Nucleolar and Ribosomal DNA Structure under Stress: Yeast Lessons for Aging and Cancer
Abstract
:1. Introduction
2. The Nucleolus as a Marker of Cancer and Aging in Metazoans
3. The Structure of the Nucleolus and the rDNA in the Yeast Saccharomyces cerevisiae
Morphological Changes of the Yeast Nucleolus during the Cell Cycle
4. Key Facts about Yeast rDNA Transcription and Ribosome Production
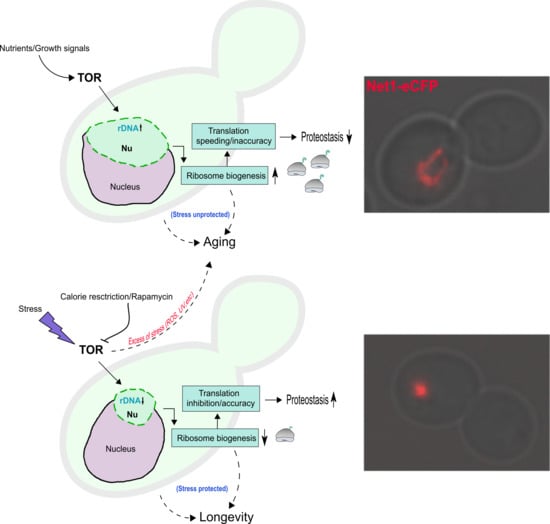
5. TORC1, Stress and the Size of the Nucleolus
TORC1, Condensin, Epigenetics and the rDNA Chromatin Compaction
6. Nucleolar Stress Remodelling and p53 Stabilization in Cancer
7. Future Perspectives in Nucleolar Stress and Health: From Yeast to Humans
Author Contributions
Funding
Conflicts of Interest
References
- Fontana, F. Traité sur le vénin de la Vipere, sur les Poisons Americains, sur le Laurier-Cerise et sur Quelques Autres Poisons Vegetaux. On y a Joint des Observations sur la Structure Primitive du Corps Animal; Florence, 1781. Available online: https://archive.org/details/traitsurlevn01font/page/n6 (accessed on 24 July 2019).
- McClintock, B. The relation of a particular chromosomal element to the development of the nucleoli in Zea mays. Z. Zellforsch. Mikrosk. Anat. 1934, 21, 294–326. [Google Scholar] [CrossRef]
- Woolford, J.L.; Baserga, S.J. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 2013, 195, 643–681. [Google Scholar] [CrossRef] [PubMed]
- Boisvert, F.M.; van Koningsbruggen, S.; Navascués, J.; Lamond, A.I. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007, 8, 574–585. [Google Scholar] [CrossRef] [PubMed]
- Lindström, M.S.; Jurada, D.; Bursac, S.; Orsolic, I.; Bartek, J.; Volarevic, S. Nucleolus as an emerging hub in maintenance of genome stability and cancer pathogenesis. Oncogene 2018, 37, 2351–2366. [Google Scholar] [CrossRef] [PubMed]
- Mayer, C.; Bierhoff, H.; Grummt, I. The nucleolus as a stress sensor: JNK2 inactivates the transcription factor TIF-IA and down-regulates rRNA synthesis. Genes Dev. 2005, 19, 933–941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boulon, S.; Westman, B.J.; Hutten, S.; Boisvert, F.M.; Lamond, A.I. The Nucleolus under Stress. Mol. Cell 2010, 40, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Grummt, I. The nucleolus—Guardian of cellular homeostasis and genome integrity. Chromosoma 2013, 122, 487–497. [Google Scholar] [CrossRef]
- Pederson, T. The plurifunctional nucleolus. Nucleic Acids Res. 1998, 26, 3871–3876. [Google Scholar] [CrossRef] [Green Version]
- Thiry, M.; Lafontaine, D.L. Birth of a nucleolus: The evolution of nucleolar compartments. Trends Cell Biol. 2005, 15, 194–199. [Google Scholar] [CrossRef]
- Hernandez-Verdun, D.; Roussel, P.; Thiry, M.; Sirri, V.; Lafontaine, D.L. The nucleolus: Structure/function relationship in RNA metabolism. Wiley Interdiscip. Rev. RNA 2010, 1, 415–431. [Google Scholar] [CrossRef]
- McDonald, R.B. Biology of Aging, 1st ed.; Garland Science; Taylor & Francis Group, LLC: New York, NY, USA, 2014. [Google Scholar]
- Bitterman, K.J.; Medvedik, O.; Sinclair, D.A. Longevity Regulation in Saccharomyces cerevisiae: Linking Metabolism, Genome Stability, and Heterochromatin. Microbiol. Mol. Biol. Rev. 2003, 67, 376–399. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Zhou, C.; Kennedy, B.K. The yeast replicative aging model. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 2690–2696. [Google Scholar] [CrossRef] [PubMed]
- Longo, V.D.; Shadel, G.S.; Kaeberlein, M.; Kennedy, B. Replicative and Chronological Aging in Saccharomyces cerevisiae. Cell Metab. 2012, 16, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Stenram, U. Nucleolar Size in the Liver of Rats Fed on High and Nonprotein Diets After Starvation. Cells Tissues Organs 1956, 26, 352–361. [Google Scholar] [CrossRef]
- Stenram, U. Nucleolar Size in the Liver of Rats Fed Diets Deficient in Essential Amino Acids. Acta Pathol. Microbiol. Scand. 1956, 38, 364–374. [Google Scholar] [CrossRef] [PubMed]
- Derenzini, M.; Trerè, D.; Pession, A.; Govoni, M.; Sirri, V.; Chieco, P. Nucleolar size indicates the rapidity of cell proliferation in cancer tissues. J. Pathol. 2000, 191, 181–186. [Google Scholar] [CrossRef]
- Pianese, G. Beitrag zur Histologie und Aetiologie der Carcinoma. Histologische und experimentelle Untersuchungen. Beitr. Pathol. Anat. Allg. Pathol. 1896, 142, 1–193. [Google Scholar]
- Delahunt, B.; Sika-Paotonu, D.; Bethwaite, P.B.; William Jordan, T.; Magi-Galluzzi, C.; Zhou, M.; Samaratunga, H.; Srigley, J.R. Grading of clear cell renal cell carcinoma should be based on nucleolar prominence. Am. J. Surg. Pathol. 2011, 35, 1134–1139. [Google Scholar] [CrossRef] [PubMed]
- Moch, H.; Cubilla, A.L.; Humphrey, P.A.; Reuter, V.E.; Ulbright, T.M. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs—Part A: Renal, Penile, and Testicular Tumours. Eur. Urol. 2016, 70, 93–105. [Google Scholar] [CrossRef]
- Kaneko, S.; Ishida, T.; Sugio, K.; Yokoyama, H.; Sugimachi, K. Nucleolar Organizer Regions as a Prognostic Indicator for Stage I Non-Small Cell Lung Cancer. Cancer Res. 1991, 51, 4008–4011. [Google Scholar]
- Pich, A.; Chiusa, L.; Margaria, E. Prognostic relevance of AgNORs in tumor pathology. Micron 2000, 31, 133–141. [Google Scholar] [CrossRef]
- Derenzini, M. The AgNORs. Micron 2000, 31, 117–120. [Google Scholar] [CrossRef]
- Donizy, P.; Biecek, P.; Halon, A.; Maciejczyk, A.; Matkowski, R. Nucleoli cytomorphology in cutaneous melanoma cells—A new prognostic approach to an old concept. Diagn. Pathol. 2017, 12, 88. [Google Scholar] [CrossRef] [PubMed]
