondii. PLoS Pathog. 2008, 4, e10. [Google Scholar] [CrossRef] [PubMed]
- Heaslip, A.T.; Dzierszinski, F.; Stein, B.; Hu, K. TgMORN1 is a key organizer for the basal complex of Toxoplasma gondii. PLoS Pathog. 2010, 6, e1000754. [Google Scholar] [CrossRef] [PubMed]
- Lorestani, A.; Sheiner, L.; Yang, K.; Robertson, S.D.; Sahoo, N.; Brooks, C.F.; Ferguson, D.J.; Striepen, B.; Gubbels, M.J. A Toxoplasma MORN1 null mutant undergoes repeated divisions but is defective in basal assembly, apicoplast division and cytokinesis. PLoS ONE 2010, 5, e12302. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, D.J.; Sahoo, N.; Pinches, R.A.; Bumstead, J.M.; Tomley, F.M.; Gubbels, M.J. MORN1 has a conserved role in asexual and sexual development across the apicomplexa. Eukaryot. Cell 2008, 7, 698–711. [Google Scholar] [CrossRef] [PubMed]
- Suvorova, E.S.; Francia, M.; Striepen, B.; White, M.W. A novel bipartite centrosome coordinates the apicomplexan cell cycle. PLoS Biol. 2015, 13, e1002093. [Google Scholar] [CrossRef] [PubMed]
- Roux, K.J.; Kim, D.I.; Raida, M.; Burke, B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012, 196, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Morriswood, B.; Havlicek, K.; Demmel, L.; Yavuz, S.; Sealey-Cardona, M.; Vidilaseris, K.; Anrather, D.; Kostan, J.; Djinovic-Carugo, K.; Roux, K.J.; et al. Novel Bilobe Components in Trypanosoma brucei Identified Using Proximity-Dependent Biotinylation. Eukaryot. Cell 2013, 12, 356–367. [Google Scholar] [CrossRef] [PubMed]
- Kwon, K.; Beckett, D. Function of a conserved sequence motif in biotin holoenzyme synthetases. Protein Sci.: Publ. Protein Soc. 2000, 9, 1530–1539. [Google Scholar] [CrossRef] [PubMed]
- Lane, M.D.; Young, D.L.; Lynen, F. The Enzymatic Synthesis of Holotranscarboxylase from Apotranscarboxylase and (+)-Biotin. I. Purification of the Apoenzyme and Synthetase; Characteristics of the Reaction. J. Biol. Chem. 1964, 239, 2858–2864. [Google Scholar] [PubMed]
- Beckett, D.; Kovaleva, E.; Schatz, P.J. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci: Publ. Protein Soc. 1999, 8, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.I.; Birendra, K.C.; Zhu, W.; Motamedchaboki, K.; Doye, V.; Roux, K.J. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc. Natl. Acad. Sci. USA 2014, 111, E2453–E2461. [Google Scholar] [CrossRef] [PubMed]
- Wirtz, E.; Leal, S.; Ochatt, C.; Cross, G.A. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999, 99, 89–101. [Google Scholar] [CrossRef]
- Zhou, Q.; Gheiratmand, L.; Chen, Y.; Lim, T.K.; Zhang, J.; Li, S.; Xia, N.; Liu, B.; Lin, Q.; He, C.Y. A comparative proteomic analysis reveals a new bi-lobe protein required for bi-lobe duplication and cell division in Trypanosoma brucei. PLoS ONE 2010, 5, e9660. [Google Scholar] [CrossRef] [PubMed]
- Andre, J.; Harrison, S.; Towers, K.; Qi, X.; Vaughan, S.; McKean, P.G.; Ginger, M.L. The tubulin cofactor C family member TBCCD1 orchestrates cytoskeletal filament formation. J. Cell Sci. 2013, 126, 5350–5356. [Google Scholar] [CrossRef] [PubMed]
