Pivotal Role of STAT3 in Shaping Glioblastoma Immune Microenvironment
Abstract
:1. Introduction
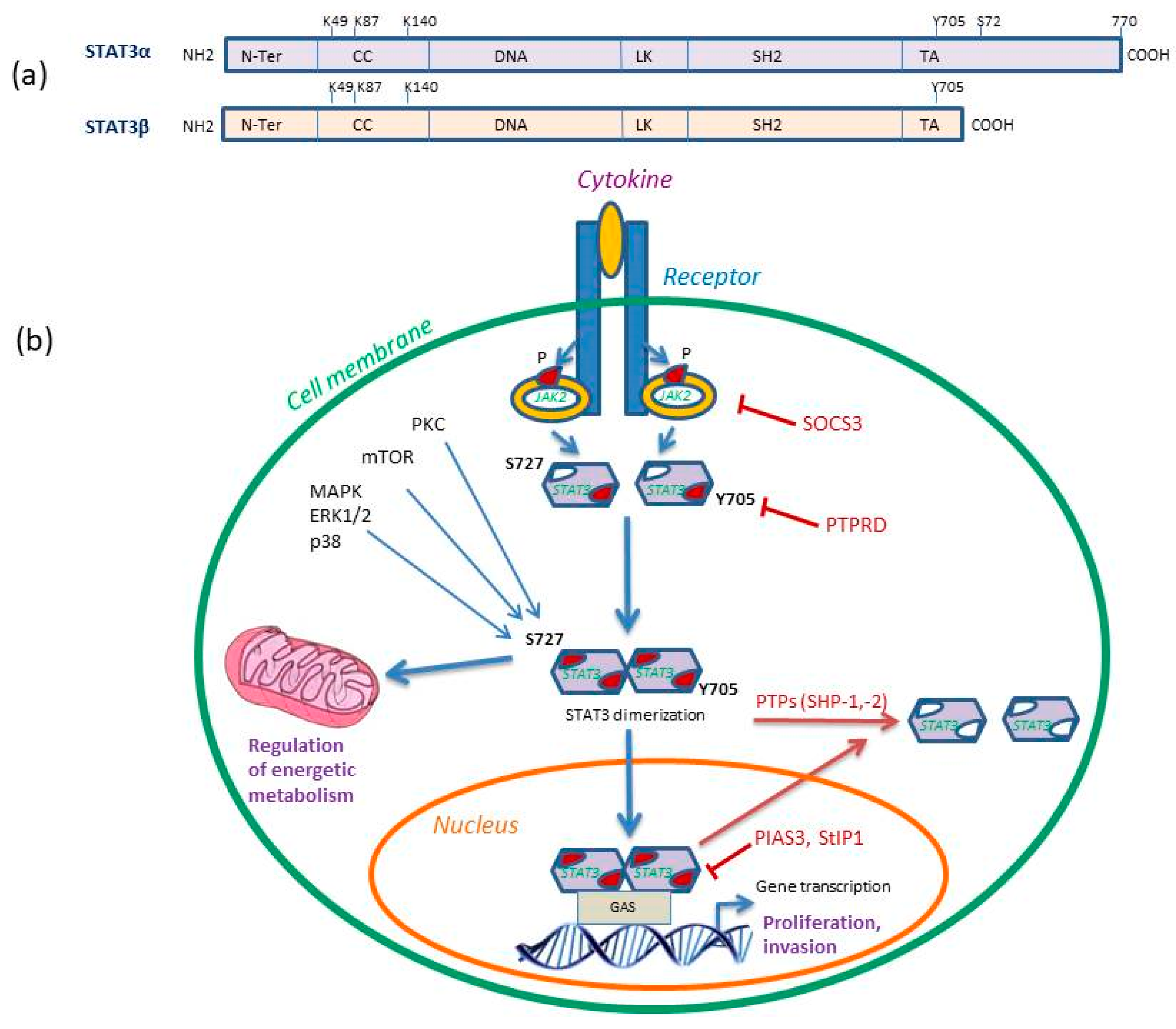
2. Description and Regulation of STAT3 Protein Signaling
3. Evidence of STAT3 Activation in Glioma Cells
4. STAT3 Regulation of Glioblastoma Immune Microenvironment
5. Role of STAT3 in Gliomas Resistance to Treatment
6. Role of STAT3 Targeting in Glioblastomas Immunotherapy
7. Conclusions and Future Directions
Funding
Conflicts of Interest
References
- Da Hora, C.C.; Schweiger, M.W.; Wurdinger, T.; Tannous, B.A. Patient-derived glioma models: From patients to dish to animals. Cells 2019, 8, 1177. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Tamimi, A.F.; Juweid, M. Epidemiology and outcome of glioblastoma. In Glioblastoma [Internet]; De Vleeschouwer, S., Ed.; Codon Publications: Brisbane, Australia, 2017; Chapter 8. PMID: 29251870. [Google Scholar]
- Gao, Y.; Li, L.; Song, L. Expression of p16 and Survivin in gliomas and their correlation with cell proliferation. Oncol. Lett. 2015, 10, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Dunn, G.P.; Rinne, M.L.; Wykosky, J.; Genovese, G.; Quayle, S.N.; Dunn, I.F.; Agarwalla, P.K.; Chheda, M.G.; Campos, B.; Wang, A.; et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012, 26, 756–784. [Google Scholar] [CrossRef] [Green Version]
- Pirozzi, C.J.; Yan, H. Improved grading of IDH-mutated astrocytic gliomas. Nat. Rev. Neurol. 2018, 14, 383–384. [Google Scholar] [CrossRef]
- Gusyatiner, O.; Hegi, M.E. Glioma epigenetics: From subclassification to novel treatment options. Semin. Cancer Biol. 2018, 51, 50–58. [Google Scholar] [CrossRef]
- Spyropoulou, A.; Piperi, C.; Adamopoulos, C.; Papavassiliou, A.G. Deregulated chromatin remodeling in the pathobiology of brain tumors. Neuromolecular Med. 2013, 15, 1–24. [Google Scholar] [CrossRef]
- Spyropoulou, A.; Gargalionis, A.; Dalagiorgou, G.; Adamopoulos, C.; Papavassiliou, K.A.; Lea, R.W.; Piperi, C.; Papavassiliou, A.G. Role of histone lysine methyltransferases SUV39H1 and SETDB1 in gliomagenesis: Modulation of cell proliferation, migration, and colony formation. Neuromolecular Med. 2014, 16, 70–82. [Google Scholar] [CrossRef]
- Meyer, M.; Reimand, J.; Lan, X.; Head, R.; Zhu, X.; Kushida, M.; Bayani, J.; Pressey, J.C.; Lionel, A.C.; Clarke, I.D.; et al. Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proc. Natl. Acad. Sci. USA 2015, 112, 851–856. [Google Scholar] [CrossRef] [Green Version]
- Gieryng, A.; Pszczolkowska, D.; Walentynowicz, K.A.; Rajan, W.D.; Kaminska, B. Immune microenvironment of gliomas. Lab. Investig. 2017, 97, 498–518. [Google Scholar] [CrossRef] [Green Version]
