Identification of Extrachromosomal Linear microDNAs Interacted with microRNAs in the Cell Nuclei
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Extrachromosomal Single-Stranded Linear DNAs and Related microRNAs
2.2. SSLmicroDNA Library Construction and Sequencing
2.3. Atomic Force Microscopy
2.4. Fluorescence in situ Hybridization
2.5. Pull-Down Assays and Quantitative Real-Time PCR
2.6. Cell Culture and Isolation of Nuclei
2.7. Sequence Analysis
2.8. Detection of the Endogenous Linear microDNAs
2.9. Transfection and Detection of Exogenous SSLmicroDNAs and Reverse Strands
3. Results
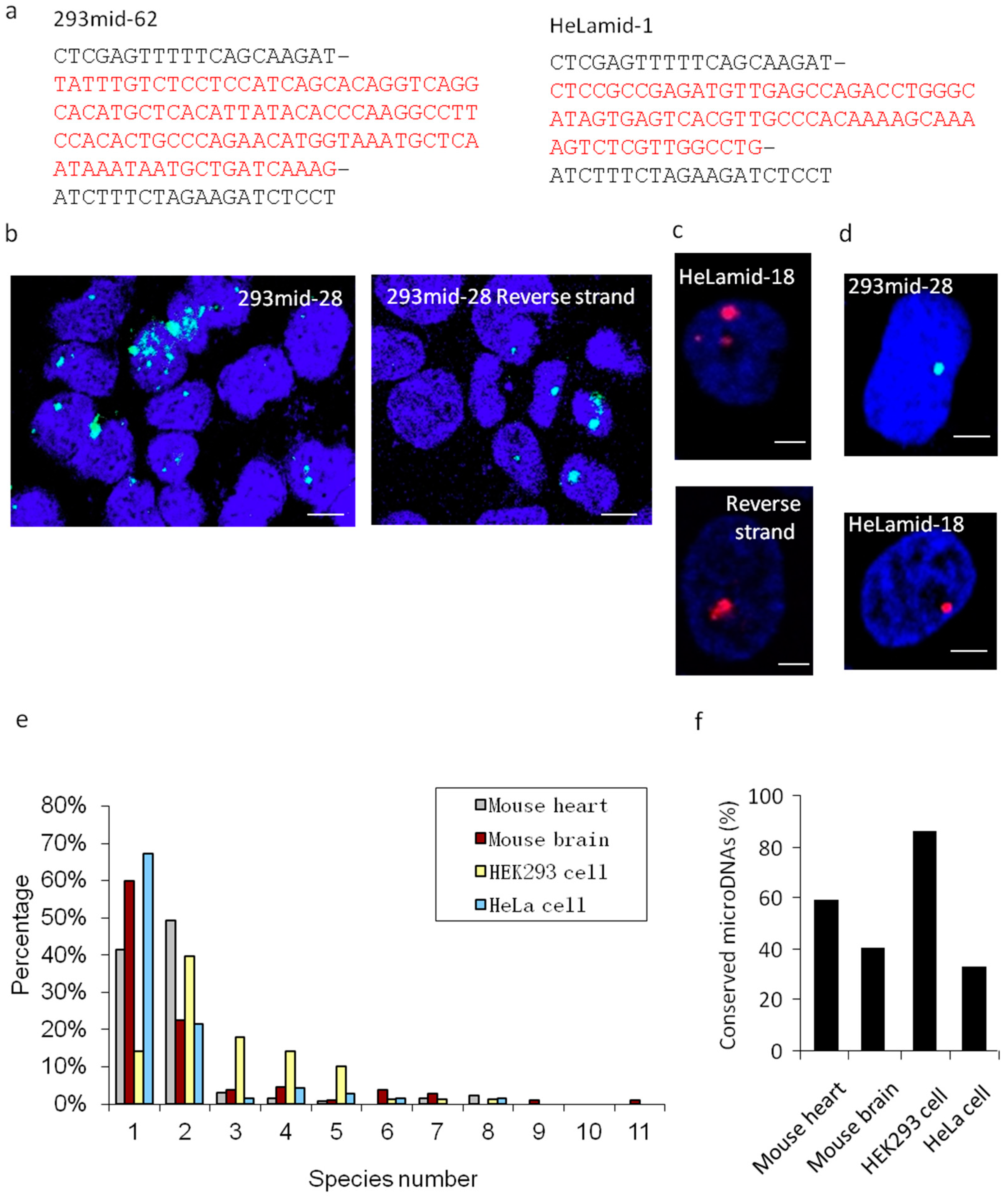
3.1. Small, Single-Stranded Linear Extrachromosomal DNAs (named SSLmicroDNAs) were Identified in Cell Nucleus
3.2. SSLmicroDNAs Exhibited a Series of Unique Features and Their Origins Hypothesis was Put Forward
3.3. Cell Nuclei- Located SSLmicroDNAs Owned a High Conservation Level
3.4. SSLmicroDNAs Interacted with microRNAs in the Cell Nucleus
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kuttler, F.; Mai, S. Formation of non-random extrachromosomal elements during development, differentiation and oncogenesis. Semin. Cancer Biol. 2007, 17, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Handa, H. Linear plasmids in plant mitochondria: Peaceful coexistences or malicious invasions? Mitochondrion 2008, 8, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, D.A.; Guarente, L. Extrachromosomal rDNA circles—A cause of aging in yeast. Cell 1997, 91, 1033–1042. [Google Scholar] [CrossRef]
- Huber, D.; Rustchenko, E. Large circular and linear rDNA plasmids in candida albicans. Yeast (Chichester Engl.) 2001, 18, 261–272. [Google Scholar] [CrossRef]
- Casjens, S.; Palmer, N.; van Vugt, R.; Huang, W.M.; Stevenson, B.; Rosa, P.; Lathigra, R.; Sutton, G.; Peterson, J.; Dodson, R.J.; et al. A bacterial genome in flux: The twelve linear and nine circular extrachromosomal dnas in an infectious isolate of the lyme disease spirochete borrelia burgdorferi. Mol. Microbiol. 2000, 35, 490–516. [Google Scholar] [CrossRef] [PubMed]
- Shadel, G.S.; Clayton, D.A. Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem. 1997, 66, 409–435. [Google Scholar] [CrossRef] [PubMed]
- Shibata, Y.; Kumar, P.; Layer, R.; Willcox, S.; Gagan, J.R.; Griffith, J.D.; Dutta, A. Extrachromosomal microdnas and chromosomal microdeletions in normal tissues. Science (New York NY) 2012, 336, 82–86. [Google Scholar] [CrossRef]
- Cox, D.; Yuncken, C.; Spriggs, A.I. Minute chromatin bodies in malignant tumours of childhood. Lancet (Lond. Engl.) 1965, 1, 55–58. [Google Scholar] [CrossRef]
- Hahn, P.J. Molecular biology of double-minute chromosomes. BioEssays 1993, 15, 477–484. [Google Scholar] [CrossRef]