- Brasseur, A.; Bayat, S.; Chua, X.L.; Zhang, Y.; Zhou, Q.; Low, B.C.; He, C.Y. The bi-lobe-associated LRRP1 regulates Ran activity in Trypanosoma brucei. J. Cell Sci. 2014, 127, 4846–4856. [Google Scholar] [CrossRef] [PubMed]
- Gheiratmand, L.; Brasseur, A.; Zhou, Q.; He, C.Y. Biochemical characterization of the bi-lobe reveals a continuous structural network linking the bi-lobe to other single-copied organelles in Trypanosoma brucei. J. Biol. Chem. 2013, 288, 3489–3499. [Google Scholar] [CrossRef] [PubMed]
- He, C.Y.; Ho, H.H.; Malsam, J.; Chalouni, C.; West, C.M.; Ullu, E.; Toomre, D.; Warren, G. Golgi duplication in Trypanosoma brucei. J. Cell biol. 2004, 165, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Ho, H.H.; He, C.Y.; de Graffenried, C.L.; Murrells, L.J.; Warren, G. Ordered assembly of the duplicating Golgi in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 2006, 103, 7676–7681. [Google Scholar] [CrossRef] [PubMed]
- Rios, R.M. The centrosome-Golgi apparatus nexus. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2014, 369, 20130462. [Google Scholar] [CrossRef] [PubMed]
- Uzawa, M.; Grams, J.; Madden, B.; Toft, D.; Salisbury, J.L. Identification of a complex between centrin and heat shock proteins in CSF-arrested Xenopus oocytes and dissociation of the complex following oocyte activation. Dev. Biol. 1995, 171, 51–59. [Google Scholar] [CrossRef] [PubMed]
- He, C.Y.; Pypaert, M.; Warren, G. Golgi duplication in Trypanosoma brucei requires Centrin2. Science 2005, 310, 1196–1198. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Gheiratmand, L.; He, C.Y. An interplay between Centrin2 and Centrin4 on the bi-lobed structure in Trypanosoma brucei. Mol. Microbiol. 2012, 83, 1153–1161. [Google Scholar] [CrossRef] [PubMed]
- Matthews, K.R.; Sherwin, T.; Gull, K. Mitochondrial genome repositioning during the differentiation of the African trypanosome between life cycle forms is microtubule mediated. J. Cell Sci. 1995, 108, 2231–2239. [Google Scholar] [PubMed]
- Silverman, J.S.; Bangs, J.D. Form and function in the trypanosomal secretory pathway. Curr. Opin. Microbiol. 2012, 15, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Bangs, J.D. Replication of the ERES:Golgi junction in bloodstream-form African trypanosomes. Mol. Microbiol. 2011, 82, 1433–1443. [Google Scholar] [CrossRef] [PubMed]
- Selvapandiyan, A.; Kumar, P.; Salisbury, J.L.; Wang, C.C.; Nakhasi, H.L. Role of centrins 2 and 3 in organelle segregation and cytokinesis in Trypanosoma brucei. PLoS ONE 2012, 7, e45288. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Hu, H.; Lun, Z.R.; Li, Z. Centrin3 in trypanosomes maintains the stability of a flagellar inner-arm dynein for cell motility. Nat. Commun. 2014, 5, 4060. [Google Scholar] [CrossRef] [PubMed]
- Allen, C.L.; Goulding, D.; Field, M.C. Clathrin-mediated endocytosis is essential in Trypanosoma brucei. EMBO J. 2003, 22, 4991–5002. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, J.K.; Wenby, R.B.; Meiselman, H.J.; Fisher, T.C. The hydrodynamic radii of macromolecules and their effect on red blood cell aggregation. Biophys. J. 2004, 87, 4259–4270. [Google Scholar] [CrossRef] [PubMed]