- Ruggero, D.; Pandolfi, P.P. Does the ribosome translate cancer? Nat. Rev. Cancer 2003, 3, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Montanaro, L.; Treré, D.; Derenzini, M. Nucleolus, ribosomes, and cancer. Am. J. Pathol. 2008, 173, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Ruggero, D. Revisiting the nucleolus: From marker to dynamic integrator of cancer signaling. Sci. Signal 2012, 5, pe38. [Google Scholar] [CrossRef] [PubMed]
- Zlotorynski, E. Ageing: Live longer with small nucleoli. Nat. Rev. Mol. Cell Biol. 2017, 18, 651. [Google Scholar] [CrossRef]
- Tiku, V.; Jain, C.; Raz, Y.; Nakamura, S.; Heestand, B.; Liu, W.; Späth, M.; Suchiman, H.E.D.; Müller, R.U.; Slagboom, P.E.; et al. Small nucleoli are a cellular hallmark of longevity. Nat. Commun. 2016, 8, 16083. [Google Scholar] [CrossRef]
- Buchwalter, A.; Hetzer, M.W. Nucleolar expansion and elevated protein translation in premature aging. Nat. Commun. 2017, 8, 328. [Google Scholar] [CrossRef]
- Sinclair, D.A.; Mills, K.; Guarente, L. Accelerated aging and nucleolar fragmentation in yeast SGS1 mutants. Science 1997, 277, 1313–1316. [Google Scholar] [CrossRef]
- Lewinska, A.; Miedziak, B.; Kulak, K.; Molon, M.; Wnuk, M. Links between nucleolar activity, rDNA stability, aneuploidy and chronological aging in the yeast Saccharomyces cerevisiae. Biogerontology 2014, 15, 289–316. [Google Scholar] [CrossRef] [PubMed]
- Bemiller, P.M.; Lee, L.H. Nucleolar changes in senescing WI-38 cells. Mech. Ageing Dev. 1978, 8, 417–427. [Google Scholar] [CrossRef]
- Tiku, V.; Antebi, A. Nucleolar Function in Lifespan Regulation. Trends Cell Biol. 2018, 28, 662–672. [Google Scholar] [CrossRef] [PubMed]
- Venema, J.; Tollervey, D. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 1999, 33, 261–311. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T. Ribosomal RNA gene repeats, their stability and cellular senescence. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2014, 90, 119–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turowski, T.W.; Tollervey, D. Cotranscriptional events in eukaryotic ribosome synthesis. Wiley Interdiscip. Rev. RNA 2015, 6, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Toussaint, M.; Levasseur, G.; Tremblay, M.; Paquette, M.; Conconi, A. Psoralen photocrosslinking, a tool to study the chromatin structure of RNA polymerase I-transcribed ribosomal genes. Biochem. Cell Biol. 2005, 83, 449–459. [Google Scholar] [CrossRef]
- Kobayashi, T.; Heck, D.J.; Nomura, M.; Horiuchi, T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: Requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998, 12, 3821–3830. [Google Scholar] [CrossRef]
- Kobayashi, T.; Horiuchi, T.; Tongaonkar, P.; Vu, L.; Nomura, M. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell 2004, 117, 441–453. [Google Scholar] [CrossRef]
- Kobayashi, T.; Ganley, A.R.D. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science 2005, 309, 1581–1584. [Google Scholar] [CrossRef]
- Houseley, J.; Tollervey, D. Repeat expansion in the budding yeast ribosomal DNA can occur independently of the canonical homologous recombination machinery. Nucleic Acids Res. 2011, 39, 8778–8791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jack, C.V.; Cruz, C.; Hull, R.M.; Keller, M.A.; Ralser, M.; Houseley, J. Regulation of ribosomal DNA amplification by the TOR pathway. Proc. Natl. Acad. Sci. USA 2015, 112, 9674–9679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iida, T.; Kobayashi, T. RNA Polymerase I Activators Count and Adjust Ribosomal RNA Gene Copy Number. Mol. Cell 2019, 73, 645–654.e13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinclair, D.A.; Guarente, L. Extrachromosomal rDNA circles—A cause of aging in yeast. Cell 1997, 91, 1033–1042. [Google Scholar] [CrossRef]
- Mekhail, K.; Seebacher, J.; Gygi, S.P.; Moazed, D. Role for perinuclear chromosome tethering in maintenance of genome stability. Nature 2008, 456, 667–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mekhail, K.; Moazed, D. The nuclear envelope in genome organization, expression and stability. Nat. Rev. Mol. Cell Biol. 2010, 11, 317–328. [Google Scholar] [CrossRef] [Green Version]
- Taddei, A.; Gasser, S.M. Structure and function in the budding yeast nucleus. Genetics 2012, 192, 107–129. [Google Scholar] [CrossRef]
- Torres-Rosell, J.; Sunjevaric, I.; de Piccoli, G.; Sacher, M.; Eckert-Boulet, N.; Reid, R.; Jentsch, S.; Rothstein, R.; Aragón, L.; Lisby, M. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat. Cell Biol. 2007, 9, 923–931. [Google Scholar] [CrossRef]
- Kobayashi, T.; Sasaki, M. Ribosomal DNA stability is supported by many ’buffer genes’—introduction to the Yeast rDNA Stability Database. FEMS Yeast Res. 2017, 17. [Google Scholar] [CrossRef]
- Campbell, J.L.; Lorenz, A.; Witkin, K.L.; Hays, T.; Loidl, J.; Cohen-Fix, O. Yeast Nuclear Envelope Subdomains with Distinct Abilities to Resist Membrane Expansion. Mol. Biol. Cell. 2006, 17, 1768–1778. [Google Scholar] [CrossRef] [Green Version]
- Marko, J.F. The liquid drop nature of nucleoli. Nucleus 2012, 3, 115–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miné-Hattab, J.; Taddei, A. Physical principles and functional consequences of nuclear compartmentalization in budding yeast. Curr. Opin. Cell Biol. 2019, 58, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Feric, M.; Vaidya, N.; Harmon, T.S.; Mitrea, D.M.; Zhu, L.; Richardson, T.M.; Kriwacki, R.W.; Pappu, R.V.; Brangwynne, C.P. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell 2016, 165, 1686–1697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hult, C.; Adalsteinsson, D.; Vasquez, P.A.; Lawrimore, J.; Bennett, M.; York, A.; Cook, D.; Yeh, E.; Forest, M.G.; Bloom, K. Enrichment of dynamic chromosomal crosslinks drive phase separation of the nucleolus. Nucleic Acids Res. 2017, 45, 11159–11173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guacci, V.; Hogan, E.; Koshland, D. Chromosome condesation and sister chromatid pairing in budding yeast. J. Cell Biol. 1994, 125, 517–530. [Google Scholar] [CrossRef] [PubMed]
