Hits Discovery on the Androgen Receptor: In Silico Approaches to Identify Agonist Compounds
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Sources
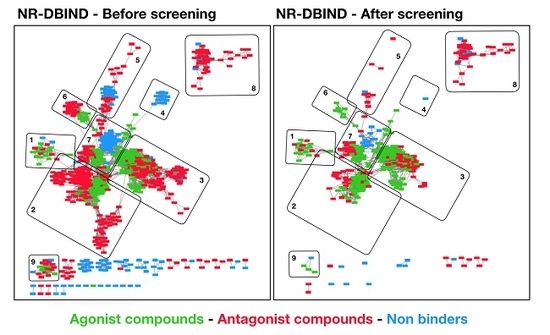
2.1.1. NR-DBIND
2.1.2. Tox21
2.2. Small Molecules Preparation
2.3. Molecule Similarity Networks and Chemical Space Analysis
2.4. Protein Preparation
2.5. Pharmacophore Modelling Protocol
2.5.1. Primary Pharmacophore Modelling
- The training set molecules were clustered using LigandScout default parameters;
- “Merged” pharmacophore models were generated, i.e., models considering features common to at least 10% of the cluster small molecules. 10 pharmacophore models were generated per cluster.
2.5.2. Pharmacophore Selection
2.5.3. Pharmacophore Models Optimization
2.6. Small Molecules Docking Protocol
3. Results
3.1. Similarity Between Agonist and Non-agonist Compounds
3.2. Pharmacophores Ability to Discriminate Agonist Compounds
3.2.1. Performance on the NR-DBIND Datasets
3.2.2. Performance on the Tox21 Dataset
3.3. Docking Performances
3.3.1. Performance on the Training Set
3.3.2. Performance on the Tox21 Set
3.4. Pharmacophore Modeling vs. Docking on the NR-DBIND Set
4. Discussion
4.1. NRs Modulators’ Tricky Structures
4.2. Recommendations on Screening Strategies
4.3. Publication Bias
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tan, M.H.E.; Li, J.; Xu, H.E.; Melcher, K.; Yong, E. Androgen receptor: Structure, role in prostate cancer and drug discovery. Acta Pharmacol. Sin. 2015, 36, 3–23. [Google Scholar] [CrossRef]
- Mohler, M.L.; Bohl, C.E.; Jones, A.; Coss, C.C.; Narayanan, R.; He, Y.; Hwang, D.J.; Dalton, J.T.; Miller, D.D. Nonsteroidal selective androgen receptor modulators (SARMs): Dissociating the anabolic and androgenic activities of the androgen receptor for therapeutic benefit. J. Med. Chem. 2009, 52, 3597–3617. [Google Scholar] [PubMed]
- Davey, R.A.; Grossmann, M. Androgen Receptor Structure, Function and Biology: From Bench to Bedside. Clin. Biochem. Rev. 2016, 37, 3–15. [Google Scholar] [PubMed]
- Cutress, M.L.; Whitaker, H.C.; Mills, I.G.; Stewart, M.; Neal, D.E. Structural basis for the nuclear import of the human androgen receptor. J. Cell Sci. 2008, 121, 957–968. [Google Scholar] [PubMed]
- Jiang, M.; Ma, Y.; Chen, C.; Fu, X.; Yang, S.; Li, X.; Yu, G.; Mao, Y.; Xie, Y.; Li, Y. Androgen-Responsive Gene Database: Integrated Knowledge on Androgen-Responsive Genes. Mol. Endocrinol. 2009, 23, 1927–1933. [Google Scholar]
- Leung, J.K.; Sadar, M.D. Non-Genomic Actions of the Androgen Receptor in Prostate Cancer. Front. Endocrinol. 2017, 8, 2. [Google Scholar]
- Fujita, K.; Nonomura, N. Role of Androgen Receptor in Prostate Cancer: A Review. World J. Mens Health 2019, 37, 288–295. [Google Scholar]
- Brooke, G.N.; Bevan, C.L. The Role of Androgen Receptor Mutations in Prostate Cancer Progression. Curr. Genom. 2009, 10, 18–25. [Google Scholar]
- Ponnusamy, S.; Sullivan, R.D.; You, D.; Zafar, N.; He Yang, C.; Thiyagarajan, T.; Johnson, D.L.; Barrett, M.L.; Koehler, N.J.; Star, M.; et al. Androgen receptor agonists increase lean mass, improve cardiopulmonary functions and extend survival in preclinical models of Duchenne muscular dystrophy. Hum. Mol. Genet. 2017, 26, 2526–2540. [Google Scholar]
