Cellular and Structural Studies of Eukaryotic Cells by Cryo-Electron Tomography
Abstract
:1. Introduction
2. Principles of Cryo-Electron Tomography
3. How to Apply Cryo-ET to Different Parts of Eukaryotic Cells
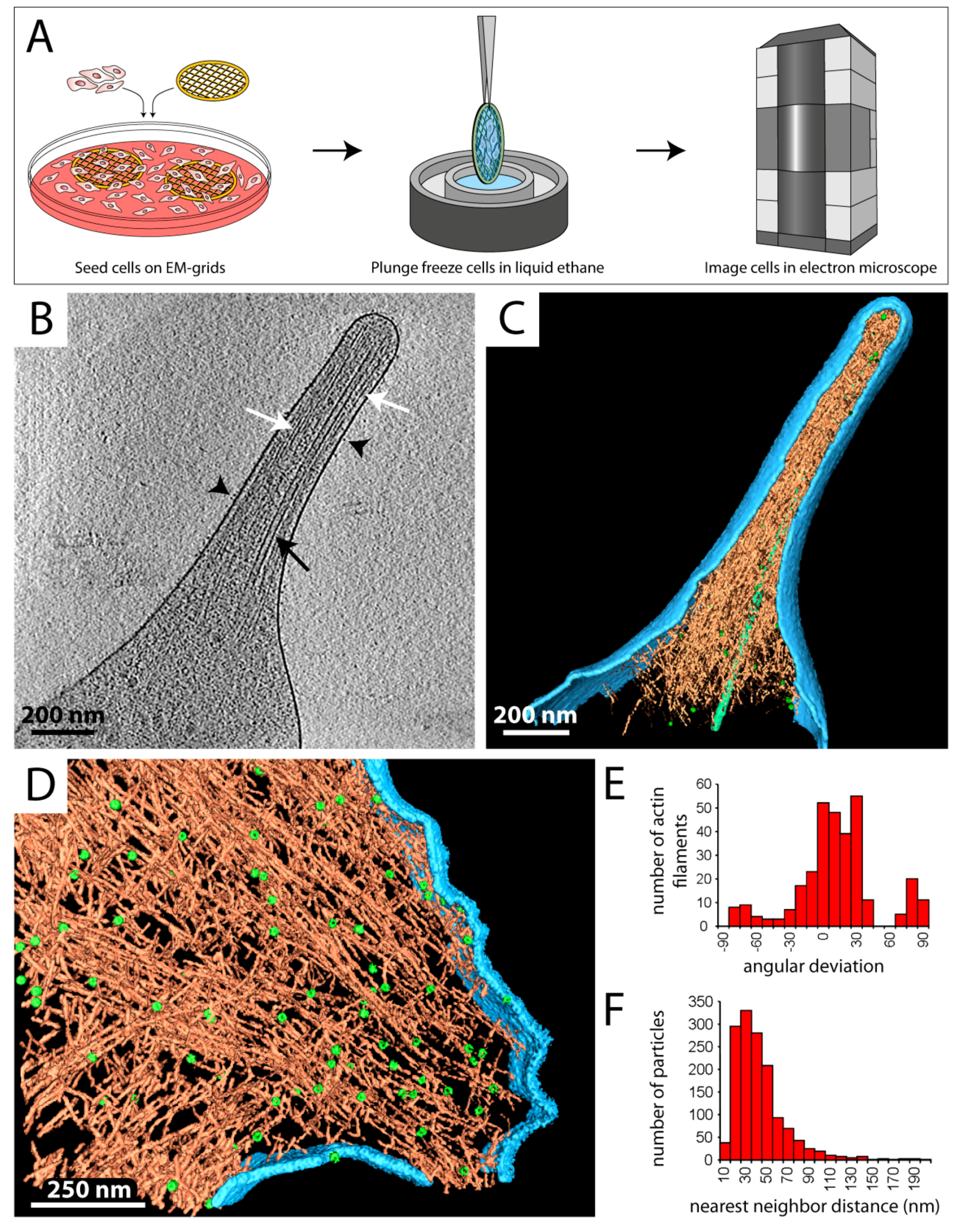
3.1. Studying Molecular Processes at the Cell Periphery
3.2. Deep into the Cell by Cryo-ET
3.2.1. Thinning Cells by Cryo-FIB Milling
3.2.2. Permeabilization and Minimal-Purification Approaches as Thinning Methods to Image the Nucleus
4. Identifying Structures of Interest within Cellular Cryo-Tomograms
4.1. Correlative Light and Electron Microscopy as a Tool to Localize Target Proteins
4.2. Gold Nanoparticles as Tags to Identify Target Proteins
5. Conclusion and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shi, Y.G. A Glimpse of Structural Biology through X-ray Crystallography. Cell 2014, 159, 995–1014. [Google Scholar] [CrossRef]
- Bonvin, A.M.; Boelens, R.; Kaptein, R. NMR analysis of protein interactions. Curr. Opin. Chem. Biol. 2005, 9, 501–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucic, V.; Rigort, A.; Baumeister, W. Cryo-electron tomography: The challenge of doing structural biology in situ. J. Cell Biol. 2013, 202, 407–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, C.V.; Sali, A.; Baumeister, W. The molecular sociology of the cell. Nature 2007, 450, 973–982. [Google Scholar] [CrossRef]
- Medalia, O.; Weber, I.; Frangakis, A.S.; Nicastro, D.; Gerisch, G.; Baumeister, W. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science 2002, 298, 1209–1213. [Google Scholar] [CrossRef] [PubMed]
- Medalia, O.; Beck, M.; Ecke, M.; Weber, I.; Neujahr, R.; Baumeister, W.; Gerisch, G. Organization of actin networks in intact filopodia. Curr. Biol. CB 2007, 17, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Al-Amoudi, A.; Diez, D.C.; Betts, M.J.; Frangakis, A.S. The molecular architecture of cadherins in native epidermal desmosomes. Nature 2007, 450, 832–837. [Google Scholar] [CrossRef]
- Engel, B.D.; Schaffer, M.; Albert, S.; Asano, S.; Plitzko, J.M.; Baumeister, W. In situ structural analysis of Golgi intracisternal protein arrays. Proc. Natl. Acad. Sci. USA 2015, 112, 11264–11269. [Google Scholar] [CrossRef] [Green Version]
- Brandt, F.; Carlson, L.A.; Hartl, F.U.; Baumeister, W.; Grunewald, K. The Three-Dimensional Organization of Polyribosomes in Intact Human Cells. Mol. Cell 2010, 39, 560–569. [Google Scholar] [CrossRef]
- Asano, S.; Fukuda, Y.; Beck, F.; Aufderheide, A.; Forster, F.; Danev, R.; Baumeister, W. Proteasomes. A molecular census of 26S proteasomes in intact neurons. Science 2015, 347, 439–442. [Google Scholar] [CrossRef]
- Mahamid, J.; Pfeffer, S.; Schaffer, M.; Villa, E.; Danev, R.; Cuellar, L.K.; Forster, F.; Hyman, A.A.; Plitzko, J.M.; Baumeister, W. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 2016, 351, 969–972. [Google Scholar] [CrossRef]
- Dobro, M.J.; Oikonomou, C.M.; Piper, A.; Cohen, J.; Guo, K.; Jensen, T.; Tadayon, J.; Donermeyer, J.; Park, Y.; Solis, B.A.; et al. Uncharacterized bacterial structures revealed by electron cryotomography. J. Bacteriol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Scheffer, M.P.; Gonzalez-Gonzalez, L.; Seybert, A.; Ratera, M.; Kunz, M.; Valpuesta, J.M.; Fita, I.; Querol, E.; Pinol, J.; Martin-Benito, J.; et al. Structural characterization of the NAP; the major adhesion complex of the human pathogen Mycoplasma genitalium. Mol. Microbiol. 2017, 105, 869–879. [Google Scholar] [CrossRef] [PubMed]