- Lavoie, B.D.; Hogan, E.; Koshland, D. In vivo requirements for rDNA chromosome condensation reveal two cell-cycle-regulated pathways for mitotic chromosome folding. Genes Dev. 2004, 18, 76–87. [Google Scholar] [CrossRef]
- Lavoie, B.D.; Hogan, E.; Koshland, D. In vivo dissection of the chromosome condensation machinery: Reversibility of condensation distinguishes contributions of condensin and cohesin. J. Cell. Biol. 2002, 156, 805–815. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, J.; Loidl, J. Behaviour of nucleolus organizing regions (NORs) and nucleoli during mitotic and meiotic divisions in budding yeast. Chromosome Res. 2004, 12, 427–438. [Google Scholar] [CrossRef]
- Machín, F.; Torres-Rosell, J.; Jarmuz, A.; Aragón, L. Spindle-independent condensation-mediated segregation of yeast ribosomal DNA in late anaphase. J. Cell Biol. 2005, 168, 209–219. [Google Scholar] [CrossRef] [Green Version]
- Miyazaki, T.; Kobayashi, T. Visualization of the dynamic behavior of ribosomal RNA gene repeats in living yeast cells. Genes Cells 2011, 16, 491–502. [Google Scholar] [CrossRef]
- Shen, D.; Skibbens, R.V. Temperature-dependent regulation of rDNA condensation in Saccharomyces cerevisiae. Cell Cycle 2017, 16, 1118–1127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, Y.; Acar, M. Live-Cell Imaging of Chromatin Condensation Dynamics by CRISPR. iScience 2018, 4, 216–235. [Google Scholar] [CrossRef] [PubMed]
- Freeman, L.; Aragon-Alcaide, L.; Strunnikov, A. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J. Cell Biol. 2000, 149, 811–824. [Google Scholar] [CrossRef] [PubMed]
- Lavoie, B.D.; Tuffo, K.M.; Oh, S.; Koshland, D.; Holm, C. Mitotic Chromosome Condensation Requires Brn1p, the Yeast Homologue of Barren. Mol. Biol. Cell. 2000, 11, 1293–1304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, W.W.; Peterson, E.A.; Yeung, M.; Lavoie, B.D. Condensin is required for chromosome arm cohesion during mitosis. Genes Dev. 2006, 20, 2973–2984. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, M.; Higuchi, T.; Katis, V.L.; Uhlmann, F. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell 2004, 117, 471–482. [Google Scholar] [CrossRef]
- D’Amours, D.; Stegmeier, F.; Amon, A. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell 2004, 117, 455–469. [Google Scholar] [CrossRef]
- Torres-Rosell, J.; Machín, F.; Jarmuz, A.; Aragón, L. Nucleolar segregation lags behind the rest of the genome and requires Cdc14 phosphatase activation by the FEAR network. Cell Cycle 2004, 3, 496–502. [Google Scholar] [CrossRef]
- Varela, E.; Shimada, K.; Laroche, T.; Leroy, D.; Gasser, S.M. Lte1, Cdc14 and MEN-controlled Cdk inactivation in yeast coordinate rDNA decompaction with late telophase progression. EMBO J. 2009, 28, 1562–1575. [Google Scholar] [CrossRef] [Green Version]
- St-Pierre, J.; Douziech, M.; Bazile, F.; Pascariu, M.; Bonneil, É.; Sauvé, V.; Ratsima, H.; D’Amours, D. Polo Kinase Regulates Mitotic Chromosome Condensation by Hyperactivation of Condensin DNA Supercoiling Activity. Mol. Cell 2009, 34, 416–426. [Google Scholar] [CrossRef]
- Miller, O.L.; Beatty, B.R. Visualization of nucleolar genes. Science 1969, 164, 955–957. [Google Scholar] [CrossRef] [PubMed]
- Laird, C.D.; Chooi, W.Y. Morphology of transcription units in Drosophila melanogaster. Chromosoma 1976, 58, 193–218. [Google Scholar] [CrossRef] [PubMed]
- Mougey, E.B.; O’Reilly, M.; Osheim, Y.; Miller, O.L.; Beyer, A.; Sollner-Webb, B. The terminal balls characteristic of eukaryotic rRNA transcription units in chromatin spreads are rRNA processing complexes. Genes Dev. 1993, 7, 1609–1619. [Google Scholar] [CrossRef] [PubMed]
- Oakes, M.; Aris, J.P.; Brockenbrough, J.S.; Wai, H.; Vu, L.; Nomura, M. Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae. J. Cell Biol. 1998, 143, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Trumtel, S.; Leger-Silvestre, I.; Gleizes, P.E.; Teulieres, F.; Gas, N. Assembly and Functional Organization of the Nucleolus: Ultrastructural Analysis of Saccharomyces cerevisiae Mutants. Mol. Biol. Cell 2000, 11, 2175–2189. [Google Scholar] [CrossRef]
- Raška, I. Oldies but goldies: Searching for Christmas trees within the nucleolar architecture. Trends Cell Biol. 2003, 13, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Granneman, S.; Baserga, S.J. Crosstalk in gene expression: Coupling and co-regulation of rDNA transcription, pre-ribosome assembly and pre-rRNA processing. Curr. Opin. Cell Biol. 2005, 17, 281–286. [Google Scholar] [CrossRef]
- Koš, M.; Tollervey, D. Yeast Pre-rRNA Processing and Modification Occur Cotranscriptionally. Mol. Cell 2010, 37, 809–820. [Google Scholar] [CrossRef]
- Warner, J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999, 24, 437–440. [Google Scholar] [CrossRef]
- Warner, J.R.; Vilardell, J.; Sohn, J.H. Economics of ribosome biosynthesis. Cold Spring Harb. Symp. Quant. Biol. 2001, 66, 567–574. [Google Scholar] [CrossRef]
- Oakes, M.; Siddiqi, I.; Vu, L.; Aris, J.; Nomura, M. Transcription Factor UAF, Expansion and Contraction of Ribosomal DNA (rDNA) Repeats, and RNA Polymerase Switch in Transcription of Yeast rDNA. Mol. Cell. Biol. 1999, 19, 8559–8569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kief, D.R.; Warner, J.R. Coordinate control of syntheses of ribosomal ribonucleic acid and ribosomal proteins during nutritional shift-up in Saccharomyces cerevisiae. Mol. Cell. Biol. 2015, 1, 1007–1015. [Google Scholar] [CrossRef] [PubMed]