- Hardman, K.D.; Ainsworth, C.F. Structure of concanavalin A at 2.4-A resolution. Biochemistry 1972, 11, 4910–4919. [Google Scholar] [CrossRef] [PubMed]
- Majorek, K.A.; Porebski, P.J.; Dayal, A.; Zimmerman, M.D.; Jablonska, K.; Stewart, A.J.; Chruszcz, M.; Minor, W. Structural and immunologic characterization of bovine, horse, and rabbit serum albumins. Mol. Immunol. 2012, 52, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Lou, Y.; Lin, W.H.; Xue, H.W. MORN motifs in plant PIPKs are involved in the regulation of subcellular localization and phospholipid binding. Cell Res. 2006, 16, 466–478. [Google Scholar] [CrossRef] [PubMed]
- Habicht, J.; Woehle, C.; Gould, S.B. Tetrahymena Expresses More than a Hundred Proteins with Lipid-binding MORN Motifs that can Differ in their Subcellular Localisations. J. Eukaryot. Microbiol. 2015, 62, 694–700. [Google Scholar] [CrossRef] [PubMed]
- Mikami, K.; Saavedra, L.; Sommarin, M. Is membrane occupation and recognition nexus domain functional in plant phosphatidylinositol phosphate kinases? Plant Signal. Behav. 2010, 5, 1241–1244. [Google Scholar] [CrossRef] [PubMed]
- Im, Y.J.; Davis, A.J.; Perera, I.Y.; Johannes, E.; Allen, N.S.; Boss, W.F. The N-terminal membrane occupation and recognition nexus domain of Arabidopsis phosphatidylinositol phosphate kinase 1 regulates enzyme activity. J. Biol. Chem. 2007, 282, 5443–5452. [Google Scholar] [CrossRef] [PubMed]
- Mikami, K.; Saavedra, L.; Hiwatashi, Y.; Uji, T.; Hasebe, M.; Sommarin, M. A dibasic amino acid pair conserved in the activation loop directs plasma membrane localization and is necessary for activity of plant type I/II phosphatidylinositol phosphate kinase. Plant Physiol. 2010, 153, 1004–1015. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.R.; Jing, C.; Walker, P.A.; Martin, S.R.; Howell, S.A.; Blackburn, G.M.; Gamblin, S.J.; Xiao, B. Crystal structure and functional analysis of the histone methyltransferase SET7/9. Cell 2002, 111, 105–115. [Google Scholar] [CrossRef]
- Jacobs, S.A.; Harp, J.M.; Devarakonda, S.; Kim, Y.; Rastinejad, F.; Khorasanizadeh, S. The active site of the SET domain is constructed on a knot. Nat. Struct. Biol. 2002, 9, 833–838. [Google Scholar] [CrossRef] [PubMed]
- Xiao, B.; Jing, C.; Wilson, J.R.; Walker, P.A.; Vasisht, N.; Kelly, G.; Howell, S.; Taylor, I.A.; Blackburn, G.M.; Gamblin, S.J. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature 2003, 421, 652–656. [Google Scholar] [CrossRef] [PubMed]
- Stachowiak, J.C.; Schmid, E.M.; Ryan, C.J.; Ann, H.S.; Sasaki, D.Y.; Sherman, M.B.; Geissler, P.L.; Fletcher, D.A.; Hayden, C.C. Membrane bending by protein-protein crowding. Nat. Cell Biol. 2012, 14, 944–949. [Google Scholar] [CrossRef] [PubMed]
- Engstler, M.; Pfohl, T.; Herminghaus, S.; Boshart, M.; Wiegertjes, G.; Heddergott, N.; Overath, P. Hydrodynamic flow-mediated protein sorting on the cell surface of trypanosomes. Cell 2007, 131, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Demmel, L.; Schmidt, K.; Lucast, L.; Havlicek, K.; Zankel, A.; Koestler, T.; Reithofer, V.; de Camilli, P.; Warren, G. The endocytic activity of the flagellar pocket in Trypanosoma brucei is regulated by an adjacent phosphatidylinositol phosphate kinase. J. Cell Sci. 2014, 127, 2351–2364. [Google Scholar] [CrossRef] [PubMed]