- Seybert, A.; Gonzalez-Gonzalez, L.; Scheffer, M.P.; Lluch-Senar, M.; Mariscal, A.M.; Querol, E.; Matthaeus, F.; Pinol, J.; Frangakis, A.S. Cryo-electron tomography analyses of terminal organelle mutants suggest the motility mechanism of Mycoplasma genitalium. Mol. Microbiol. 2018, 108, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Bock, D.; Medeiros, J.M.; Tsao, H.F.; Penz, T.; Weiss, G.L.; Aistleitner, K.; Horn, M.; Pilhofer, M. In situ architecture, function, and evolution of a contractile injection system. Science 2017, 357, 713–717. [Google Scholar] [CrossRef] [PubMed]
- Pilhofer, M.; Aistleitner, K.; Ladinsky, M.S.; Konig, L.; Horn, M.; Jensen, G.J. Architecture and host interface of environmental chlamydiae revealed by electron cryotomography. Environ. Microbiol. 2014, 16, 417–429. [Google Scholar] [CrossRef]
- Henderson, G.P.; Jensen, G.J. Three-dimensional structure of Mycoplasma pneumoniae’s attachment organelle and a model for its role in gliding motility. Mol. Microbiol. 2006, 60, 376–385. [Google Scholar] [CrossRef]
- Riedel, C.; Vasishtan, D.; Siebert, C.A.; Whittle, C.; Lehmann, M.J.; Mothes, W.; Grunewald, K. Native structure of a retroviral envelope protein and its conformational change upon interaction with the target cell. J. Struct. Biol. 2017, 197, 172–180. [Google Scholar] [CrossRef] [Green Version]
- Zeev-Ben-Mordehai, T.; Vasishtan, D.; Hernandez Duran, A.; Vollmer, B.; White, P.; Prasad Pandurangan, A.; Siebert, C.A.; Topf, M.; Grunewald, K. Two distinct trimeric conformations of natively membrane-anchored full-length herpes simplex virus 1 glycoprotein B. Proc. Natl. Acad. Sci. USA 2016, 113, 4176–4181. [Google Scholar] [CrossRef]
- Wan, W.; Kolesnikova, L.; Clarke, M.; Koehler, A.; Noda, T.; Becker, S.; Briggs, J.A.G. Structure and assembly of the Ebola virus nucleocapsid. Nature 2017, 551, 394–397. [Google Scholar] [CrossRef] [Green Version]
- Mattei, S.; Glass, B.; Hagen, W.J.H.; Krausslich, H.G.; Briggs, J.A.G. The structure and flexibility of conical HIV-1 capsids determined within intact virions. Science 2016, 354, 1434–1437. [Google Scholar] [CrossRef] [PubMed]
- Schur, F.K.; Hagen, W.J.; Rumlova, M.; Ruml, T.; Muller, B.; Krausslich, H.G.; Briggs, J.A. Structure of the immature HIV-1 capsid in intact virus particles at 8.8 A resolution. Nature 2015, 517, 505–508. [Google Scholar] [CrossRef] [PubMed]
- Irobalieva, R.N.; Martins, B.; Medalia, O. Cellular structural biology as revealed by cryo-electron tomography. J. Cell Sci. 2016, 129, 469–476. [Google Scholar] [CrossRef] [Green Version]
- Dubrovsky, A.; Sorrentino, S.; Harapin, J.; Sapra, K.T.; Medalia, O. Developments in cryo-electron tomography for in situ structural analysis. Arch. Biochem. Biophys. 2015, 581, 78–85. [Google Scholar] [CrossRef]
- Engel, B.D.; Schaffer, M.; Kuhn Cuellar, L.; Villa, E.; Plitzko, J.M.; Baumeister, W. Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo-electron tomography. Elife 2015, 4. [Google Scholar] [CrossRef] [Green Version]
- Albert, S.; Schaffer, M.; Beck, F.; Mosalaganti, S.; Asano, S.; Thomas, H.F.; Plitzko, J.M.; Beck, M.; Baumeister, W.; Engel, B.D. Proteasomes tether to two distinct sites at the nuclear pore complex. Proc. Natl. Acad. Sci. USA 2017, 114, 13726–13731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schermelleh, L.; Heintzmann, R.; Leonhardt, H. A guide to super-resolution fluorescence microscopy. J. Cell Biol. 2010, 190, 165–175. [Google Scholar] [CrossRef] [Green Version]
- Kopek, B.G.; Paez-Segala, M.G.; Shtengel, G.; Sochacki, K.A.; Sun, M.G.; Wang, Y.; Xu, C.S.; van Engelenburg, S.B.; Taraska, J.W.; Looger, L.L.; et al. Diverse protocols for correlative super-resolution fluorescence imaging and electron microscopy of chemically fixed samples. Nat. Protoc. 2017, 12, 916–946. [Google Scholar] [CrossRef] [Green Version]