- Kief, D.R.; Warner, J.R. Hierarchy of elements regulating synthesis of ribosomal proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 2015, 1, 1016–1023. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, P.; Nishikawa, J.L.; Breitkreutz, B.J.; Tyers, M. Systematic identification of pathways that couple cell growth and division in yeast. Science 2002, 297, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, P.; Rupeš, I.; Sharom, J.R.; Schneper, L.; Broach, J.R.; Tyers, M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004, 18, 2491–2505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albert, B.; Tomassetti, S.; Gloor, Y.; Dilg, D.; Mattarocci, S.; Kubik, S.; Hafner, L.; Shore, D. Sfp1 regulates transcriptional networks driving cell growth and division through multiple promoter-binding modes. Genes Dev. 2019, 33, 288–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weston, L.; Greenwood, J.; Nurse, P. Genome-wide screen for cell growth regulators in fission yeast. J. Cell Sci. 2017, 130, 2049–2055. [Google Scholar] [CrossRef] [Green Version]
- Marion, R.M.; Regev, A.; Segal, E.; Barash, Y.; Koller, D.; Friedman, N.; O’Shea, E.K. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc. Natl. Acad. Sci. USA 2004, 101, 14315–14322. [Google Scholar] [CrossRef] [Green Version]
- Urban, J.; Soulard, A.; Huber, A.; Lippman, S.; Mukhopadhyay, D.; Deloche, O.; Wanke, V.; Anrather, D.; Ammerer, G.; Riezman, H.; et al. Sch9 Is a Major Target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 2007, 26, 663–674. [Google Scholar] [CrossRef]
- Lempiäinen, H.; Uotila, A.; Urban, J.; Dohnal, I.; Ammerer, G.; Loewith, R.; Shore, D. Sfp1 Interaction with TORC1 and Mrs6 Reveals Feedback Regulation on TOR Signaling. Mol. Cell 2009, 33, 704–716. [Google Scholar] [CrossRef] [Green Version]
- Polymenis, M.; Schmidt, E.V. Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes Dev. 1997, 11, 2522–2531. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.E.; Soulard, A.; Hall, M.N. TOR regulates ribosomal protein gene expression via PKA and the Forkhead Transcription Factor FHL1. Cell 2004, 119, 969–979. [Google Scholar] [CrossRef] [PubMed]
- Loewith, R.; Hall, M.N. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 2011, 189, 1177–1201. [Google Scholar] [CrossRef] [PubMed]
- Jacinto, E.; Loewith, R.; Schmidt, A.; Lin, S.; Rüegg, M.A.; Hall, A.; Hall, M.N. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 2004, 6, 1122–1128. [Google Scholar] [CrossRef] [PubMed]
- Rispal, D.; Eltschinger, S.; Stahl, M.; Vaga, S.; Bodenmiller, B.; Abraham, Y.; Filipuzzi, I.; Movva, N.R.; Aebersold, R.; Helliwell, S.B.; et al. Target of rapamycin complex 2 regulates actin polarization and endocytosis via multiple pathways. J. Biol. Chem. 2015, 290, 14963–14978. [Google Scholar] [CrossRef] [PubMed]
- Crespo, J.L.; Hall, M.N. Elucidating TOR Signaling and Rapamycin Action: Lessons from Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2002, 66, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Loewith, R.; Jacinto, E.; Wullschleger, S.; Lorberg, A.; Crespo, J.L.; Bonenfant, D.; Oppliger, W.; Jenoe, P.; Hall, M.N. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 2002, 10, 457–468. [Google Scholar] [CrossRef]
- Reinke, A.; Anderson, S.; McCaffery, J.M.; Yates, J.; Aronova, S.; Chu, S.; Fairclough, S.; Iverson, C.; Wedaman, K.P.; Powers, T. TOR Complex 1 Includes a Novel Component, Tco89p (YPL180w), and Cooperates with Ssd1p to Maintain Cellular Integrity in Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 14752–14762. [Google Scholar] [CrossRef] [Green Version]
- Zurita-Martinez, S.A.; Puria, R.; Pan, X.; Boeke, J.D.; Cardenas, M.E. Efficient Tor signaling requires a functional class C Vps protein complex in Saccharomyces cerevisiae. Genetics 2007, 176, 2139–2150. [Google Scholar] [CrossRef]
- Martin, D.E.; Powers, T.; Hall, M.N. Regulation of ribosome biogenesis: Where is TOR? Cell Metab. 2006, 4, 259–260. [Google Scholar] [CrossRef] [Green Version]
- Mayer, C.; Grummt, I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 2006, 25, 6384–6391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powers, T.; Walter, P. Regulation of Ribosome Biogenesis by the Rapamycin-sensitive TOR-signaling Pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 1999, 10, 987–1000. [Google Scholar] [CrossRef] [PubMed]
- Berger, A.B.; Decourty, L.; Badis, G.; Nehrbass, U.; Jacquier, A.; Gadal, O. Hmo1 Is Required for TOR-Dependent Regulation of Ribosomal Protein Gene Transcription. Mol. Cell. Biol. 2007, 27, 8015–8026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Y.; Zheng, X.F. Sch9 partially mediates TORC1 signaling to control ribosomal RNA synthesis. Cell Cycle 2009, 8, 4085–4090. [Google Scholar] [CrossRef] [PubMed]
- Laferté, A.; Favry, E.; Sentenac, A.; Riva, M.; Carles, C.; Chédin, S. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev. 2006, 20, 2030–2040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chédin, S.; Laferté, A.; Hoang, T.; Lafontaine, D.L.; Riva, M.; Carles, C. Is ribosome synthesis controlled by Pol I transcription? Cell Cycle 2007, 6, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Lempiäinen, H.; Shore, D. Growth control and ribosome biogenesis. Curr. Opin. Cell. Biol. 2009, 21, 855–863. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Tsang, C.K.; Watkins, M.; Bertram, P.G.; Zheng, X.F. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature 2006, 442, 1058–1061. [Google Scholar] [CrossRef]