- Cozzoli, A.; Capogrosso, R.F.; Sblendorio, V.T.; Dinardo, M.M.; Jagerschmidt, C.; Namour, F.; Camerino, G.M.; De Luca, A. GLPG0492, a novel selective androgen receptor modulator, improves muscle performance in the exercised-mdx mouse model of muscular dystrophy. Pharmacol. Res. 2013, 72, 9–24. [Google Scholar]
- Narayanan, R.; Mohler, M.L.; Bohl, C.E.; Miller, D.D.; Dalton, J.T. Selective androgen receptor modulators in preclinical and clinical development. Nucl. Recept. Signal. 2008, 6, e010. [Google Scholar] [CrossRef] [PubMed]
- Di Donato, M.; Bilancio, A.; D’Amato, L.; Claudiani, P.; Oliviero, M.A.; Barone, M.V.; Auricchio, A.; Appella, E.; Migliaccio, A.; Auricchio, F.; et al. Cross-talk between androgen receptor/filamin A and TrkA regulates neurite outgrowth in PC12 cells. Mol. Biol. Cell 2015, 26, 2858–2872. [Google Scholar] [CrossRef] [PubMed]
- Tanrikulu, Y.; Krüger, B.; Proschak, E. The holistic integration of virtual screening in drug discovery. Drug Discov. Today 2013, 18, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Réau, M.; Lagarde, N.; Zagury, J.-F.; Montes, M. Nuclear Receptors Database Including Negative Data (NR-DBIND): A Database Dedicated to Nuclear Receptors Binding Data Including Negative Data and Pharmacological Profile. J. Med. Chem. 2018, 62, 2894–2904. [Google Scholar] [CrossRef]
- Wolber, G.; Seidel, T.; Bendix, F.; Langer, T. Molecule-pharmacophore superpositioning and pattern matching in computational drug design. Drug Discov. Today 2008, 13, 23–29. [Google Scholar] [CrossRef]
- Wolber, G.; Langer, T. LigandScout: 3-D Pharmacophores Derived from Protein-Bound Ligands and Their Use as Virtual Screening Filters. J. Chem. Inf. Model. 2005, 45, 160–169. [Google Scholar] [CrossRef]
- Korb, O.; Stützle, T.; Exner, T.E. Empirical scoring functions for advanced protein-ligand docking with PLANTS. J. Chem. Inf. Model. 2009, 49, 84–96. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- AID 743053—qHTS Assay to Identify Small Molecule Agonists of the Androgen Receptor (AR) Signaling Pathway: Summary—PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/743053 (accessed on 29 July 2019).
- Huang, R.; Xia, M.; Cho, M.-H.; Sakamuru, S.; Shinn, P.; Houck, K.A.; Dix, D.J.; Judson, R.S.; Witt, K.L.; Kavlock, R.J.; et al. Chemical Genomics Profiling of Environmental Chemical Modulation of Human Nuclear Receptors. Environ. Health Perspect. 2011, 119, 1142–1148. [Google Scholar] [CrossRef]
- RDKit: Open-Source Cheminformatics. Available online: www.rdkit.org (accessed on 13 November 2019).
- Cytoscape: An Open Source Platform for Complex Network Analysis and Visualization. Available online: https://cytoscape.org/ (accessed on 30 July 2019).
- Lagarde, N.; Zagury, J.-F.; Montes, M. Importance of the pharmacological profile of the bound ligand in enrichment on nuclear receptors: Toward the use of experimentally validated decoy ligands. J. Chem. Inf. Model. 2014, 54, 2915–2944. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Ben Nasr, N.; Guillemain, H.; Lagarde, N.; Zagury, J.-F.; Montes, M. Multiple structures for virtual ligand screening: Defining binding site properties-based criteria to optimize the selection of the query. J. Chem. Inf. Model. 2013, 53, 293–311. [Google Scholar] [CrossRef] [PubMed]
- Empereur-Mot, C.; Guillemain, H.; Latouche, A.; Zagury, J.-F.; Viallon, V.; Montes, M. Predictiveness curves in virtual screening. J. Cheminform. 2015, 7, 52. [Google Scholar] [CrossRef] [PubMed]