- Giannuzzi, L.A.; Stevie, F.A. Introduction to Focused Ion Beams; Springer Science + Business Media Inc.: New York, NY, USA, 2005. [Google Scholar]
- Turgay, Y.; Eibauer, M.; Goldman, A.E.; Shimi, T.; Khayat, M.; Ben-Harush, K.; Dubrovsky-Gaupp, A.; Sapra, K.T.; Goldman, R.D.; Medalia, O. The molecular architecture of lamins in somatic cells. Nature 2017, 543, 261–264. [Google Scholar] [CrossRef] [Green Version]
- Dubochet, J.; McDowall, A.W.; Menge, B.; Schmid, E.N.; Lickfeld, K.G. Electron microscopy of frozen-hydrated bacteria. J. Bacteriol. 1983, 155, 381–390. [Google Scholar]
- Dubochet, J.; Adrian, M.; Chang, J.J.; Homo, J.C.; Lepault, J.; McDowall, A.W.; Schultz, P. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 1988, 21, 129–228. [Google Scholar] [CrossRef] [PubMed]
- Dubochet, J.; Mcdowall, A.W. Vitrification of Pure Water for Electron-Microscopy. J. Microsc.-Oxf. 1981, 124, Rp3–Rp4. [Google Scholar] [CrossRef]
- Studer, D.; Humbel, B.M.; Chiquet, M. Electron microscopy of high pressure frozen samples: Bridging the gap between cellular ultrastructure and atomic resolution. Histochem. Cell. Biol. 2008, 130, 877–889. [Google Scholar] [CrossRef] [PubMed]
- Frank, J. Electron Tomography: Methods for Three-Dimensional Visualization of Structures in the Cell; Springer: Berlin, Germany, 2006. [Google Scholar] [CrossRef]
- Kremer, J.R.; Mastronarde, D.N.; McIntosh, J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996, 116, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Kunz, M.; Frangakis, A.S. Super-sampling SART with ordered subsets. J. Struct. Biol. 2014, 188, 107–115. [Google Scholar] [CrossRef]
- Radermacher, M. Three-dimensional reconstruction of single particles from random and nonrandom tilt series. J. Electron. Microsc. Tech. 1988, 9, 359–394. [Google Scholar] [CrossRef] [PubMed]
- Andersen, A.H.; Kak, A.C. Simultaneous algebraic reconstruction technique (SART): A superior implementation of the art algorithm. Ultrason. Imaging 1984, 6, 81–94. [Google Scholar] [CrossRef]
- Briggs, J.A. Structural biology in situ—The potential of subtomogram averaging. Curr. Opin. Struct. Biol. 2013, 23, 261–267. [Google Scholar] [CrossRef]
- Forster, F.; Hegerl, R. Structure determination in situ by averaging of tomograms. Methods Cell Biol. 2007, 79, 741–767. [Google Scholar] [CrossRef]
- Forster, F.; Medalia, O.; Zauberman, N.; Baumeister, W.; Fass, D. Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography. Proc. Natl. Acad. Sci. USA 2005, 102, 4729–4734. [Google Scholar] [CrossRef] [Green Version]
- Schur, F.K.M.; Hagen, W.J.H.; de Marco, A.; Briggs, J.A.G. Determination of protein structure at 8.5 angstrom resolution using cryo-electron tomography and sub-tomogram averaging. J. Struct. Biol. 2013, 184, 394–400. [Google Scholar] [CrossRef]
- Turonova, B.; Schur, F.K.M.; Wan, W.; Briggs, J.A.G. Efficient 3D-CTF correction for cryo-electron tomography using NovaCTF improves subtomogram averaging resolution to 3.4 angstrom. J. Struct. Biol. 2017, 199, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Hutchings, J.; Stancheva, V.; Miller, E.A.; Zanetti, G. Subtomogram averaging of COPII assemblies reveals how coat organization dictates membrane shape. Nat. Commun. 2018, 9, 4154. [Google Scholar] [CrossRef] [PubMed]
- Bharat, T.A.M.; Russo, C.J.; Lowe, J.; Passmore, L.A.; Scheres, S.H.W. Advances in Single-Particle Electron Cryomicroscopy Structure Determination applied to Sub-tomogram Averaging. Structure 2015, 23, 1743–1753. [Google Scholar] [CrossRef] [Green Version]