- Claypool, J.A. Tor Pathway Regulates Rrn3p-dependent Recruitment of Yeast RNA Polymerase I to the Promoter but Does Not Participate in Alteration of the Number of Active Genes. Mol. Biol. Cell 2003, 15, 946–956. [Google Scholar] [CrossRef]
- Philippi, A.; Steinbauer, R.; Reiter, A.; Fath, S.; Leger-Silvestre, I.; Milkereit, P.; Griesenbeck, J.; Tschochner, H. TOR-dependent reduction in the expression level of Rrn3p lowers the activity of the yeast RNA Pol I machinery, but does not account for the strong inhibition of rRNA production. Nucleic Acids Res. 2010, 38, 5315–5326. [Google Scholar] [CrossRef] [Green Version]
- Blattner, C.; Jennebach, S.; Herzog, F.; Mayer, A.; Cheung, A.C.; Witte, G.; Lorenzen, K.; Hopfner, K.P.; Heck, A.J.; Aebersold, R.; et al. Molecular basis of Rrn3-regulated RNA polymerase I initiation and cell growth. Genes Dev. 2011, 25, 2093–2105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Betz, C.; Hall, M.N. Where is mTOR and what is it doing there? J. Cell Biol. 2013, 203, 563–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Y.; Tsang, C.K.; Zheng, X.F. Mechanisms of regulation of RNA polymerase III-dependent transcription by TORC1. EMBO J. 2009, 28, 2220–2230. [Google Scholar] [CrossRef] [PubMed]
- Filer, D.; Thompson, M.A.; Takhaveev, V.; Dobson, A.J.; Kotronaki, I.; Green, J.W.; Heinemann, M.; Tullet, J.M.; Alic, N. RNA polymerase III limits longevity downstream of TORC1. Nature 2017, 552, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Barbet, N.C.; Schneider, U.; Helliwell, S.B.; Stansfield, I.; Tuite, M.F.; Hall, M.N. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 1996, 7, 25–42. [Google Scholar] [CrossRef]
- Hara, K.; Yonezawa, K.; Kozlowski, M.T.; Sugimoto, T.; Andrabi, K.; Weng, Q.P.; Kasuga, M.; Nishimoto, I.; Avruch, J. Regulation of eIF-4E BP1 phosphorylation by mTOR. J. Biol. Chem. 1997, 272, 26457–26463. [Google Scholar] [CrossRef] [PubMed]
- Hara, K.; Yonezawa, K.; Weng, Q.P.; Kozlowski, M.T.; Belham, C.; Avruch, J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 1998, 273, 14484–14494. [Google Scholar] [CrossRef]
- Berset, C.; Trachsel, H.; Altmann, M. The TOR (target of rapamycin) signal transduction pathway regulates the stability of translation initiation factor eIF4G in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2002, 95, 4264–4269. [Google Scholar] [CrossRef]
- Thoreen, C.C.; Chantranupong, L.; Keys, H.R.; Wang, T.; Gray, N.S.; Sabatini, D.M. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 2012, 485, 109–113. [Google Scholar] [CrossRef]
- Xie, J.; de Souza Alves, V.; von der Haar, T.; O’Keefe, L.; Lenchine, R.V.; Jensen, K.B.; Liu, R.; Coldwell, M.J.; Wang, X.; Proud, C.G. Regulation of the Elongation Phase of Protein Synthesis Enhances Translation Accuracy and Modulates Lifespan. Curr. Biol. 2019, 29, 737–749.e5. [Google Scholar] [CrossRef] [Green Version]
- Hansen, M.; Taubert, S.; Crawford, D.; Libina, N.; Lee, S.J.; Kenyon, C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell 2007, 6, 95–110. [Google Scholar] [CrossRef] [PubMed]
- Stout, G.J.; Stigter, E.C.; Essers, P.B.; Mulder, K.W.; Kolkman, A.; Snijders, D.S.; van Den Broek, N.J.; Betist, M.C.; Korswagen, H.C.; MacInnes, A.W.; et al. Insulin/IGF-1-mediated longevity is marked by reduced protein metabolism. Mol. Syst. Biol. 2013, 9, 679. [Google Scholar] [CrossRef] [PubMed]
- Sherman, M.Y.; Qian, S.B. Less is more: Improving proteostasis by translation slow down. Trends Biochem. Sci. 2013, 38, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Conn, C.S.; Qian, S.B. mTOR signaling in protein homeostasis: Less is more? Cell Cycle 2011, 10, 1940–1947. [Google Scholar] [CrossRef] [PubMed]
- Conn, C.S.; Qian, S.B. Nutrient signaling in protein homeostasis: An increase in quantity at the expense of quality. Sci. Signal 2013, 6, ra24. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, S.; Cuervo, A.M. Proteostasis and aging. Nat. Med. 2015, 21, 1406–1415. [Google Scholar] [CrossRef]
- Dennis, P.B.; Fumagalli, S.; Thomas, G. Target of rapamycin (TOR): Balancing the opposing forces of protein synthesis and degradation. Curr. Opin. Genet. Dev. 1999, 9, 49–54. [Google Scholar] [CrossRef]
- Qian, S.B.; Zhang, X.; Sun, J.; Bennink, J.R.; Yewdell, J.W.; Patterson, C. mTORC1 links protein quality and quantity control by sensing chaperone availability. J. Biol. Chem. 2010, 285, 27385–27395. [Google Scholar] [CrossRef]
- Perić, M.; Lovrić, A.; Šarić, A.; Musa, M.; Bou Dib, P.; Rudan, M.; Nikolić, A.; Sobočanec, S.; Mikecin, A.M.; Dennerlein, S.; et al. TORC1-mediated sensing of chaperone activity alters glucose metabolism and extends lifespan. Aging Cell 2017, 16, 994–1005. [Google Scholar] [CrossRef]
- Hatakeyama, R.; Péli-Gulli, M.P.; Hu, Z.; Jaquenoud, M.; Garcia Osuna, G.M.; Sardu, A.; Dengjel, J.; de Virgilio, C. Spatially Distinct Pools of TORC1 Balance Protein Homeostasis. Mol. Cell 2019, 73, 325–338. [Google Scholar] [CrossRef]
- Tsang, C.K.; Bertram, P.G.; Ai, W.; Drenan, R.; Zheng, X.F. Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. EMBO J. 2003, 22, 6045–6056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ha, C.W.; Huh, W.K. Rapamycin increases rDNA stability by enhancing association of Sir2 with rDNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2011, 39, 1336–1350. [Google Scholar] [CrossRef] [PubMed]
- Matos-Perdomo, E.; Machín, F. The ribosomal DNA metaphase loop of Saccharomyces cerevisiae gets condensed upon heat stress in a Cdc14-independent TORC1-dependent manner. Cell Cycle 2018, 17, 200–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Webster, D.L.; Watson, K. Ultrastructural changes in yeast following heat shock and recovery. Yeast 1993, 9, 1165–1175. [Google Scholar] [CrossRef] [PubMed]