- Matias, D.; Balça-Silva, J.; da Graça, G.C.; Wanjiru, C.M.; Macharia, L.W.; Nascimento, C.P.; Roque, N.R.; Coelho-Aguiar, J.M.; Pereira, C.M.; Dos Santos, M.F.; et al. Microglia/astrocytes–glioblastoma crosstalk: Crucial molecular mechanisms and microenvironmental factors. Front. Cell Neurosci. 2018, 12, 235. [Google Scholar] [CrossRef] [PubMed]
- Rébé, C.; Ghiringhelli, F. STAT3, a master regulator of anti-tumor immune response. Cancers 2019, 11, 1280. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.; Kurzrock, R.; Kim, Y.; Woessner, R.; Younes, A.; Nemunaitis, J.; Fowler, N.; Zhou, T.; Schmidt, J.; Jo, M.; et al. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci. Transl. Med. 2015, 7, 314ra185. [Google Scholar] [CrossRef] [PubMed]
- De la Iglesia, N.; Konopka, G.; Puram, S.V.; Chan, J.A.; Bachoo, R.M.; You, M.J.; Levy, D.E.; Depinho, R.A.; Bonni, A. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev. 2008, 22, 449–462. [Google Scholar] [CrossRef] [Green Version]
- Pencik, J.; Schlederer, M.; Gruber, W.; Unger, C.; Walker, S.M.; Chalaris, A.; Marié, I.J.; Hassler, M.R.; Javaheri, T.; Aksoy, O. STAT3 regulated ARF expression suppresses prostate cancer metastasis. Nat. Commun. 2015, 6, 7736. [Google Scholar] [CrossRef] [Green Version]
- Mostofa, A.G.; Punganuru, S.R.; Madala, H.R.; Al-Obaide, M.; Srivenugopal, K.S. The process and regulatory components of inflammation in brain oncogenesis. Biomolecules 2017, 7, 34. [Google Scholar] [CrossRef]
- Ouédraogo, Z.G.; Biau, J.; Kemeny, J.L.; Morel, L.; Verrelle, P.; Chautard, E. Role of STAT3 in genesis and progression of human malignant gliomas. Mol. Neurobiol. 2017, 54, 5780–5797. [Google Scholar] [CrossRef]
- Chang, N.; Ahn, S.H.; Kong, D.S.; Lee, H.W.; Nam, D.H. The role of STAT3 in glioblastoma progression through dual influences on tumor cells and the immune microenvironment. Mol. Cell Endocrinol. 2017, 451, 53–65. [Google Scholar] [CrossRef]
- Majoros, A.; Platanitis, E.; Kernbauer-Hölzl, E.; Rosebrock, F.; Müller, M.; Decker, T. Canonical and non-canonical aspects of JAK-STAT signaling: lessons from interferons for cytokine responses. Front. Immunol. 2017, 8, 29. [Google Scholar] [CrossRef]
- Lidia, A.; Valeria, P. Nucleus, mitochondrion, or reticulum? STAT3 à la carte. Int. J. Mol. Sci. 2018, 19, 2820. [Google Scholar] [CrossRef]