- Danev, R.; Buijsse, B.; Khoshouei, M.; Plitzko, J.M.; Baumeister, W. Volta potential phase plate for in-focus phase contrast transmission electron microscopy. Proc. Natl. Acad. Sci. USA 2014, 111, 15635–15640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukuda, Y.; Laugks, U.; Lucic, V.; Baumeister, W.; Danev, R. Electron cryotomography of vitrified cells with a Volta phase plate. J. Struct. Biol. 2015, 190, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Grimm, R.; Singh, H.; Rachel, R.; Typke, D.; Zillig, W.; Baumeister, W. Electron tomography of ice-embedded prokaryotic cells. Biophys. J. 1998, 74, 1031–1042. [Google Scholar] [CrossRef]
- Mattila, P.K.; Lappalainen, P. Filopodia: Molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 2008, 9, 446–454. [Google Scholar] [CrossRef]
- Aramaki, S.; Mayanagi, K.; Jin, M.; Aoyama, K.; Yasunaga, T. Filopodia formation by crosslinking of F-actin with fascin in two different binding manners. Cytoskeleton (Hoboken) 2016, 73, 365–374. [Google Scholar] [CrossRef]
- Sorrentino, S.; Studt, J.D.; Medalia, O.; Tanuj Sapra, K. Roll, adhere, spread and contract: Structural mechanics of platelet function. Eur. J. Cell Biol. 2015, 94, 129–138. [Google Scholar] [CrossRef]
- Sorrentino, S.; Studt, J.D.; Horev, M.B.; Medalia, O.; Sapra, K.T. Toward correlating structure and mechanics of platelets. Cell Adhes. Migr. 2016, 10, 568–575. [Google Scholar] [CrossRef] [PubMed]
- Elad, N.; Volberg, T.; Patla, I.; Hirschfeld-Warneken, V.; Grashoff, C.; Spatz, J.P.; Fassler, R.; Geiger, B.; Medalia, O. The role of integrin-linked kinase in the molecular architecture of focal adhesions. J. Cell Sci. 2013, 126, 4099–4107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patla, I.; Volberg, T.; Elad, N.; Hirschfeld-Warneken, V.; Grashoff, C.; Fassler, R.; Spatz, J.P.; Geiger, B.; Medalia, O. Dissecting the molecular architecture of integrin adhesion sites by cryo-electron tomography. Nat. Cell Biol. 2010, 12, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Woodward, C.L.; Mendonca, L.M.; Jensen, G.J. Direct visualization of vaults within intact cells by electron cryo-tomography. Cell Mol. Life Sci. 2015, 72, 3401–3409. [Google Scholar] [CrossRef] [Green Version]
- Carlson, L.A.; de Marco, A.; Oberwinkler, H.; Habermann, A.; Briggs, J.A.G.; Krausslich, H.G.; Grunewald, K. Cryo Electron Tomography of Native HIV-1 Budding Sites. PLoS Pathog. 2010, 6. [Google Scholar] [CrossRef]
- Stauffer, S.; Rahman, S.A.; de Marco, A.; Carlson, L.A.; Glass, B.; Oberwinkler, H.; Herold, N.; Briggs, J.A.G.; Muller, B.; Grunewald, K.; et al. The Nucleocapsid Domain of Gag Is Dispensable for Actin Incorporation into HIV-1 and for Association of Viral Budding Sites with Cortical F-Actin. J. Virol. 2014, 88, 7893–7903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, Y.W.; Rettberg, L.A.; Treuner-Lange, A.; Iwasa, J.; Sogaard-Andersen, L.; Jensen, G.J. Architecture of the type IVa pilus machine. Science 2016, 351, aad2001. [Google Scholar] [CrossRef]
- Chang, Y.W.; Rettberg, L.A.; Ortega, D.R.; Jensen, G.J. In vivo structures of an intact type VI secretion system revealed by electron cryotomography. EMBO Rep. 2017, 18, 1090–1099. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Norris, S.J.; Liu, J. Molecular architecture of the bacterial flagellar motor in cells. Biochemistry 2014, 53, 4323–4333. [Google Scholar] [CrossRef]
- Krause, D.C.; Chen, S.Y.; Shi, J.; Jensen, A.J.; Sheppard, E.S.; Jensen, G.J. Electron cryotomography of Mycoplasma pneumoniae mutants correlates terminal organelle architectural features and function. Mol. Microbiol. 2018, 108, 306–318. [Google Scholar] [CrossRef]