- Takahara, T.; Maeda, T. Transient Sequestration of TORC1 into Stress Granules during Heat Stress. Mol. Cell 2012, 47, 242–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matos-Perdomo, E.; Machín, F. TORC1, stress and the nucleolus. Aging 2018, 10, 857–858. [Google Scholar] [CrossRef] [PubMed]
- Kos-Braun, I.C.; Jung, I.; Koš, M. Tor1 and CK2 kinases control a switch between alternative ribosome biogenesis pathways in a growth-dependent manner. PLoS Biol. 2017, 15, e2000245. [Google Scholar] [CrossRef] [PubMed]
- Steffen, K.K.; MacKay, V.L.; Kerr, E.O.; Tsuchiya, M.; Hu, D.; Fox, L.A.; Dang, N.; Johnston, E.D.; Oakes, J.A.; Tchao, B.N.; et al. Yeast Life Span Extension by Depletion of 60S Ribosomal Subunits Is Mediated by Gcn4. Cell 2008, 133, 292–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golam Mostofa, M.; Rahman, M.A.; Koike, N.; Yeasmin, A.M.; Islam, N.; Waliullah, T.M.; Hosoyamada, S.; Shimobayashi, M.; Kobayashi, T.; Hall, M.N.; et al. CLIP and cohibin separate rDNA from nucleolar proteins destined for degradation by nucleophagy. J. Cell Biol. 2018, 217, 2675–2690. [Google Scholar] [CrossRef] [Green Version]
- Papandreou, M.E.; Tavernarakis, N. Nucleophagy: From homeostasis to disease. Cell Death Differ 2019, 26, 630–639. [Google Scholar] [CrossRef]
- Tsang, C.K.; Li, H.; Zheng, X.F. Nutrient starvation promotes condensin loading to maintain rDNA stability. EMBO J. 2007, 26, 448–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rohde, J.R.; Cardenas, M.E. The Tor Pathway Regulates Gene Expression by Linking Nutrient Sensing to Histone Acetylation. Mol. Cell. Biol. 2003, 23, 629–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsang, C.K.; Wei, Y.; Zheng, X.S. Compacting DNA During the Interphase. Cell Cycle 2007, 6, 2213–2218. [Google Scholar] [CrossRef] [PubMed]
- Tsang, C.K.; Zheng, X.F. Opposing role of condensin and radiation-sensitive gene RAD52 in ribosomal DNA stability regulation. J. Biol. Chem. 2009, 284, 21908–21919. [Google Scholar] [CrossRef] [PubMed]
- Gadal, O.; Labarre, S.; Boschiero, C.; Thuriaux, P. Hmo1, an HMG-box protein, belongs to the yeast ribosomal DNA transcription system. EMBO J. 2002, 21, 5498–5507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merz, K.; Hondele, M.; Goetze, H.; Gmelch, K.; Stoeckl, U.; Griesenbeck, J. Actively transcribed rRNA genes in S. cerevisiae are organized in a specialized chromatin associated with the high-mobility group protein Hmo1 and are largely devoid of histone molecules. Genes Dev. 2008, 22, 1190–1204. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Workman, J.J.; Strahl, B.D.; Laribee, R.N. Histone H3 and TORC1 prevent organelle dysfunction and cell death by promoting nuclear retention of HMGB proteins. Epigenet. Chromatin 2016, 9, 34. [Google Scholar] [CrossRef] [Green Version]
- Xiao, L.; Kamau, E.; Donze, D.; Grove, A. Expression of yeast high mobility group protein HMO1 is regulated by TOR signaling. Gene 2011, 489, 55–62. [Google Scholar] [CrossRef]
- Wang, D.; Mansisidor, A.; Prabhakar, G.; Hochwagen, A. Condensin and Hmo1 Mediate a Starvation-Induced Transcriptional Position Effect within the Ribosomal DNA Array. Cell Rep. 2016, 14, 1010–1017. [Google Scholar] [CrossRef] [Green Version]
- Ryu, H.Y.; Ahn, S.H. Yeast histone H3 lysine 4 demethylase Jhd2 regulates mitotic ribosomal DNA condensation. BMC Biol. 2014, 12, 75. [Google Scholar] [CrossRef]
- De los Santos-Velázquez, A.I.; de Oya, I.G.; Manzano-López, J.; Monje-Casas, F. Late rDNA Condensation Ensures Timely Cdc14 Release and Coordination of Mitotic Exit Signaling with Nucleolar Segregation. Curr. Biol. 2017, 27, 3248–3263.e5. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.D.; Strunnikov, A.V. Cdc14p/FEAR Pathway Controls Segregation of Nucleolus in S. cerevisiae by Facilitating Condensin Targeting to rDNA Chromatin in Anaphase. Cell Cycle 2004, 3, 960–967. [Google Scholar] [CrossRef] [PubMed]
- Machín, F.; Torres-Rosell, J.; De Piccoli, G.; Carballo, J.A.; Cha, R.S.; Jarmuz, A.; Aragón, L. Transcription of ribosomal genes can cause nondisjunction. J. Cell Biol. 2006, 173, 893–903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clemente-Blanco, A.; Mayán-Santos, M.; Schneider, D.A.; Machín, F.; Jarmuz, A.; Tschochner, H.; Aragón, L. Cdc14 inhibits transcription by RNA polymerase I during anaphase. Nature 2009, 458, 219–222. [Google Scholar] [CrossRef]
- Turowski, T.W.; Lebaron, S.; Zhang, E.; Peil, L.; Dudnakova, T.; Petfalski, E.; Granneman, S.; Rappsilber, J.; Tollervey, D. Rio1 mediates ATP-dependent final maturation of 40S ribosomal subunits. Nucleic Acids Res. 2014, 42, 12189–12199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iacovella, M.G.; Bremang, M.; Basha, O.; Giacò, L.; Carotenuto, W.; Golfieri, C.; Szakal, B.; Maschio, M.D.; Infantino, V.; Beznoussenko, G.V.; et al. Integrating Rio1 activities discloses its nutrient-activated network in Saccharomyces cerevisiae. Nucleic Acids Res. 2018, 46, 7586–7611. [Google Scholar] [CrossRef] [PubMed]
- Iacovella, M.G.; Golfieri, C.; Massari, L.F.; Busnelli, S.; Pagliuca, C.; Dal Maschio, M.; Infantino, V.; Visintin, R.; Mechtler, K.; Ferreira-Cerca, S.; et al. Rio1 promotes rDNA stability and downregulates RNA polymerase I to ensure rDNA segregation. Nat. Commun. 2015, 6, 6643. [Google Scholar] [CrossRef]
- Girke, P.; Seufert, W. Compositional reorganization of the nucleolus in budding yeast mitosis. Mol. Biol. Cell 2019, 30, 591–606. [Google Scholar] [CrossRef]
- Laribee, R.N. Transcriptional and Epigenetic Regulation by the Mechanistic Target of Rapamycin Complex 1 Pathway. J. Mol. Biol. 2018, 430, 4874–4890. [Google Scholar] [CrossRef]