- Bowman, T.; Broome, M.A.; Sinibaldi, D.; Wharton, W.; Pledger, W.J.; Sedivy, J.M.; Irby, R.; Yeatman, T.; Courtneidge, S.A.; Jove, R. Stat3-mediated Myc expression is required for Src transformation and PDGF-induced mitogenesis. Proc. Natl. Acad. Sci. USA 2001, 98, 7319–7324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, F.; Zhang, Q.; Chen, W.; Han, C.; He, Y.; Ran, Q.; Yao, S. IL-6 increases SDCBP expression, cell proliferation, and cell invasion by activating JAK2/STAT3 in human glioma cells. Am. J. Transl. Res. 2017, 9, 4617–4626. [Google Scholar] [PubMed]
- Jimenez-Pascual, A.; Siebzehnrubl, F.A. Fibroblast growth factor receptor functions in glioblastoma. Cells 2019, 8, 715. [Google Scholar] [CrossRef] [PubMed]
- Carrasco-Garcia, E.; Martinez-Lacaci, I.; Mayor-López, L.; Tristante, E.; Carballo-Santana, M.; García-Morales, P.; Ventero Martin, M.P.; Fuentes-Baile, M.; Rodriguez-Lescure, Á.; Saceda, M. PDGFR and IGF-1R inhibitors induce a G2/M arrest and subsequent cell death in human glioblastoma cell lines. Cells 2018, 7, 131. [Google Scholar] [CrossRef]
- Liu, L.H.; Li, H.; Cheng, X.X.; Kong, Q.Y.; Chen, X.Y.; Wu, M.L.; Li, Y.; Liu, J.; Li, C. Correlative analyses of the expression levels of PIAS3, p-SHP2, SOCS1 and SOCS3 with STAT3 activation in human astrocytomas. Mol. Med. Rep. 2017, 15, 847–852. [Google Scholar] [CrossRef]
- Zhang, X.; Guo, A.; Yu, J.; Possemato, A.; Chen, Y.; Zheng, W.; Polakiewicz, R.D.; Kinzler, K.W.; Vogelstein, B.; Velculescu, V.E.; et al. Identification of STAT3 as a substrate of receptor protein tyrosine phosphatase T. Proc. Natl. Acad. Sci. USA 2007, 104, 4060–4064. [Google Scholar] [CrossRef] [Green Version]
- Yeh, J.E.; Frank, D.A. STAT3-interacting proteins as modulators of transcription factor function: Implications to targeted cancer therapy. Chem. Med. Chem. 2016, 11, 795–801. [Google Scholar] [CrossRef]
- Doucette, T.A.; Kong, L.Y.; Yang, Y.; Ferguson, S.D.; Yang, J.; Wei, J.; Qiao, W.; Fuller, G.N.; Bhat, K.P.; Aldape, K.; et al. Signal transducer and activator of transcription 3 promotes angiogenesis and drives malignant progression in glioma. Neuro Oncol. 2012, 14, 1136–1145. [Google Scholar] [CrossRef] [Green Version]
- Schaefer, L.K.; Ren, Z.; Fuller, G.N.; Schaefer, T.S. Constitutive activation of Stat3alpha in brain tumors: Localization to tumor endothelial cells and activation by the endothelial tyrosine kinase receptor (VEGFR-2). Oncogene 2002, 21, 2058–2065. [Google Scholar] [CrossRef]
- Brantley, E.C.; Nabors, L.B.; Gillespie, G.Y.; Choi, Y.H.; Palmer, C.A.; Harrison, K.; Roarty, K.; Benveniste, E.N. Loss of protein inhibitors of activated STAT-3 expression in glioblastoma multiforme tumors: Implications for STAT-3 activation and gene expression. Clin. Cancer Res. 2008, 14, 4694–4704. [Google Scholar] [CrossRef]