- Di Paolo, G.; de Camilli, P. Phosphoinositides in cell regulation and membrane dynamics. Nature 2006, 443, 651–657. [Google Scholar] [CrossRef] [PubMed]
- Ziolkowska, N.E.; Christiano, R.; Walther, T.C. Organized living: Formation mechanisms and functions of plasma membrane domains in yeast. Trends Cell Biol. 2012, 22, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Douglas, L.M.; Konopka, J.B. Fungal membrane organization: The eisosome concept. Annu. Rev. Microbiol. 2014, 68, 377–393. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Heuser, J.E.; Roth, R.; Goodenough, U. Eisosome Ultrastructure and Evolution in Fungi, Microalgae, and Lichens. Eukaryot. Cell 2015, 14, 1017–1042. [Google Scholar] [CrossRef] [PubMed]
- Walther, T.C.; Brickner, J.H.; Aguilar, P.S.; Bernales, S.; Pantoja, C.; Walter, P. Eisosomes mark static sites of endocytosis. Nature 2006, 439, 998–1003. [Google Scholar] [CrossRef] [PubMed]
- Stradalova, V.; Stahlschmidt, W.; Grossmann, G.; Blazikova, M.; Rachel, R.; Tanner, W.; Malinsky, J. Furrow-like invaginations of the yeast plasma membrane correspond to membrane compartment of Can1. J. Cell Sci. 2009, 122, 2887–2894. [Google Scholar] [CrossRef] [PubMed]
- Grossmann, G.; Malinsky, J.; Stahlschmidt, W.; Loibl, M.; Weig-Meckl, I.; Frommer, W.B.; Opekarova, M.; Tanner, W. Plasma membrane microdomains regulate turnover of transport proteins in yeast. J. Cell Biol. 2008, 183, 1075–1088. [Google Scholar] [CrossRef] [PubMed]
- Olivera-Couto, A.; Grana, M.; Harispe, L.; Aguilar, P.S. The eisosome core is composed of BAR domain proteins. Mol. Biol. Cell 2011, 22, 2360–2372. [Google Scholar] [CrossRef] [PubMed]
- Ziolkowska, N.E.; Karotki, L.; Rehman, M.; Huiskonen, J.T.; Walther, T.C. Eisosome-driven plasma membrane organization is mediated by BAR domains. Nat. Struct. Mol. Biol. 2011, 18, 854–856. [Google Scholar] [CrossRef] [PubMed]
- Karotki, L.; Huiskonen, J.T.; Stefan, C.J.; Ziolkowska, N.E.; Roth, R.; Surma, M.A.; Krogan, N.J.; Emr, S.D.; Heuser, J.; Grunewald, K.; et al. Eisosome proteins assemble into a membrane scaffold. J. Cell Biol. 2011, 195, 889–902. [Google Scholar] [CrossRef] [PubMed]
- Moreira, K.E.; Schuck, S.; Schrul, B.; Frohlich, F.; Moseley, J.B.; Walther, T.C.; Walter, P. Seg1 controls eisosome assembly and shape. J. Cell Biol. 2012, 198, 405–420. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morriswood, B. Form, Fabric, and Function of a Flagellum-Associated Cytoskeletal Structure. Cells 2015, 4, 726-747. https://doi.org/10.3390/cells4040726
Morriswood B. Form, Fabric, and Function of a Flagellum-Associated Cytoskeletal Structure. Cells. 2015; 4(4):726-747. https://doi.org/10.3390/cells4040726
Chicago/Turabian StyleMorriswood, Brooke. 2015. "Form, Fabric, and Function of a Flagellum-Associated Cytoskeletal Structure" Cells 4, no. 4: 726-747. https://doi.org/10.3390/cells4040726
APA StyleMorriswood, B. (2015). Form, Fabric, and Function of a Flagellum-Associated Cytoskeletal Structure. Cells, 4(4), 726-747. https://doi.org/10.3390/cells4040726