- Workman, J.J.; Chen, H.; Nicholas Laribee, R. Saccharomyces cerevisiae TORC1 controls histone acetylation by signaling through the sit4/PP6 phosphatase to regulate sirtuin deacetylase nuclear accumulation. Genetics 2016, 203, 1733–1746. [Google Scholar] [CrossRef]
- Anderson, R.M.; Bitterman, K.J.; Wood, J.G.; Medvedik, O.; Cohen, H.; Lin, S.S.; Manchester, J.K.; Gordon, J.I.; Sinclair, D.A. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J. Biol. Chem. 2002, 277, 18881–18890. [Google Scholar] [CrossRef] [PubMed]
- Medvedik, O.; Lamming, D.W.; Kim, K.D.; Sinclair, D.A. MSN2 and MSN4 link calorie restriction and TOR to sirtuin-mediated lifespan extension in Saccharomyces cerevisiae. PLoS Biol. 2007, 5, 2330–2341. [Google Scholar] [CrossRef] [PubMed]
- Machín, F.; Paschos, K.; Jarmuz, A.; Torres-Rosell, J.; Pade, C.; Aragón, L. Condensin Regulates rDNA Silencing by Modulating Nucleolar Sir2p. Curr. Biol. 2004, 14, 125–130. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.D.; Butylin, P.; Strunnikov, A. Condensin function in mitotic nucleolar segregation is regulated by rDNA transcription. Cell Cycle 2006, 5, 2260–2267. [Google Scholar] [CrossRef]
- Wilkins, B.J.; Rall, N.A.; Ostwal, Y.; Kruitwagen, T.; Hiragami-Hamada, K.; Winkler, M.; Barral, Y.; Fischle, W.; Neumann, H. A Cascade of Histone Modifications Induces Chromatin Condensation in Mitosis. Science 2014, 343, 77–80. [Google Scholar] [CrossRef] [Green Version]
- Kruitwagen, T.; Denoth-Lippuner, A.; Wilkins, B.J.; Neumann, H.; Barral, Y. Axial contraction and short-range compaction of chromatin synergistically promote mitotic chromosome condensation. eLife 2015, 4, e10396. [Google Scholar] [CrossRef]
- Zhiteneva, A.; Bonfiglio, J.J.; Makarov, A.; Colby, T.; Vagnarelli, P.; Schirmer, E.C.; Matic, I.; Earnshaw, W.C. Mitotic post-translational modifications of histones promote chromatin compaction in vitro. Open Biol. 2017, 7, 170076. [Google Scholar] [CrossRef]
- Sinha, D.; Shogren-Knaak, M.A. Role of direct interactions between the histone H4 tail and the H2A core in long range nucleosome contacts. J. Biol. Chem. 2010, 285, 16572–16581. [Google Scholar] [CrossRef]
- Zhang, R.; Erler, J.; Langowski, J. Histone Acetylation Regulates Chromatin Accessibility: Role of H4K16 in Inter-nucleosome Interaction. Biophys. J. 2017, 112, 450–459. [Google Scholar] [CrossRef]
- Dang, W.; Steffen, K.K.; Perry, R.; Dorsey, J.A.; Johnson, F.B.; Shilatifard, A.; Kaeberlein, M.; Kennedy, B.K.; Berger, S.L. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature 2009, 459, 802–807. [Google Scholar] [CrossRef]
- Molina-Serrano, D.; Schiza, V.; Demosthenous, C.; Stavrou, E.; Oppelt, J.; Kyriakou, D.; Liu, W.; Zisser, G.; Bergler, H.; Dang, W.; et al. Loss of Nat4 and its associated histone H4 N-terminal acetylation mediates calorie restriction-induced longevity. EMBO Rep. 2016, 17, 1829–1843. [Google Scholar] [CrossRef] [PubMed]
- Schiza, V.; Molina-Serrano, D.; Kyriakou, D.; Hadjiantoniou, A.; Kirmizis, A. N-alpha-terminal Acetylation of Histone H4 Regulates Arginine Methylation and Ribosomal DNA Silencing. PLoS Genet. 2013, 9, e1003805. [Google Scholar] [CrossRef] [PubMed]
- Birch, J.L.; Tan, B.C.; Panov, K.I.; Panova, T.B.; Andersen, J.S.; Owen-Hughes, T.A.; Russell, J.; Lee, S.C.; Zomerdijk, J.C. FACT facilitates chromatin transcription by RNA polymerases I and III. EMBO J. 2009, 28, 854–865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, J.M.; French, S.L.; Osheim, Y.N.; Li, M.; Hall, L.; Beyer, A.L.; Smith, J.S. Rpd3- and Spt16-Mediated Nucleosome Assembly and Transcriptional Regulation on Yeast Ribosomal DNA Genes. Mol. Cell. Biol. 2013, 33, 2748–2759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beckwith, S.L.; Schwartz, E.K.; García-Nieto, P.E.; King, D.A.; Gowans, G.J.; Wong, K.M.; Eckley, T.L.; Paraschuk, A.P.; Peltan, E.L.; Lee, L.R.; et al. The INO80 chromatin remodeler sustains metabolic stability by promoting TOR signaling and regulating histone acetylation. PLoS Genet. 2018, 14, e1007216. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Fan, M.; Pfeffer, L.M.; Laribee, R.N. The histone H3 lysine 56 acetylation pathway is regulated by target of rapamycin (TOR) signaling and functions directly in ribosomal RNA biogenesis. Nucleic Acids Res. 2012, 40, 6534–6546. [Google Scholar] [CrossRef] [Green Version]
- Neumüller, R.A.; Gross, T.; Samsonova, A.A.; Vinayagam, A.; Buckner, M.; Founk, K.; Hu, Y.; Sharifpoor, S.; Rosebrock, A.P.; Andrews, B.; et al. Conserved regulators of nucleolar size revealed by global phenotypic analyses. Sci. Signal 2013, 6, ra70. [Google Scholar] [CrossRef]
- Srivastava, R.; Srivastava, R.; Ahn, S.H. The Epigenetic Pathways to Ribosomal DNA Silencing. Microbiol. Mol. Biol. Rev. 2016, 80, 545–563. [Google Scholar] [CrossRef] [Green Version]
- Gartenberg, M.R.; Smith, J.S. The nuts and bolts of transcriptionally silent chromatin in Saccharomyces cerevisiae. Genetics 2016, 203, 1563–1599. [Google Scholar] [CrossRef]
- Xu, H.H.; Su, T.; Xue, Y. Histone H3 N-terminal acetylation sites especially K14 are important for rDNA silencing and aging. Sci. Rep. 2016, 6, 21900. [Google Scholar] [CrossRef]
- Molina-Serrano, D.; Kyriakou, D.; Kirmizis, A. Histone Modifications as an Intersection Between Diet and Longevity. Front. Genet. 2019, 10, 192. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.M.; Bitterman, K.J.; Wood, J.G.; Medvedik, O.; Sinclair, D.A. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature 2003, 423, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Kaeberlein, M.; Kirkland, K.T.; Fields, S.; Kennedy, B.K. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004, 2, e296. [Google Scholar] [CrossRef] [PubMed]