- Qin, Z.; Lin, W.T.; Zhu, S.W.; Franco, A.T.; Liu, J. Imaging the Motility and Chemotaxis Machineries in Helicobacter pylori by Cryo-Electron Tomography. J. Bacteriol. 2017, 199. [Google Scholar] [CrossRef]
- Pilhofer, M.; Jensen, G.J. The bacterial cytoskeleton: More than twisted filaments. Curr. Opin. Cell Biol. 2013, 25, 125–133. [Google Scholar] [CrossRef]
- Wolf, S.G.; Mutsafi, Y.; Dadosh, T.; Ilani, T.; Lansky, Z.; Horowitz, B.; Rubin, S.; Elbaum, M.; Fass, D. 3D visualization of mitochondrial solid-phase calcium stores in whole cells. Elife 2017, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Amoudi, A.; Norlen, L.P.; Dubochet, J. Cryo-electron microscopy of vitreous sections of native biological cells and tissues. J. Struct. Biol. 2004, 148, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.E.; Leith, A.; Mannella, C.A.; Frank, J.; Marko, M. Towards high-resolution three-dimensional imaging of native mammalian tissue: Electron tomography of frozen-hydrated rat liver sections. J. Struct. Biol. 2006, 153, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.E.; Marko, M.; Frank, J.; Mannella, C.A. Electron tomographic analysis of frozen-hydrated tissue sections. J. Struct. Biol. 2002, 138, 63–73. [Google Scholar] [CrossRef]
- Norlen, L.; Al-Amoudi, A.; Dubochet, J. A cryotransmission electron microscopy study of skin barrier formation. J. Investig. Dermatol. 2003, 120, 555–560. [Google Scholar] [CrossRef]
- Norlen, L.; Al-Amoudi, A. Stratum corneum keratin structure, function, and formation: The cubic rod-packing and membrane templating model. J. Investig. Dermatol. 2004, 123, 715–732. [Google Scholar] [CrossRef]
- Al-Amoudi, A.; Frangakis, A.S. Three-dimensional visualization of the molecular architecture of cell-cell junctions in situ by cryo-electron tomography of vitreous sections. Methods Mol. Biol. 2013, 961, 97–117. [Google Scholar] [CrossRef]
- Richter, K. Cutting artefacts on ultrathin cryosections of biological bulk specimens. Micron 1994, 25, 297–308. [Google Scholar] [CrossRef]
- Richter, K.; Gnagi, H.; Dubochet, J. A model for cryosectioning based on the morphology of vitrified ultrathin sections. J. Microsc. 1991, 163, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.J.; Mcdowall, A.W.; Lepault, J.; Freeman, R.; Walter, C.A.; Dubochet, J. Freezing, Sectioning and Observation Artifacts of Frozen Hydrated Sections for Electron-Microscopy. J. Microsc. 1983, 132, 109–123. [Google Scholar] [CrossRef]
- Al-Amoudi, A.; Studer, D.; Dubochet, J. Cutting artefacts and cutting process in vitreous sections for cryo-electron microscopy. J. Struct. Biol. 2005, 150, 109–121. [Google Scholar] [CrossRef]
- Marko, M.; Hsieh, C.; Schalek, R.; Frank, J.; Mannella, C. Focused-ion-beam thinning of frozen-hydrated biological specimens for cryo-electron microscopy. Nat. Methods 2007, 4, 215–217. [Google Scholar] [CrossRef] [PubMed]
- Heymann, J.B.; Cardone, G.; Winkler, D.C.; Steven, A.C. Computational resources for cryo-electron tomography in Bsoft. J. Struct. Biol. 2008, 161, 232–242. [Google Scholar] [CrossRef] [Green Version]
- Amat, F.; Moussavi, F.; Comolli, L.R.; Elidan, G.; Downing, K.H.; Horowitz, M. Markov random field based automatic image alignment for electron tomography. J. Struct. Biol. 2008, 161, 260–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medeiros, J.M.; Bock, D.; Pilhofer, M. Imaging bacteria inside their host by cryo-focused ion beam milling and electron cryotomography. Curr. Opin. Microbiol. 2018, 43, 62–68. [Google Scholar] [CrossRef]