- Lin, G.-S.; Chen, Y.-P.; Lin, Z.-X.; Wang, X.-F.; Zheng, Z.-Q.; Chen, L. STAT3 serine 727 phosphorylation influences clinical outcome in glioblastoma. Int. J. Clin. Exp. Pathol. 2014, 7, 3141–3149. [Google Scholar] [PubMed]
- Tu, Y.; Zhong, Y.; Fu, J.; Cao, Y.; Fu, G.; Tian, X.; Wang, B. Activation of JAK/STAT signal pathway predicts poor prognosis of patients with gliomas. Med. Oncol. 2011, 28, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Luwor, R.B.; Stylli, S.S.; Kaye, A.H. The role of Stat3 in glioblastoma multiforme. J. Clin. Neurosci. 2013, 20, 907–911. [Google Scholar] [CrossRef] [PubMed]
- Puram, S.V.; Yeung, C.M.; Jahani-Asl, A.; Lin, C.; de la Iglesia, N.; Konopka, G.; Jackson-Grusby, L.; Bonni, A. STAT3-iNOS signaling mediates EGFRvIII-induced glial proliferation and transformation. J. Neurosci. 2012, 32, 7806–7818. [Google Scholar] [CrossRef]
- Wang, H.; Lathia, J.D.; Wu, Q.; Wang, J.; Li, Z.; Heddleston, J.M.; Eyler, C.E.; Elderbroom, J.; Gallagher, J.; Schuschu, J.; et al. Targeting interleukin 6 signaling suppresses glioma stem cell survival and tumor growth. Stem Cells Dayt. Ohio 2009, 27, 2393–2404. [Google Scholar] [CrossRef]
- Xu, Y.; Li, Z.; Zhang, C.; Ji, Y.; Chen, F. Knockdown of PKCε expression inhibits growth, induces apoptosis and decreases invasiveness of human glioma cells partially through Stat3. J. Mol. Neurosci. 2014, 55, 21–31. [Google Scholar] [CrossRef]
- Chumbalkar, V.; Latha, K.; Hwang, Y.-H.; Maywald, R.; Hawley, L.; Sawaya, R.; Diao, L.; Baggerly, K.; Cavenee, W.K.; Furnari, F.B.; et al. Analysis of phosphotyrosine signaling in glioblastoma identifies STAT5 as a novel downstream target of ΔEGFR. J. Proteome Res. 2011, 10, 1343–1352. [Google Scholar] [CrossRef]
- Veeriah, S.; Brennan, C.; Meng, S.; Singh, B.; Fagin, J.A.; Solit, D.B.; Paty, P.B.; Rohle, D.; Vivanco, I.; Chmielecki, J.; et al. The tyrosine phosphatase PTPRD is a tumor suppressor that is frequently inactivated and mutated in glioblastoma and other human cancers. Proc. Natl. Acad. Sci. USA 2009, 106, 9435–9440. [Google Scholar] [CrossRef] [Green Version]
- Yamini, B. NF-κB, mesenchymal differentiation and glioblastoma. Cells 2018, 7, 125. [Google Scholar] [CrossRef]
- Tan, M.S.Y.; Sandanaraj, E.; Chong, Y.K.; Lim, S.W.; Koh, L.W.H.; Ng, W.H.; Tan, N.S.; Tan, P.; Ang, B.T.; Tang, C. A STAT3-based gene signature stratifies glioma patients for targeted therapy. Nat. Commun. 2019, 10, 3601. [Google Scholar] [CrossRef]
- Haybaeck, J.; Obrist, P.; Schindler, C.U.; Spizzo, G.; Doppler, W. STAT-1 expression in human glioblastoma and peritumoral tissue. Anticancer Res. 2007, 27, 3829–3835. [Google Scholar] [PubMed]
