Monitoring Autophagy in the Model Green Microalga Chlamydomonas reinhardtii
Abstract
:1. Introduction
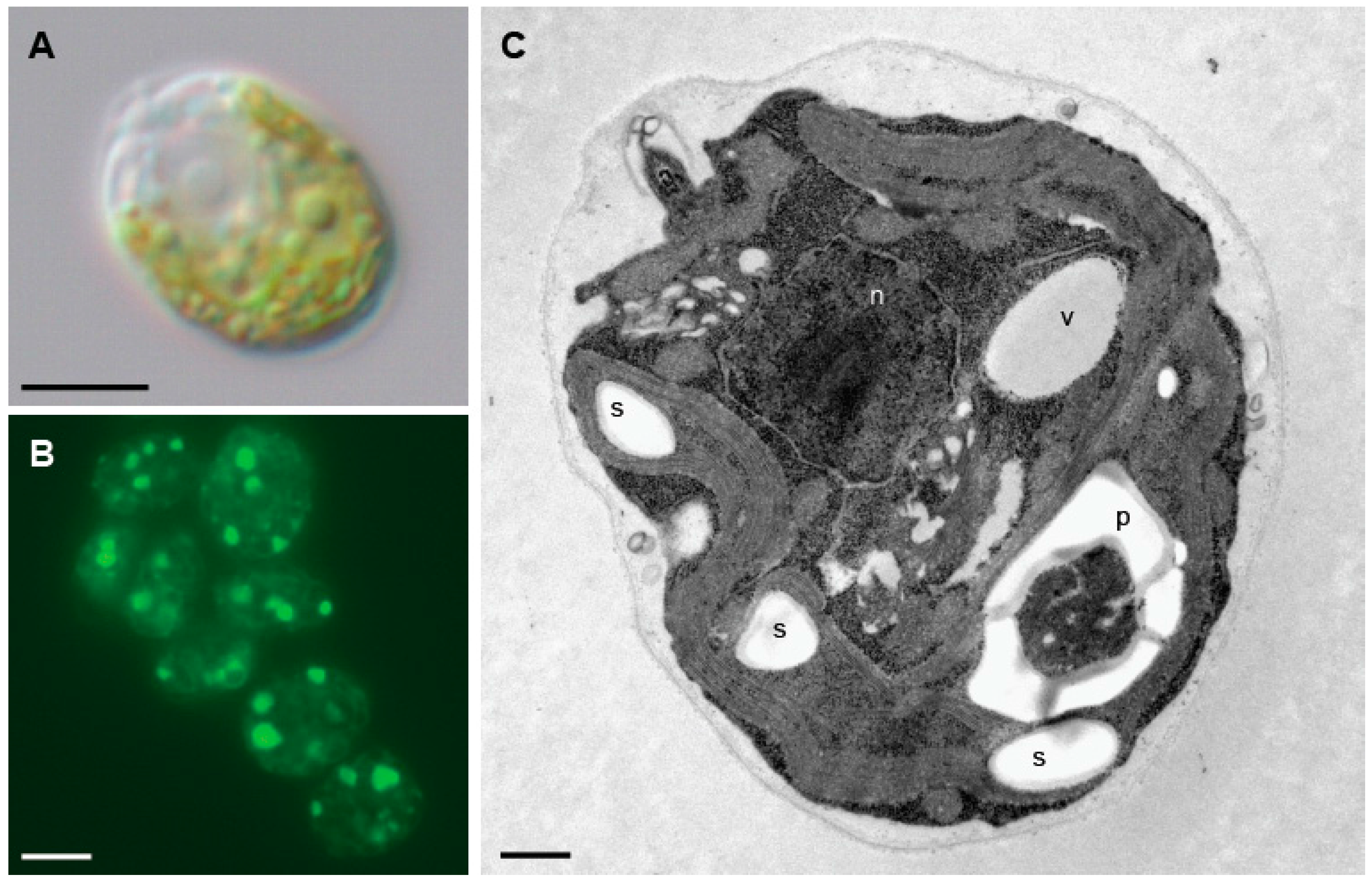
2. Chlamydomonas as a Photosynthetic Model System to Study Autophagy
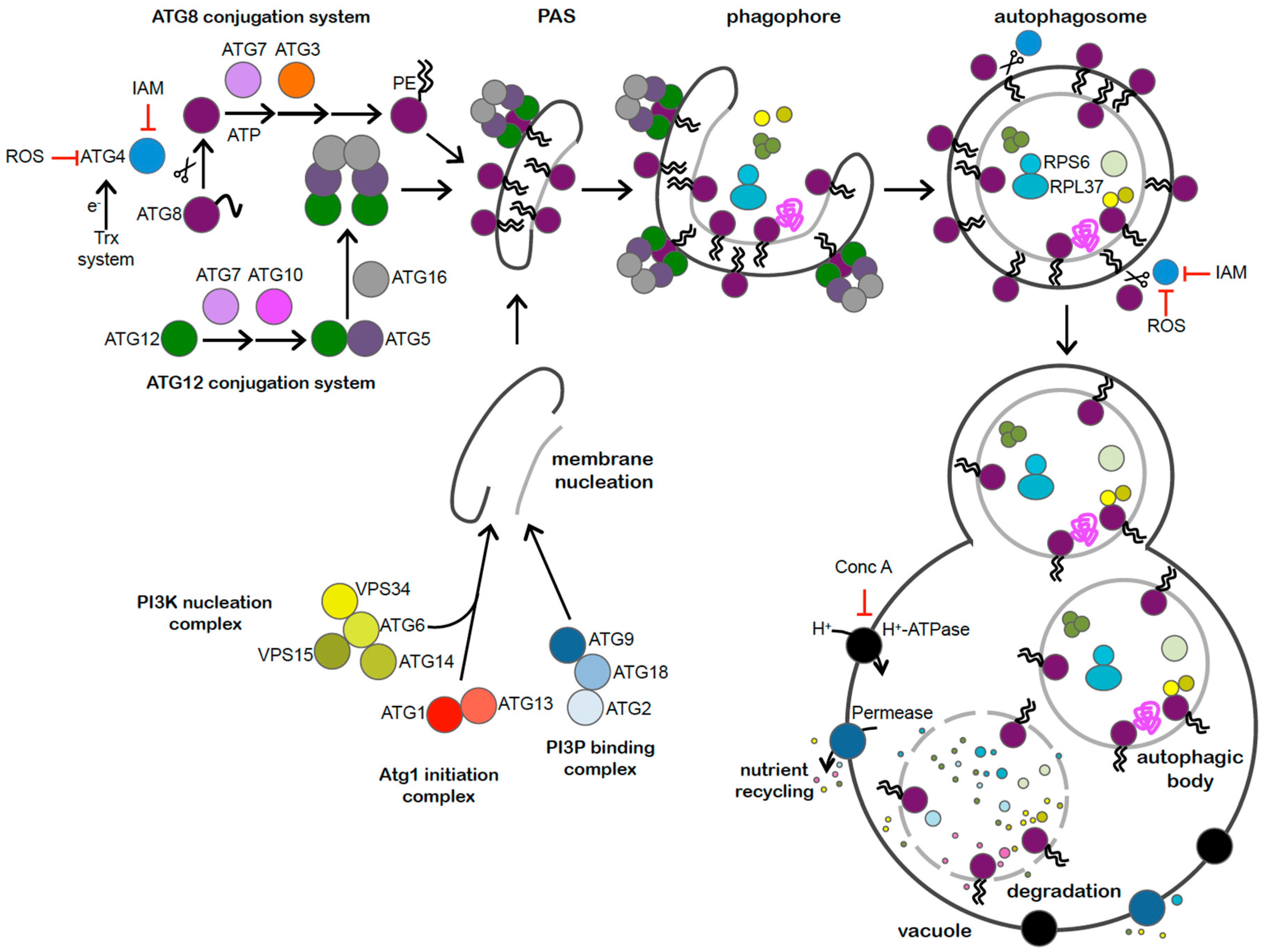
3. Methods for Monitoring Autophagy in Chlamydomonas
3.1. ATG8 Lipidation
3.2. Autophagic Flux
3.3. Cellular Localization of ATG8
3.4. ATG4 Proteolytic Assay
3.5. Transcriptional Activation of ATG Genes
4. Perspectives
Acknowledgments
Conflicts of Interest
References
- He, C.; Klionsky, D.J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009, 43, 67–93. [Google Scholar] [CrossRef] [PubMed]
- Nakatogawa, H.; Suzuki, K.; Kamada, Y.; Ohsumi, Y. Dynamics and diversity in autophagy mechanisms: Lessons from yeast. Nat. Rev. Mol. Cell Biol. 2009, 10, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Tsukada, M.; Ohsumi, Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993, 333, 169–174. [Google Scholar] [CrossRef]
- Bassham, D.C.; Laporte, M.; Marty, F.; Moriyasu, Y.; Ohsumi, Y.; Olsen, L.J.; Yoshimoto, K. Autophagy in development and stress responses of plants. Autophagy 2006, 2, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Troya, S.; Perez-Perez, M.E.; Florencio, F.J.; Crespo, J.L. The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy 2008, 4, 851–865. [Google Scholar] [CrossRef] [PubMed]
- Shemi, A.; Ben-Dor, S.; Vardi, A. Elucidating the composition and conservation of the autophagy pathway in photosynthetic eukaryotes. Autophagy 2015, 11, 701–715. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.R.; Vierstra, R.D. Autophagic recycling: Lessons from yeast help define the process in plants. Curr. Opin. Plant Biol. 2005, 8, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011, 27, 107–132. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Klionsky, D.J. Autophagosome formation: Core machinery and adaptations. Nat. Cell Biol. 2007, 9, 1102–1109. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Acevedo Arozena, A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, E.H. Chlamydomonas as a Model Organism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 363–406. [Google Scholar] [CrossRef] [PubMed]
- Goodenough, U.W. Green yeast. Cell 1992, 70, 533–538. [Google Scholar] [CrossRef]