- Harapin, J.; Bormel, M.; Sapra, K.T.; Brunner, D.; Kaech, A.; Medalia, O. Structural analysis of multicellular organisms with cryo-electron tomography. Nat. Methods 2015, 12, 634–636. [Google Scholar] [CrossRef]
- Hsieh, C.; Schmelzer, T.; Kishchenko, G.; Wagenknecht, T.; Marko, M. Practical workflow for cryo focused-ion-beam milling of tissues and cells for cryo-TEM tomography. J. Struct. Biol. 2014, 185, 32–41. [Google Scholar] [CrossRef] [Green Version]
- Rigort, A.; Bauerlein, F.J.; Villa, E.; Eibauer, M.; Laugks, T.; Baumeister, W.; Plitzko, J.M. Focused ion beam micromachining of eukaryotic cells for cryoelectron tomography. Proc. Natl. Acad. Sci. USA 2012, 109, 4449–4454. [Google Scholar] [CrossRef] [Green Version]
- Kvam, E.; Goldfarb, D.S. Nucleus-vacuole junctions in yeast: Anatomy of a membrane contact site. Biochem. Soc. Trans. 2006, 34, 340–342. [Google Scholar] [CrossRef] [PubMed]
- Hagen, C.; Dent, K.C.; Zeev-Ben-Mordehai, T.; Grange, M.; Bosse, J.B.; Whittle, C.; Klupp, B.G.; Siebert, C.A.; Vasishtan, D.; Bauerlein, F.J.B.; et al. Structural Basis of Vesicle Formation at the Inner Nuclear Membrane. Cell 2015, 163, 1692–1701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Strunk, K.; Zhao, G.P.; Gray, J.L.; Zhang, P.J. 3D structure determination of native mammalian cells using cryo-FIB and cryo-electron tomography. J. Struct. Biol. 2012, 180, 318–326. [Google Scholar] [CrossRef] [Green Version]
- Schaffer, M.; Mahamid, J.; Engel, B.D.; Laugks, T.; Baumeister, W.; Plitzko, J.M. Optimized cryo-focused ion beam sample preparation aimed at in situ structural studies of membrane proteins. J. Struct. Biol. 2017, 197, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Eibauer, M.; Pellanda, M.; Turgay, Y.; Dubrovsky, A.; Wild, A.; Medalia, O. Structure and gating of the nuclear pore complex. Nat. Commun. 2015, 6, 7532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Appen, A.; Kosinski, J.; Sparks, L.; Ori, A.; DiGuilio, A.L.; Vollmer, B.; Mackmull, M.T.; Banterle, N.; Parca, L.; Kastritis, P.; et al. In situ structural analysis of the human nuclear pore complex. Nature 2015, 526, 140–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beck, M.; Lucic, V.; Forster, F.; Baumeister, W.; Medalia, O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature 2007, 449, 611–615. [Google Scholar] [CrossRef]
- Zwerger, M.; Eibauer, M.; Medalia, O. Insights into the gate of the nuclear pore complex. Nucleus 2016, 7, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P. Correlative cryo-electron tomography and optical microscopy of cells. Curr. Opin. Struct. Biol. 2013, 23, 763–770. [Google Scholar] [CrossRef] [Green Version]
- Sahl, S.J.; Hell, S.W.; Jakobs, S. Fluorescence nanoscopy in cell biology. Nat. Rev. Mol. Cell Biol. 2017, 18, 685–701. [Google Scholar] [CrossRef]
- Vangindertael, J.; Camacho, R.; Sempels, W.; Mizuno, H.; Dedecker, P.; Janssen, K.P.F. An introduction to optical super-resolution microscopy for the adventurous biologist. Methods Appl. Fluores 2018, 6. [Google Scholar] [CrossRef]
- Jun, S.; Ke, D.; Debiec, K.; Zhao, G.; Meng, X.; Ambrose, Z.; Gibson, G.A.; Watkins, S.C.; Zhang, P. Direct visualization of HIV-1 with correlative live-cell microscopy and cryo-electron tomography. Structure 2011, 19, 1573–1581. [Google Scholar] [CrossRef] [PubMed]
- Sartori, A.; Gatz, R.; Beck, F.; Rigort, A.; Baumeister, W.; Plitzko, J.M. Correlative microscopy: Bridging the gap between fluorescence light microscopy and cryo-electron tomography. J. Struct. Biol. 2007, 160, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Arnold, J.; Mahamid, J.; Lucic, V.; de Marco, A.; Fernandez, J.J.; Laugks, T.; Mayer, T.; Hyman, A.A.; Baumeister, W.; Plitzko, J.M. Site-Specific Cryo-focused Ion Beam Sample Preparation Guided by 3D Correlative Microscopy. Biophys. J. 2016, 110, 860–869. [Google Scholar] [CrossRef]