- De Groot, J.; Liang, J.; Kong, L.Y.; Wei, J.; Piao, Y.; Fuller, G.; Qiao, W.; Heimberger, A.B. Modulating antiangiogenic resistance by inhibiting the signal transducer and activator of transcription 3 pathway in glioblastoma. Oncotarget 2012, 3, 1036–1048. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, G.; Deng, L.; Liu, Q.; Dai, J.; Shen, J.; Zhang, J. IL-6 augments the invasiveness of U87MG human glioblastoma multiforme cells via up-regulation of MMP-2 and fascin-1. Oncol. Rep. 2010, 23, 1553–1559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, J.; Zhou, A.; Wu, Y.; Morris, S.A.; Lin, K.; Amin, S.; Verhaak, R.; Fuller, G.; Xie, K.; Heimberger, A.B.; et al. miR-182-5p induced by STAT3 activation promotes glioma tumorigenesis. Cancer Res. 2016, 76, 4293–4304. [Google Scholar] [CrossRef] [PubMed]
- Liebelt, B.D.; Shingu, T.; Zhou, X.; Ren, J.; Shin, S.A.; Hu, J. Glioma stem cells: Signaling, microenvironment, and therapy. Stem Cells Int. 2016, 2016, 7849890. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Ye, H.; Qi, Z.; Mo, L.; Yue, Q.; Baral, A.; Hoon, D.S.B.; Vera, J.C.; Heiss, J.D.; Chen, C.C.; et al. B7-H4(B7x)-mediated cross-talk between glioma-initiating cells and macrophages via the IL6/JAK/STAT3 pathway lead to poor prognosis in glioma patients. Clin. Cancer Res. 2016, 22, 2778–2790. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Li, A.; Ahn, S.; Song, H.; Zhang, W. Inositol Polyphosphate-5-Phosphatase F (INPP5F) inhibits STAT3 activity and suppresses gliomas tumorigenicity. Sci. Rep. 2014, 4, 7330. [Google Scholar] [CrossRef]
- Kim, E.; Kim, M.; Woo, D.H.; Shin, Y.; Shin, J.; Chang, N.; Oh, Y.T.; Kim, H.; Rheey, J.; Nakano, I.; et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell 2013, 23, 839–852. [Google Scholar] [CrossRef]
- Su, Y.L.; Banerjee, S.; White, S.V.; Kortylewski, M. STAT3 in tumor-associated myeloid cells: Multitasking to disrupt immunity. Int. J. Mol. Sci. 2018, 19, E1803. [Google Scholar] [CrossRef]
- Martinez, F.O.; Gordon, S. The M1 and M2 paradigm of macrophage activation: Time for reassessment. F1000Prime Rep. 2014, 6, 13. [Google Scholar] [CrossRef]
- Wolfle, S.J.; Strebovsky, J.; Bartz, H.; Sahr, A.; Arnold, C.; Kaiser, C.; Dalpke, A.H.; Heeg, K. PD-L1 expression on tolerogenic APCs is controlled by STAT-3. Eur. J. Immunol. 2011, 41, 413–424. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, S.D.; Srinivasan, V.M.; Heimberger, A.B. The role of STAT3 in tumor-mediated immune suppression. J. Neuro-Oncol. 2015, 123, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, C.M.; Holowka, T.; Sun, J.; Blagih, J.; Amiel, E.; DeBerardinis, R.J.; Cross, J.R.; Jung, E.; Thompson, C.B.; Jones, R.G.; et al. Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood 2010, 115, 4742–4749. [Google Scholar] [CrossRef] [PubMed]