- Grossman, A.R.; Lohr, M.; Im, C.S. Chlamydomonas reinhardtii in the landscape of pigments. Annu. Rev. Genet. 2004, 38, 119–173. [Google Scholar] [CrossRef] [PubMed]
- Rochaix, J.D. Chlamydomonas reinhardtii as the photosynthetic yeast. Annu. Rev. Genet. 1995, 29, 209–230. [Google Scholar] [CrossRef] [PubMed]
- Sager, R.; Palade, G.E. Structure and development of the chloroplast in Chlamydomonas. I. The normal green cell. J. Biophys. Biochem. Cytol. 1957, 3, 463–488. [Google Scholar] [CrossRef] [PubMed]
- Merchant, S.S.; Prochnik, S.E.; Vallon, O.; Harris, E.H.; Karpowicz, S.J.; Witman, G.B.; Terry, A.; Salamov, A.; Fritz-Laylin, L.K.; Marechal-Drouard, L.; et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 2007, 318, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Blaby, I.K.; Blaby-Haas, C.E.; Tourasse, N.; Hom, E.F.; Lopez, D.; Aksoy, M.; Grossman, A.; Umen, J.; Dutcher, S.; Porter, M.; et al. The Chlamydomonas genome project: A decade on. Trends Plant Sci. 2014, 19, 672–680. [Google Scholar] [CrossRef] [PubMed]
- Merchant, S.S.; Kropat, J.; Liu, B.; Shaw, J.; Warakanont, J. TAG, you’re it! Chlamydomonas as a reference organism for understanding algal triacylglycerol accumulation. Curr. Opin. Biotechnol. 2012, 23, 352–363. [Google Scholar] [CrossRef] [PubMed]
- Goodson, C.; Roth, R.; Wang, Z.T.; Goodenough, U. Structural correlates of cytoplasmic and chloroplast lipid body synthesis in Chlamydomonas reinhardtii and stimulation of lipid body production with acetate boost. Eukaryot. Cell 2011, 10, 1592–1606. [Google Scholar] [CrossRef] [PubMed]
- Perez-Perez, M.E.; Crespo, J.L. Autophagy in the model alga Chlamydomonas reinhardtii. Autophagy 2010, 6, 562–563. [Google Scholar] [CrossRef] [PubMed]
- Perez-Perez, M.E.; Florencio, F.J.; Crespo, J.L. Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol. 2010, 152, 1874–1888. [Google Scholar] [CrossRef] [PubMed]
- Perez-Perez, M.E.; Couso, I.; Crespo, J.L. Carotenoid deficiency triggers autophagy in the model green alga Chlamydomonas reinhardtii. Autophagy 2012, 8, 376–388. [Google Scholar] [CrossRef] [PubMed]