- Shimizu, N.; Ochi, T.; Itonaga, K. Replication timing of amplified genetic regions relates to intranuclear localization but not to genetic activity or g/r band. Exp. Cell Res. 2001, 268, 201–210. [Google Scholar] [CrossRef]
- Turner, K.M.; Deshpande, V.; Beyter, D.; Koga, T.; Rusert, J.; Lee, C.; Li, B.; Arden, K.; Ren, B.; Nathanson, D.A.; et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature 2017, 543, 122–125. [Google Scholar] [CrossRef] [PubMed]
- Starling, S. Cancer genomics: Ecdetect hunts extrachromosomal DNA. Nat. Rev. Genet. 2017, 18, 212. [Google Scholar] [CrossRef] [PubMed]
- Dillon, L.W.; Kumar, P.; Shibata, Y.; Wang, Y.H.; Willcox, S.; Griffith, J.D.; Pommier, Y.; Takeda, S.; Dutta, A. Production of extrachromosomal microdnas is linked to mismatch repair pathways and transcriptional activity. Cell Rep. 2015, 11, 1749–1759. [Google Scholar] [CrossRef] [PubMed]
- Cara, A.; Reitz, M.S., Jr. New insight on the role of extrachromosomal retroviral DNA. Leukemia 1997, 11, 1395–1399. [Google Scholar] [CrossRef] [PubMed]
- Matuszewski, B.; Ciechomski, K.; Kloc, M. Extrachromosomal rDNA and polarity of pro-oocytes during ovary development in creophilus maxillosus (coleoptera, staphylinidae). Folia Histochem. Cytobiol. 1999, 37, 179–190. [Google Scholar] [PubMed]
- McDermott, P.; Connolly, V.; Kavanagh, T.A. The mitochondrial genome of a cytoplasmic male sterile line of perennial ryegrass (Lolium perenne L.) contains an integrated linear plasmid-like element. TAG. Theor. Appl. Genet. Theor. Angew. Genetik 2008, 117, 459–470. [Google Scholar] [CrossRef]
- Rose, K.; Fetzner, S. Identification of linear plasmid pam1 in the flavonoid degrading strain actinoplanes missouriensis(t) (dsm 43046). Plasmid 2006, 55, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Palmer, N.; Fraser, C.; Casjens, S. Distribution of twelve linear extrachromosomal dnas in natural isolates of lyme disease spirochetes. J. Bacteriol. 2000, 182, 2476–2480. [Google Scholar] [CrossRef]
- Sun, T.; Dong, Y.H.; Du, W.; Shi, C.Y.; Wang, K.; Tariq, M.A.; Wang, J.X.; Li, P.F. The role of micrornas in myocardial infarction: From molecular mechanism to clinical application. Int. J. Mol. Sci. 2017, 18, 745. [Google Scholar] [CrossRef]
- Wang, K.; Sun, T.; Li, N.; Wang, Y.; Wang, J.X.; Zhou, L.Y.; Long, B.; Liu, C.Y.; Liu, F.; Li, P.F. Mdrl lncrna regulates the processing of mir-484 primary transcript by targeting mir-361. PLoS Genet. 2014, 10, e1004467. [Google Scholar] [CrossRef]