- Netherby, C.S.; Messmer, M.N.; Burkard-Mandel, L.; Colligan, S.; Miller, A.; Cortes Gomez, E.; Wang, J.; Nemeth, M.J.; Abrams, S.I. The granulocyte progenitor stage is a key target of IRF8-mediated regulation of myeloid-derived suppressor cell production. J. Immunol. 2017, 198, 4129–4139. [Google Scholar] [CrossRef] [PubMed]
- Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Bronte, V. Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol. 2012, 12, 253–268. [Google Scholar] [CrossRef] [Green Version]
- Vasquez-Dunddel, D.; Pan, F.; Zeng, Q.; Gorbounov, M.; Albesiano, E.; Fu, J.; Blosser, R.L.; Tam, A.J.; Bruno, T.; Zhang, H.; et al. STAT3 regulates arginase-I in myeloid-derived suppressor cells from cancer patients. J. Clin. Investig. 2013, 123, 1580–1589. [Google Scholar] [CrossRef] [Green Version]
- Lau, J.; Ilkhanizadeh, S.; Wang, S.; Miroshnikova, Y.A.; Salvatierra, N.A.; Wong, R.A.; Schmidt, C.; Weaver, V.M.; Weiss, W.A.; Persson, A.I. STAT3 blockade inhibits radiation-induced malignant progression in glioma. Cancer Res. 2015, 75, 4302–4311. [Google Scholar] [CrossRef]
- Yang, Y.P.; Chang, Y.L.; Huang, P.I.; Chiou, G.Y.; Tseng, L.M.; Chiou, S.H.; Chen, M.H.; Chen, M.T.; Shih, Y.H.; Chang, C.H.; et al. Resveratrol suppresses tumorigenicity and enhances radiosensitivity in primary glioblastoma tumor initiating cells by inhibiting the STAT3 axis. J. Cell Physiol. 2012, 227, 976–993. [Google Scholar] [CrossRef]
- Ouédraogo, Z.G.; Müller-Barthélémy, M.; Kemeny, J.-L.; Dedieu, V.; Biau, J.; Khalil, T.; Raoelfils, L.I.; Granzotto, A.; Pereira, B.; Beaudoin, C.; et al. STAT3 serine 727 phosphorylation: A relevant target to radiosensitize human glioblastoma. Brain Pathol. Zur. Switz. 2016, 26, 18–30. [Google Scholar] [CrossRef]
- Lee, E.-S.; Ko, K.-K.; Joe, Y.A.; Kang, S.G.; Hong, Y.K. Inhibition of STAT3 reverses drug resistance acquired in temozolomide-resistant human glioma cells. Oncol. Lett. 2011, 2, 115–121. [Google Scholar] [CrossRef]
- Rosenberg, S.A.; Yang, J.C.; Restifo, N.P. Cancer immunotherapy: Moving beyond current vaccines. Nat. Med. 2004, 10, 909e915. [Google Scholar] [CrossRef] [PubMed]
- Austin, J.W.; Lu, P.; Majumder, P.; Ahmed, R.; Boss, J.M. STAT3, STAT4, NFATc1, and CTCF regulate PD-1 through multiple novel regulatory regions in murine T cells. J. Immunol. 2014, 192, 4876–4886. [Google Scholar] [CrossRef] [PubMed]