- Rastinejad, F.; Huang, P.; Chandra, V.; Khorasanizadeh, S. Understanding Nuclear Receptor Form and Function Using Structural Biology. J. Mol. Endocrinol. 2013, 51, T1–T21. [Google Scholar] [CrossRef]
- Togashi, M.; Borngraeber, S.; Sandler, B.; Fletterick, R.J.; Webb, P.; Baxter, J.D. Conformational adaptation of nuclear receptor ligand binding domains to agonists: Potential for novel approaches to ligand design. J. Steroid Biochem. Mol. Biol. 2005, 93, 127–137. [Google Scholar]
- Spencer, T.A.; Li, D.; Russel, J.S.; Collins, J.L.; Bledsoe, R.K.; Consler, T.G.; Moore, L.B.; Galardi, C.M.; McKee, D.D.; Moore, J.T.; et al. Pharmacophore Analysis of the Nuclear Oxysterol Receptor LXRα. J. Med. Chem. 2001, 44, 886–897. [Google Scholar]
- Lewis, S.N.; Garcia, Z.; Hontecillas, R.; Bassaganya-Riera, J.; Bevan, D.R. Pharmacophore modeling improves virtual screening for novel peroxisome proliferator-activated receptor-gamma ligands. J. Comput. Aided Mol. Des. 2015, 29, 421–439. [Google Scholar] [CrossRef]
- Lagarde, N.; Delahaye, S.; Zagury, J.-F.; Montes, M. Discriminating agonist and antagonist ligands of the nuclear receptors using 3D-pharmacophores. J. Cheminform. 2016, 8, 43. [Google Scholar]
- Brzozowski, A.M.; Pike, A.C.; Dauter, Z.; Hubbard, R.E.; Bonn, T.; Engström, O.; Ohman, L.; Greene, G.L.; Gustafsson, J.A.; Carlquist, M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature 1997, 389, 753–758. [Google Scholar] [CrossRef]
- Shiau, A.K.; Barstad, D.; Radek, J.T.; Meyers, M.J.; Nettles, K.W.; Katzenellenbogen, B.S.; Katzenellenbogen, J.A.; Agard, D.A.; Greene, G.L. Structural characterization of a subtype-selective ligand reveals a novel mode of estrogen receptor antagonism. Nat. Struct. Biol. 2002, 9, 359–364. [Google Scholar]
- Mysinger, M.M.; Carchia, M.; Irwin, J.J.; Shoichet, B.K. Directory of Useful Decoys, Enhanced (DUD-E): Better Ligands and Decoys for Better Benchmarking. J. Med. Chem. 2012, 55, 6582–6594. [Google Scholar] [PubMed]
- Roy, A. Early Probe and Drug Discovery in Academia: A Minireview. High Throughput 2018, 7, 4. [Google Scholar]





| Set | pKi-Pharmacophores | pIC50-Pharmacophores | pKi-pIC50-Pharmacophores | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sens | Spec | EF | Sens | Spec | EF | Sens | Spec | EF | ||
| NR-DBIND | pKi | Train: 0.95 Test: 0.80 | Train: 0.88 Test: 0.89 | Train: 2.44 Test: 2.03 | 0.23 | 0.87 | 1.22 | - | - | - |
| pIC50 | 0.61 | 0.62 | 1.43 | Train: 0.98 Test: 0.74 | Train: 0.94 Test: 0.91 | Train: 3.45 Test: 3.08 | - | - | - | |
| pKi-pIC50 | 0.78 | 0.73 | 1.90 | 0.53 | 0.89 | 2.33 | 0.92 | 0.68 | 1.88 | |
| Tox21 | Tox21 (<1 µM) | 0.01 | 0.90 | 0.06 | 0.23 | 0.95 | 2.53 | 0.24 | 0.88 | 1.15 |
| Tox21 (<100 nM) | 0.01 | 0.90 | 0.09 | 0.27 | 0.95 | 3.47 | 0.29 | 0.88 | 1.57 | |
| Tox21 (<10 nM) | 0.00 | 0.90 | 0.00 | 0.24 | 0.95 | 3.44 | 0.24 | 0.88 | 1.45 | |
| PLANTS | AutoDock VINA | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Max | Pdb | Min | Mean | Std | Max | Pdb | Min | Mean | Std | ||
| AR | Single | 0.69 | 1xow | 0.43 | 0.56 | 0.06 | 0.72 | 2pir | 0.52 | 0.66 | 0.05 |
| ensemble_2 | 0.69 | 1xow, 2ama | 0.47 | 0.56 | 0.06 | 0.76 | 1t5z, 2piw | 0.6 | 0.71 | 0.03 | |
| ensemble_3 | 0.68 | 1xow, 1xj7, 2ama | 0.46 | 0.56 | 0.05 | 0.78 | 2piq, 2pip, 2amb | 0.64 | 0.73 | 0.02 | |