- Thum, T.; Gross, C.; Fiedler, J.; Fischer, T.; Kissler, S.; Bussen, M.; Galuppo, P.; Just, S.; Rottbauer, W.; Frantz, S.; et al. Microrna-21 contributes to myocardial disease by stimulating map kinase signalling in fibroblasts. Nature 2008, 456, 980–984. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. Micrornas: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Zhang, X.; Li, H.; Burnett, J.C.; Rossi, J.J. The role of antisense long noncoding rna in small rna-triggered gene activation. RNA (New York NY) 2014, 20, 1916–1928. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Saetrom, P.; Snove, O., Jr.; Rossi, J.J. Microrna-directed transcriptional gene silencing in mammalian cells. Proc. Natl. Acad. Sci. USA 2008, 105, 16230–16235. [Google Scholar] [CrossRef] [PubMed]
- Kalsotra, A.; Wang, K.; Li, P.F.; Cooper, T.A. Micrornas coordinate an alternative splicing network during mouse postnatal heart development. Genes Dev. 2010, 24, 653–658. [Google Scholar] [CrossRef] [PubMed]
- Allo, M.; Buggiano, V.; Fededa, J.P.; Petrillo, E.; Schor, I.; de la Mata, M.; Agirre, E.; Plass, M.; Eyras, E.; Elela, S.A.; et al. Control of alternative splicing through sirna-mediated transcriptional gene silencing. Nat. Struct. Mol. Biol. 2009, 16, 717–724. [Google Scholar] [CrossRef] [PubMed]
- Zisoulis, D.G.; Kai, Z.S.; Chang, R.K.; Pasquinelli, A.E. Autoregulation of microrna biogenesis by let-7 and argonaute. Nature 2012, 486, 541–544. [Google Scholar] [CrossRef]
- Faas, F.G.; Rieger, B.; van Vliet, L.J.; Cherny, D.I. DNA deformations near charged surfaces: Electron and atomic force microscopy views. Biophys. J. 2009, 97, 1148–1157. [Google Scholar] [CrossRef]
- Rowley, J.D.; Diaz, M.O.; Espinosa, R., 3rd; Patel, Y.D.; van Melle, E.; Ziemin, S.; Taillon-Miller, P.; Lichter, P.; Evans, G.A.; Kersey, J.H.; et al. Mapping chromosome band 11q23 in human acute leukemia with biotinylated probes: Identification of 11q23 translocation breakpoints with a yeast artificial chromosome. Proc. Natl. Acad. Sci. USA 1990, 87, 9358–9362. [Google Scholar] [CrossRef]
- Fu, H.; Le, S.; Chen, H.; Muniyappa, K.; Yan, J. Force and atp hydrolysis dependent regulation of reca nucleoprotein filament by single-stranded DNA binding protein. Nucleic Acids Res. 2013, 41, 924–932. [Google Scholar] [CrossRef]
- Ha, T.; Kozlov, A.G.; Lohman, T.M. Single-molecule views of protein movement on single-stranded DNA. Annu. Rev. Biophys. 2012, 41, 295–319. [Google Scholar] [CrossRef] [PubMed]
- Segal, E.; Fondufe-Mittendorf, Y.; Chen, L.; Thastrom, A.; Field, Y.; Moore, I.K.; Wang, J.P.; Widom, J. A genomic code for nucleosome positioning. Nature 2006, 442, 772–778. [Google Scholar] [CrossRef] [PubMed]
- Gaasenbeek, M.; Howarth, K.; Rowan, A.J.; Gorman, P.A.; Jones, A.; Chaplin, T.; Liu, Y.; Bicknell, D.; Davison, E.J.; Fiegler, H.; et al. Combined array-comparative genomic hybridization and single-nucleotide polymorphism-loss of heterozygosity analysis reveals complex changes and multiple forms of chromosomal instability in colorectal cancers. Cancer Res. 2006, 66, 3471–3479. [Google Scholar] [CrossRef] [PubMed]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone h3k27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 2010, 107, 21931–21936. [Google Scholar] [CrossRef] [PubMed]