- Baskin, R.; Park, S.O.; Keseru, G.M.; Bisht, K.S.; Wamsley, H.L.; Sayeski, P.P. The Jak2 small molecule inhibitor, G6, reduces the tumorigenic potential of T98G glioblastoma cells in vitro and in vivo. PLoS ONE 2014, 9, e105568. [Google Scholar] [CrossRef] [PubMed]
- Mukthavaram, R.; Ouyang, X.; Saklecha, R.; Jiang, P.; Nomura, N.; Pingle, S.C.; Guo, F.; Makale, M.; Kesari, S. Effect of the JAK2/STAT3 inhibitor SAR317461 on human glioblastoma tumorspheres. J. Transl. Med. 2015, 13, 269. [Google Scholar] [CrossRef]
- Hussain, S.F.; Kong, L.Y.; Jordan, J.; Conrad, C.; Madden, T.; Fokt, I.; Priebe, W.; Heimberger, A.B. A novel small molecule inhibitor of signal transducers and activators of transcription 3 reverses immune tolerance in malignant glioma patients. Cancer Res. 2007, 67, 9630–9636. [Google Scholar] [CrossRef]
- Fujiwara, Y.; Komohara, Y.; Kudo, R.; Tsurushima, K.; Ohnishi, K.; Ikeda, T.; Takeya, M. Oleanolic acid inhibits macrophage differentiation into the M2 phenotype and glioblastoma cell proliferation by suppressing the activation of STAT3. Oncol. Rep. 2011, 26, 1533–1537. [Google Scholar] [CrossRef]
- Dai, Y.; Jiao, H.; Teng, G.; Wang, W.; Zhang, R.; Wang, Y.; Hebbard, L.; George, J.; Qiao, L. Embelin reduces colitis-associated tumorigenesis through limiting IL-6/STAT3 signaling. Mol. Cancer Ther. 2014, 13, 1206–1216. [Google Scholar] [CrossRef]
- Michaud-Levesque, J.; Bousquet-Gagnon, N.; Beliveau, R. Quercetin abrogates IL-6/STAT3 signaling and inhibits glioblastoma cell line growth and migration. Exp. Cell Res. 2012, 318, 925–935. [Google Scholar] [CrossRef]
- Hossain, D.M.S.; Moreira, D.; Zhang, Q.; Nechaev, S.; Swiderski, P.; Kortylewski, M. TLR9-targeted siRNA delivery in vivo. Methods Mol. Biol. 2016, 1364, 183–196. [Google Scholar] [CrossRef]
- Zhang, Q.; Hossain, D.M.S.; Duttagupta, P.; Moreira, D.; Zhao, X.; Won, H.; Buettner, R.; Nechaev, S.; Majka, M.; Zhang, B.; et al. Serum-resistant CpG-STAT3 decoy for targeting survival and immune checkpoint signaling in acute myeloid leukemia. Blood 2016, 127, 1687–1700. [Google Scholar] [CrossRef]
- Stackhouse, C.T.; Rowland, J.R.; Shevin, R.S.; Singh, R.; Gillespie, G.Y.; Willey, C.D. A novel assay for profiling GBM cancer model heterogeneity and drug screening. Cells 2019, 8, E702. [Google Scholar] [CrossRef] [PubMed]

| WHO Grade Type | Tumor Type | Clinical Characteristics | |
|---|---|---|---|
| Low Grade | Grade I | - Ganglioglioma - Gangliocytoma - Craniopharyngioma - Pilocytic astrocytoma - Meningioma - Subependymoma - Choroid plexus papilloma - Pineocytoma | - Benign - Non-infiltrative - Slow proliferation - Possible curable by surgery - Long-term survival |