- Crespo, J.L.; Diaz-Troya, S.; Florencio, F.J. Inhibition of target of rapamycin signaling by rapamycin in the unicellular green alga Chlamydomonas reinhardtii. Plant Physiol. 2005, 139, 1736–1749. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Tian, D.; Kong, X.; Chen, Q.; E, F.A.; Hu, X.; Jia, A. The role of nitric oxide signalling in response to salt stress in Chlamydomonas reinhardtii. Planta 2016, 244, 651–669. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.P.; Horst, I.; Duong, G.H.; Tomsett, E.V.; Litvinenko, A.C.; Howe, C.J.; Smith, A.G. Triacylglyceride Production and Autophagous Responses in Chlamydomonas reinhardtii Depend on Resource Allocation and Carbon Source. Eukaryot. Cell 2014, 13, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Goodenough, U.; Blaby, I.; Casero, D.; Gallaher, S.D.; Goodson, C.; Johnson, S.; Lee, J.H.; Merchant, S.S.; Pellegrini, M.; Roth, R.; et al. The path to triacylglyceride obesity in the sta6 strain of Chlamydomonas reinhardtii. Eukaryot. Cell 2014, 13, 591–613. [Google Scholar] [CrossRef] [PubMed]
- Perez-Martin, M.; Blaby-Haas, C.E.; Perez-Perez, M.E.; Andres-Garrido, A.; Blaby, I.K.; Merchant, S.S.; Crespo, J.L. Activation of Autophagy by Metals in Chlamydomonas reinhardtii. Eukaryot. Cell 2015, 14, 964–973. [Google Scholar] [CrossRef] [PubMed]
- Perez-Martin, M.; Perez-Perez, M.E.; Lemaire, S.D.; Crespo, J.L. Oxidative Stress Contributes to Autophagy Induction in Response to Endoplasmic Reticulum Stress in Chlamydomonas reinhardtii. Plant Physiol. 2014, 166, 997–1008. [Google Scholar] [CrossRef] [PubMed]
- Perez-Perez, M.E.; Zaffagnini, M.; Marchand, C.H.; Crespo, J.L.; Lemaire, S.D. The yeast autophagy protease Atg4 is regulated by thioredoxin. Autophagy 2014, 10, 1953–1964. [Google Scholar] [CrossRef] [PubMed]
- Scherz-Shouval, R.; Shvets, E.; Fass, E.; Shorer, H.; Gil, L.; Elazar, Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007, 26, 1749–1760. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Contento, A.L.; Nguyen, P.Q.; Bassham, D.C. Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 2007, 143, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Perez-Perez, M.E.; Lemaire, S.D.; Crespo, J.L. Control of Autophagy in Chlamydomonas Is Mediated through Redox-Dependent Inactivation of the ATG4 Protease. Plant Physiol. 2016, 172, 2219–2234. [Google Scholar] [CrossRef] [PubMed]