| PLANTS | AutoDock VINA | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Max | pdb | sens = 0.25 score spec EF | sens = 0.5 score spec EF | sens = 0.75 score spec EF | max | PDB | sens = 0.25 score spec EF | sens = 0.5 score spec EF | sens = 0.75 score spec EF | |
| Single | 0.69 | 1xow | −89.29 | −84.88 | −81.36 | 0.72 | 2pir | −9.20 | −8.7 | −8.20 |
| 0.901 | 0.757 | 0.578 | 0.913 | 0.770 | 0.611 | |||||
| 1.78 | 1.59 | 1.46 | 1.90 | 1.64 | 1.53 | |||||
| ensemble_2 | 0.69 | 1xow, 2ama | −90.03 | −85.38 | −81.70 | 0.76 | 1t5z, 2piw | −9.10 | −8.6 | −8.10 |
| 0.898 | 0.741 | 0.548 | 0.932 | 0.813 | 0.646 | |||||
| 1.75 | 1.54 | 1.40 | 2.11 | 1.83 | 1.62 | |||||
| ensemble_3 | 0.68 | 1xow, 1xj7, 2ama | −90.54 | −85.98 | −81.81 | 0.78 | 2piq, 2pip, 2amb | −9.40 | −8.9 | −8.5 |
| 0.891 | 0.741 | 0.532 | 0.923 | 0.825 | 0.707 | |||||
| 1.69 | 1.54 | 1.37 | 2.01 | 1.89 | 1.79 | |||||
| PLANTS | AutoDock VINA | |||||||
|---|---|---|---|---|---|---|---|---|
| Pdb | P (Active) = 0.5 score sens-spec NR-DBIND Tox21 | P (Active) = 0.66 score sens-spec NR-DBIND Tox21 | P (Active) = 0.75 score sens-spec NR-DBIND Tox21 | PDB | P (Active) = 0.5 score sens-spec NR-DBIND Tox21 | P (Active) = 0.66 score sens-spec NR-DBIND Tox21 | P (Active) = 0.75 score sens-spec NR-DBIND Tox21 | |
| Single | 1xow | −93.66 0.129–0.963 26 actives | - | - | 2pir | −9.50 0.170–0.93939 act/ 36 ina 22 act/ 49 ina | −10.30 0.009–0.9916 act/ 7 ina 6 act/ 9 ina | - |
| ensemble_2 | 1xow, 2ama | −93.92 0.138–0.959 32 actives | - | - | 1t5z, 2piw | −9.1 0.290–0.918 65 act/ 49 ina 28 act/ 75 ina | −9.8 0.058–0.981 13 act/ 12 ina 15 act/ 25 ina | −10.30 0.027–0.995 7 act/ 3 ina 7 act/ 14 ina |
| ensemble_3 | 1xow, 1xj7, 2ama | −94.71 0.116–0.964 26 actives | - | - | 2piq, 2pip, 2amb | −9.40 0.277–0.915 62 act/ 51 ina 25 act/ 69 ina | −10.0 0.085–0.981 19 act/ 12 ina 17 act/ 34 ina | −10.40 0.022–0.990 7 act/ 6 ina 4 act/ 17 ina |
| PLANTS | AutoDock VINA | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AUC <1µM (<100 nM/ <10 nM) | pdb | sens = 0.25 score spec EF | sens = 0.5 score spec EF | sens = 0.75 score spec EF | AUC <1µM (<100 nM/<10 nM) | PDB | sens = 0.25 score spec EF | sens = 0.5 score spec EF | sens = 0.75 score spec EF | |
| Single | 0.65 (0.66/0.7) | 1xow | −79.38 0.77 1.08 | −74.96 0.645 1.4 | −70.55 0.520 1.55 | 0.82 (0.83/0.89) | 2pir | −9.4 0.99 17.13 | −8.2 0.92 5.72 | −7.00 0.7 2.27 |
| ensemble_2 | 0.78 (0.8/0.85) | 1xow, 2ama | −87.97 0.9 12.6 | −83.36 0.83 2.8 | −77.33 0.69 2.39 | 0.8 (0.82/0.89) | 1t5z, 2piw | −9.3 0.99 19.22 | −7.2 0.81 2.55 | −6.5 0.63 2.00 |
| ensemble_3 | 0.79 (0.81/0.84) | 1xow, 1xj7, 2ama | −88.29 0.9 2.43 | −84.32 0.83 2.9 | −78.59 0.715 2.56 | 0.81 (0.84/0.90) | 2piq, 2pip, 2amb | −9.4 0.99 15.98 | −8.00 0.88 3.87 | 7.00 0.680 2.29 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Réau, M.; Lagarde, N.; Zagury, J.-F.; Montes, M. Hits Discovery on the Androgen Receptor: In Silico Approaches to Identify Agonist Compounds. Cells 2019, 8, 1431. https://doi.org/10.3390/cells8111431
Réau M, Lagarde N, Zagury J-F, Montes M. Hits Discovery on the Androgen Receptor: In Silico Approaches to Identify Agonist Compounds. Cells. 2019; 8(11):1431. https://doi.org/10.3390/cells8111431
Chicago/Turabian StyleRéau, Manon, Nathalie Lagarde, Jean-François Zagury, and Matthieu Montes. 2019. "Hits Discovery on the Androgen Receptor: In Silico Approaches to Identify Agonist Compounds" Cells 8, no. 11: 1431. https://doi.org/10.3390/cells8111431