| Grade II | - Diffuse Astrocytoma - Pure oligodendroglioma - Pineocytoma - Atypical meningioma | - Partly infiltrative - Relative slow proliferation - Can recur as higher grade | |
| High Grade | Grade III | - Anaplastic astrocytoma - Anaplastic oligodendroglioma - Anaplastic ependymoma - Anaplastic meningioma | - Malignant - Infiltrative - Can recur as high grade |
| Grade IV | - Gliobastoma multiforme (GB) - Ependymoblastoma - Medulloblastoma - Pineoblastoma | - Most malignant - Highly infiltrative - Rapid proliferation - Aggressive - Quick recurrence - Necrosis |
| Drug/Approach | Target | STAT3 Inhibition | GB Effect | Reference |
|---|---|---|---|---|
| Sorafenib | JAK1/2 | Upstream kinase inhibition | Reduce cell proliferation, increase apoptosis of GB cells | [18,19] |
| AG490 | JAK2 | Upstream kinase inhibition | Decrease migration and angiogenesis of GB cells | [18,19] |
| G6 | JAK2 | Upstream kinase inhibition | Increase apoptosis, reduces invasion | [64] |
| G5-7 | JAK2 | Upstream kinase inhibition | Reduce cell growth, decreases angiogenesis | [18,19] |
| SAR317461 | JAK2 | Upstream kinase inhibition | Induces autophagy | [65] |
| WP1066 | JAK2 | Upstream kinase inhibition | Induction of cytokines release (IL-2, IL-4, IL-12, and IL-15) that stimulate T cell effector function to overcome immunosuppression | [66] |
| Oleanolic acid | STAT3 | Blocks STAT3 phosphorylation | Suppresses the M2 polarization of TAMs by reducing IL-10 secretion | [67] |
| Embelin | STAT3 | Blocks STAT3 phosphorylation by increasing SHP2 activity | Limits IL-6/STAT3 activation and the Th17 immune response in GBs | [68] |
| Quercetin | STAT3 | Blocks STAT3 phosphorylation | Inhibitor of the IL-6/STAT3 signaling pathway in GB cells | [69] |
| CpG oligodeoxynucleotides (ODN)containing conjugates of STAT3 inhibitors with synthetic TLR9 agonists | STAT3 | Block of STAT3 phosphorylation | Reduce the tolerogenic effects of TME in vivo | [70,71] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piperi, C.; Papavassiliou, K.A.; Papavassiliou, A.G. Pivotal Role of STAT3 in Shaping Glioblastoma Immune Microenvironment. Cells 2019, 8, 1398. https://doi.org/10.3390/cells8111398
Piperi C, Papavassiliou KA, Papavassiliou AG. Pivotal Role of STAT3 in Shaping Glioblastoma Immune Microenvironment. Cells. 2019; 8(11):1398. https://doi.org/10.3390/cells8111398
Chicago/Turabian StylePiperi, Christina, Kostas A. Papavassiliou, and Athanasios G. Papavassiliou. 2019. "Pivotal Role of STAT3 in Shaping Glioblastoma Immune Microenvironment" Cells 8, no. 11: 1398. https://doi.org/10.3390/cells8111398