- Johnnidis, J.B.; Harris, M.H.; Wheeler, R.T.; Stehling-Sun, S.; Lam, M.H.; Kirak, O.; Brummelkamp, T.R.; Fleming, M.D.; Camargo, F.D. Regulation of progenitor cell proliferation and granulocyte function by microrna-223. Nature 2008, 451, 1125–1129. [Google Scholar] [CrossRef] [PubMed]
- Sloan, R.D.; Wainberg, M.A. The role of unintegrated DNA in HIV infection. Retrovirology 2011, 8, 52. [Google Scholar] [CrossRef] [PubMed]
- Schnepp, B.C.; Jensen, R.L.; Chen, C.L.; Johnson, P.R.; Clark, K.R. Characterization of adeno-associated virus genomes isolated from human tissues. J. Virol. 2005, 79, 14793–14803. [Google Scholar] [CrossRef] [PubMed]
- Schneider, W.M.; Wu, D.T.; Amin, V.; Aiyer, S.; Roth, M.J. Mulv in mutants responsive to hdac inhibitors enhance transcription from unintegrated retroviral DNA. Virology 2012, 426, 188–196. [Google Scholar] [CrossRef]
- Weiss, G.; Schaible, U.E. Macrophage defense mechanisms against intracellular bacteria. Immunol. Rev. 2015, 264, 182–203. [Google Scholar] [CrossRef]
- Lecellier, C.H.; Dunoyer, P.; Arar, K.; Lehmann-Che, J.; Eyquem, S.; Himber, C.; Saib, A.; Voinnet, O. A cellular microrna mediates antiviral defense in human cells. Science (New York NY) 2005, 308, 557–560. [Google Scholar] [CrossRef]
- Stingele, J.; Schwarz, M.S.; Bloemeke, N.; Wolf, P.G.; Jentsch, S. A DNA-dependent protease involved in DNA-protein crosslink repair. Cell 2014, 158, 327–338. [Google Scholar] [CrossRef] [PubMed]






| The Number of Clones | The Number of Sequenced Clones | The Number of Clones Containing Sequences | The Number of Clones Containing SSLmicroDNAs | Rate | |
|---|---|---|---|---|---|
| Mouse Hearts | 3140 | 470 | 165 | 128 | 27.23% |
| Mouse Brains | 3692 | 966 | 274 | 107 | 11.08% |
| HEK293 Cells | 4900 | 702 | 642 | 78 | 12.15% |
| HeLa Cells | 4700 | 2020 | 331 | 70 | 3.47% |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, T.; Wang, K.; Liu, C.; Wang, Y.; Wang, J.; Li, P. Identification of Extrachromosomal Linear microDNAs Interacted with microRNAs in the Cell Nuclei. Cells 2019, 8, 111. https://doi.org/10.3390/cells8020111
Sun T, Wang K, Liu C, Wang Y, Wang J, Li P. Identification of Extrachromosomal Linear microDNAs Interacted with microRNAs in the Cell Nuclei. Cells. 2019; 8(2):111. https://doi.org/10.3390/cells8020111
Chicago/Turabian StyleSun, Teng, Kun Wang, Cuiyun Liu, Yin Wang, Jianxun Wang, and Peifeng Li. 2019. "Identification of Extrachromosomal Linear microDNAs Interacted with microRNAs in the Cell Nuclei" Cells 8, no. 2: 111. https://doi.org/10.3390/cells8020111