- Couso, I.; Perez-Perez, M.E.; Martinez-Force, E.; Kim, H.-S.; He, Y.; Umen, J.G.; Crespo, J.L. Autophagic flux is needed for the synthesis of triacylglycerols and ribosomal protein turnover in Chlamydomonas. J. Exp. Bot. 2017, in press. [Google Scholar] [CrossRef] [PubMed]
- Ramundo, S.; Casero, D.; Muhlhaus, T.; Hemme, D.; Sommer, F.; Crevecoeur, M.; Rahire, M.; Schroda, M.; Rusch, J.; Goodenough, U.; et al. Conditional Depletion of the Chlamydomonas Chloroplast ClpP Protease Activates Nuclear Genes Involved in Autophagy and Plastid Protein Quality Control. Plant Cell 2014, 26, 2201–2222. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, C.; Garcia, I.; Moreno, I.; Perez-Perez, M.E.; Crespo, J.L.; Romero, L.C.; Gotor, C. Cysteine-generated sulfide in the cytosol negatively regulates autophagy and modulates the transcriptional profile in Arabidopsis. Plant Cell 2012, 24, 4621–4634. [Google Scholar] [CrossRef] [PubMed]
- Laureano-Marin, A.M.; Moreno, I.; Romero, L.C.; Gotor, C. Negative Regulation of Autophagy by Sulfide Is Independent of Reactive Oxygen Species. Plant Physiol. 2016, 171, 1378–1391. [Google Scholar] [CrossRef] [PubMed]
- Drose, S.; Bindseil, K.U.; Bowman, E.J.; Siebers, A.; Zeeck, A.; Altendorf, K. Inhibitory effect of modified bafilomycins and concanamycins on P- and V-type adenosinetriphosphatases. Biochemistry 1993, 32, 3902–3906. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, R.; Patena, W.; Gang, S.S.; Blum, S.R.; Ivanova, N.; Yue, R.; Robertson, J.M.; Lefebvre, P.A.; Fitz-Gibbon, S.T.; et al. An Indexed, Mapped Mutant Library Enables Reverse Genetics Studies of Biological Processes in Chlamydomonas reinhardtii. Plant Cell 2016, 28, 367–387. [Google Scholar] [CrossRef] [PubMed]
- Merkulova, E.A.; Guiboileau, A.; Naya, L.; Masclaux-Daubresse, C.; Yoshimoto, K. Assessment and optimization of autophagy monitoring methods in Arabidopsis roots indicate direct fusion of autophagosomes with vacuoles. Plant Cell Physiol. 2014, 55, 715–726. [Google Scholar] [CrossRef] [PubMed]
- Takatsuka, C.; Inoue, Y.; Matsuoka, K.; Moriyasu, Y. 3-Methyladenine inhibits autophagy in tobacco culture cells under sucrose starvation conditions. Plant Cell Physiol. 2004, 45, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Chung, T.; Suttangkakul, A.; Vierstra, R.D. The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol. 2009, 149, 220–234. [Google Scholar] [CrossRef] [PubMed]