- Wolff, G.; Hagen, C.; Grunewald, K.; Kaufmann, R. Towards correlative super-resolution fluorescence and electron cryo-microscopy. Biol. Cell 2016, 108, 245–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rigort, A.; Bauerlein, F.J.; Leis, A.; Gruska, M.; Hoffmann, C.; Laugks, T.; Bohm, U.; Eibauer, M.; Gnaegi, H.; Baumeister, W.; et al. Micromachining tools and correlative approaches for cellular cryo-electron tomography. J. Struct. Biol. 2010, 172, 169–179. [Google Scholar] [CrossRef]
- Schwartz, C.L.; Sarbash, V.I.; Ataullakhanov, F.I.; Mcintosh, J.R.; Nicastro, D. Cryo-fluorescence microscopy facilitates correlations between light and cryo-electron microscopy and reduces the rate of photobleaching. J. Microsc. 2007, 227, 98–109. [Google Scholar] [CrossRef] [Green Version]
- Ando, T.; Bhamidimarri, S.P.; Brending, N.; Colin-York, H.; Collinson, L.; De Jonge, N.; de Pablo, P.J.; Debroye, E.; Eggeling, C.; Franck, C.; et al. The 2018 correlative microscopy techniques roadmap. J. Phys. D Appl. Phys. 2018, 51. [Google Scholar] [CrossRef]
- Chang, Y.W.; Chen, S.; Tocheva, E.I.; Treuner-Lange, A.; Lobach, S.; Sogaard-Andersen, L.; Jensen, G.J. Correlated cryogenic photoactivated localization microscopy and cryo-electron tomography. Nat. Methods 2014, 11, 737–739. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Xue, Y.; Zhao, W.; Chen, Y.; Fan, C.; Gu, L.; Zhang, Y.; Zhang, X.; Sun, L.; Huang, X.; et al. Three-dimensional super-resolution protein localization correlated with vitrified cellular context. Sci. Rep. 2015, 5, 13017. [Google Scholar] [CrossRef] [Green Version]
- Hampton, C.M.; Strauss, J.D.; Ke, Z.; Dillard, R.S.; Hammonds, J.E.; Alonas, E.; Desai, T.M.; Marin, M.; Storms, R.E.; Leon, F.; et al. Correlated fluorescence microscopy and cryo-electron tomography of virus-infected or transfected mammalian cells. Nat. Protoc. 2017, 12, 150–167. [Google Scholar] [CrossRef] [PubMed]
- Lucic, V.; Kossel, A.H.; Yang, T.; Bonhoeffer, T.; Baumeister, W.; Sartori, A. Multiscale imaging of neurons grown in culture: From light microscopy to cryo-electron tomography. J. Struct. Biol. 2007, 160, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Van Driel, L.F.; Valentijn, J.A.; Valentijn, K.M.; Koning, R.I.; Koster, A.J. Tools for correlative cryo-fluorescence microscopy and cryo-electron tomography applied to whole mitochondria in human endothelial cells. Eur. J. Cell Biol. 2009, 88, 669–684. [Google Scholar] [CrossRef] [PubMed]
- Schellenberger, P.; Kaufmann, R.; Siebert, C.A.; Hagen, C.; Wodrich, H.; Grunewald, K. High-precision correlative fluorescence and electron cryo microscopy using two independent alignment markers. Ultramicroscopy 2014, 143, 41–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schorb, M.; Briggs, J.A. Correlated cryo-fluorescence and cryo-electron microscopy with high spatial precision and improved sensitivity. Ultramicroscopy 2014, 143, 24–32. [Google Scholar] [CrossRef] [Green Version]
- Faulk, W.P.; Taylor, G.M. An immunocolloid Method for Electron Microscope. Immunochemistry 1971, 8, 1081–1083. [Google Scholar]
- Brisson, A.R.; Tan, S.; Linares, R.; Gounou, C.; Arraud, N. Extracellular vesicles from activated platelets: A semiquantitative cryo-electron microscopy and immuno-gold labeling study. Platelets 2017, 28, 263–271. [Google Scholar] [CrossRef]
- Pante, N.; Fahrenkrog, B. Exploring nuclear pore complex molecular architecture by immuno-electron microscopy using Xenopus oocytes. Methods Cell Biol. 2014, 122, 81–98. [Google Scholar] [CrossRef]
- Chen, R.H.C.; Li, Q.; Snidal, C.A.; Gardezi, S.R.; Stanley, E.F. The Calcium Channel C-Terminal and Synaptic Vesicle Tethering: Analysis by Immuno-Nanogold Localization. Front. Cell Neurosci. 2017, 11, 85. [Google Scholar] [CrossRef]
- Renno, W.M. Post-embedding double-gold labeling immunoelectron microscopic co-localization of neurotransmitters in the rat brain. Neurobiology (Bp) 2001, 9, 91–106. [Google Scholar] [CrossRef]
- Roos, N.; Cyrklaff, M.; Cudmore, S.; Blasco, R.; Krijnse-Locker, J.; Griffiths, G. A novel immunogold cryoelectron microscopic approach to investigate the structure of the intracellular and extracellular forms of vaccinia virus. EMBO J. 1996, 15, 2343–2355. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Strauss, J.D.; Ke, Z.; Alonas, E.; Dillard, R.S.; Hampton, C.M.; Lamb, K.M.; Hammonds, J.E.; Santangelo, P.J.; Spearman, P.W.; et al. Native immunogold labeling of cell surface proteins and viral glycoproteins for cryo-electron microscopy and cryo-electron tomography applications. J. Histochem. Cytochem. 2015, 63, 780–792. [Google Scholar] [CrossRef] [PubMed]
- Shimi, T.; Kittisopikul, M.; Tran, J.; Goldman, A.E.; Adam, S.A.; Zheng, Y.; Jaqaman, K.; Goldman, R.D. Structural organization of nuclear lamins A, C, B1, and B2 revealed by superresolution microscopy. Mol. Biol. Cell 2015, 26, 4075–4086. [Google Scholar] [CrossRef] [Green Version]
- Dahan, I.; Sorrentino, S.; Boujemaa-Paterski, R.; Medalia, O. Tiopronin-Protected Gold Nanoparticles as a Potential Marker for Cryo-EM and Tomography. Structure 2018, 26, 1408–1413. [Google Scholar] [CrossRef] [PubMed]
- Pluckthun, A. Designed ankyrin repeat proteins (DARPins): Binding proteins for research, diagnostics, and therapy. Annu. Rev. Pharmacol. Toxicol. 2015, 55, 489–511. [Google Scholar] [CrossRef] [PubMed]
- Plitzko, J.M.; Schuler, B.; Selenko, P. Structural Biology outside the box-inside the cell. Curr. Opin. Struct. Biol. 2017, 46, 110–121. [Google Scholar] [CrossRef]
- Beck, M.; Baumeister, W. Cryo-Electron Tomography: Can it Reveal the Molecular Sociology of Cells in Atomic Detail? Trends Cell Biol. 2016, 26, 825–837. [Google Scholar] [CrossRef]
- Mulvihill, E.; van Pee, K.; Mari, S.A.; Muller, D.J.; Yildiz, O. Directly Observing the Lipid-Dependent Self-Assembly and Pore-Forming Mechanism of the Cytolytic Toxin Listeriolysin O. Nano Lett. 2015, 15, 6965–6973. [Google Scholar] [CrossRef]
- Pfreundschuh, M.; Alsteens, D.; Wieneke, R.; Zhang, C.; Coughlin, S.R.; Tampe, R.; Kobilka, B.K.; Muller, D.J. Identifying and quantifying two ligand-binding sites while imaging native human membrane receptors by AFM. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef]
- Harkiolaki, M.; Darrow, M.C.; Spink, M.C.; Kosior, E.; Dent, K.; Duke, E. Cryo-soft X-ray tomography: Using soft X-rays to explore the ultrastructure of whole cells. Emerg. Top. Life Sci. 2018. [Google Scholar] [CrossRef]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weber, M.S.; Wojtynek, M.; Medalia, O. Cellular and Structural Studies of Eukaryotic Cells by Cryo-Electron Tomography. Cells 2019, 8, 57. https://doi.org/10.3390/cells8010057
Weber MS, Wojtynek M, Medalia O. Cellular and Structural Studies of Eukaryotic Cells by Cryo-Electron Tomography. Cells. 2019; 8(1):57. https://doi.org/10.3390/cells8010057
Chicago/Turabian StyleWeber, Miriam Sarah, Matthias Wojtynek, and Ohad Medalia. 2019. "Cellular and Structural Studies of Eukaryotic Cells by Cryo-Electron Tomography" Cells 8, no. 1: 57. https://doi.org/10.3390/cells8010057