- Doelling, J.H.; Walker, J.M.; Friedman, E.M.; Thompson, A.R.; Vierstra, R.D. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 2002, 277, 33105–33114. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.R.; Doelling, J.H.; Suttangkakul, A.; Vierstra, R.D. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 2005, 138, 2097–2110. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, K.; Hanaoka, H.; Sato, S.; Kato, T.; Tabata, S.; Noda, T.; Ohsumi, Y. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 2004, 16, 2967–2983. [Google Scholar] [CrossRef] [PubMed]
- Schmollinger, S.; Muhlhaus, T.; Boyle, N.R.; Blaby, I.K.; Casero, D.; Mettler, T.; Moseley, J.L.; Kropat, J.; Sommer, F.; Strenkert, D.; et al. Nitrogen-Sparing Mechanisms in Chlamydomonas Affect the Transcriptome, the Proteome, and Photosynthetic Metabolism. Plant Cell 2014, 26, 1410–1435. [Google Scholar] [CrossRef] [PubMed]
- Svenning, S.; Lamark, T.; Krause, K.; Johansen, T. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 2011, 7, 993–1010. [Google Scholar] [CrossRef] [PubMed]
- Zientara-Rytter, K.; Lukomska, J.; Moniuszko, G.; Gwozdecki, R.; Surowiecki, P.; Lewandowska, M.; Liszewska, F.; Wawrzynska, A.; Sirko, A. Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors. Autophagy 2011, 7, 1145–1158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Perez, M.E.; Lemaire, S.D.; Crespo, J.L. Reactive oxygen species and autophagy in plants and algae. Plant Physiol. 2012, 160, 156–164. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Pérez, M.E.; Couso, I.; Heredia-Martínez, L.G.; Crespo, J.L. Monitoring Autophagy in the Model Green Microalga Chlamydomonas reinhardtii. Cells 2017, 6, 36. https://doi.org/10.3390/cells6040036
Pérez-Pérez ME, Couso I, Heredia-Martínez LG, Crespo JL. Monitoring Autophagy in the Model Green Microalga Chlamydomonas reinhardtii. Cells. 2017; 6(4):36. https://doi.org/10.3390/cells6040036
Chicago/Turabian StylePérez-Pérez, María Esther, Inmaculada Couso, Luis G. Heredia-Martínez, and José L. Crespo. 2017. "Monitoring Autophagy in the Model Green Microalga Chlamydomonas reinhardtii" Cells 6, no. 4: 36. https://doi.org/10.3390/cells6040036