- Fabrizio, P.; Gattazzo, C.; Battistella, L.; Wei, M.; Cheng, C.; McGrew, K.; Longo, V.D. Sir2 blocks extreme life-span extension. Cell 2005, 123, 655–667. [Google Scholar] [CrossRef] [PubMed]
- Lamming, D.W.; Latorre-Esteves, M.; Medvedik, O.; Wong, S.N.; Tsang, F.A.; Wang, C.; Lin, S.J.; Sinclair, D.A. Cell biology: HST2 mediates SIR2-independent life-span extension by calorie restriction. Science 2005, 309, 1861–1864. [Google Scholar] [CrossRef] [PubMed]
- Rine, J. Twists in the tale of the aging yeast. Science 2005, 310, 1124–1125. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, M.; Dang, N.; Kerr, E.O.; Hu, D.; Steffen, K.K.; Oakes, J.A.; Kennedy, B.K.; Kaeberlein, M. Sirtuin-independent effects of nicotinamide on lifespan extension from calorie restriction in yeast. Aging Cell 2006, 5, 505–514. [Google Scholar] [CrossRef]
- Smith, D.L.; McClure, J.M.; Matecic, M.; Smith, J.S. Calorie restriction extends the chronological lifespan of Saccharomyces cerevisiae independently of the Sirtuins. Aging Cell 2007, 6, 649–662. [Google Scholar] [CrossRef]
- Kaeberlein, M.; Powers, R.W. Sir2 and calorie restriction in yeast: A skeptical perspective. Ageing Res. Rev. 2007, 6, 128–140. [Google Scholar] [CrossRef]
- Longo, V.D. Dietary restriction: Standing up for sirtuins—Response. Science 2010, 329, 1013–1014. [Google Scholar] [CrossRef]
- Imai, S.I.; Guarente, L. It takes two to tango: NAD+ and sirtuins in aging/longevity control. NPJ Aging Mech. Dis. 2016, 2, 16017. [Google Scholar] [CrossRef] [PubMed]
- Riesen, M.; Morgan, A. Calorie restriction reduces rDNA recombination independently of rDNA silencing. Aging Cell 2009, 8, 624–632. [Google Scholar] [CrossRef] [Green Version]
- Ha, C.W.; Huh, W.K. The implication of Sir2 in replicative aging and senescence in Saccharomyces cerevisiae. Aging 2011, 3, 319–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potapova, T.A.; Gerton, J.L. Ribosomal DNA and the nucleolus in the context of genome organization. Chromosome Res. 2019, 27, 109–127. [Google Scholar] [CrossRef] [PubMed]
- Pich, A.; Margaria, E.; Chiusa, L.; Ponti, R.; Geuna, M. DNA ploidy and p53 expression correlate with survival and cell proliferative activity in male breast carcinoma. Hum. Pathol. 1996, 27, 676–682. [Google Scholar] [CrossRef]
- Coric, M.; Ladika-Davidovic, B.; Bumber, Z.; Danic, D.; Vuletic, L.B.; Seiwerth, S. Prognostic significance of DNA cytometry in combination with AgNOR investigation. Acta Otolaryngol. 2007, 127, 1332–1337. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Lemos, B. Ribosomal DNA copy number amplification and loss in human cancers is linked to tumor genetic context, nucleolus activity, and proliferation. PLoS Genet. 2017, 13, e1006994. [Google Scholar] [CrossRef] [PubMed]
- Mayer, C.; Grummt, I. Cellular stress and nucleolar function. Cell Cycle 2005, 4, 1036–1038. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Yi, H.; Chen, C.; Yan, S.; Yao, H.; He, G.; Li, G.; Jiang, Y.; Deng, T.; Deng, X. Nucleolar stress: Is there a reverse version? J. Cancer 2018, 9, 3723–3727. [Google Scholar] [CrossRef] [PubMed]
- Shav-Tal, Y.; Blechman, J.; Darzacq, X.; Montagna, C.; Dye, B.T.; Patton, J.G.; Singer, R.H.; Zipori, D. Dynamic Sorting of Nuclear Components into Distinct Nucleolar Caps during Transcriptional Inhibition. Mol. Biol. Cell 2005, 16, 2395–2413. [Google Scholar] [CrossRef] [Green Version]
- Larsen, D.H.; Stucki, M. Nucleolar responses to DNA double-strand breaks. Nucleic Acids Res. 2016, 44, 538–544. [Google Scholar] [CrossRef] [PubMed]
- Korsholm, L.M.; Gál, Z.; Lin, L.; Quevedo, O.; Ahmad, D.A.; Dulina, E.; Luo, Y.; Bartek, J.; Larsen, D.H. Double-strand breaks in ribosomal RNA genes activate a distinct signaling and chromatin response to facilitate nucleolar restructuring and repair. Nucleic Acid Res. 2019, gkz518. [Google Scholar] [CrossRef] [PubMed]
- Hein, N.; Hannan, K.M.; George, A.J.; Sanij, E.; Hannan, R.D. The nucleolus: An emerging target for cancer therapy. Trends Mol. Med. 2013, 19, 643–654. [Google Scholar] [CrossRef] [PubMed]
- Quin, J.E.; Devlin, J.R.; Cameron, D.; Hannan, K.M.; Pearson, R.B.; Hannan, R.D. Targeting the nucleolus for cancer intervention. Biochim. Biophys. Acta-Mol. Basis Dis. 2014, 1842, 802–816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drygin, D.; Siddiqui-Jain, A.; O’Brien, S.; Schwaebe, M.; Lin, A.; Bliesath, J.; Ho, C.B.; Proffitt, C.; Trent, K.; Whitten, J.P.; et al. Anticancer activity of CX-3543: A direct inhibitor of rRNA biogenesis. Cancer Res. 2009, 69, 7653–7661. [Google Scholar] [CrossRef] [PubMed]
- Drygin, D.; Lin, A.; Bliesath, J.; Ho, C.B.; O’Brien, S.E.; Proffitt, C.; Omori, M.; Haddach, M.; Schwaebe, M.K.; Siddiqui-Jain, A.; et al. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res. 2011, 71, 1418–1430. [Google Scholar] [CrossRef] [PubMed]
- Haddach, M.; Schwaebe, M.K.; Michaux, J.; Nagasawa, J.; O’Brien, S.E.; Whitten, J.P.; Pierre, F.; Kerdoncuff, P.; Darjania, L.; Stansfield, R.; et al. Discovery of CX-5461, the first direct and selective inhibitor of RNA polymerase I, for cancer therapeutics. ACS Med. Chem. Lett. 2012, 3, 602–606. [Google Scholar] [CrossRef]
- Wust, P.; Hildebrandt, B.; Sreenivasa, G.; Rau, B.; Gellermann, J.; Riess, H.; Felix, R.; Schlag, P. Hyperthermia in combined treatment of cancer. Lancet Oncol. 2002, 3, 487–497. [Google Scholar] [CrossRef]
- Mirzaei, H.; Suarez, J.A.; Longo, V.D. Protein and amino acid restriction, aging and disease: From yeast to humans. Trends Endocrinol. Metab. 2014, 25, 558–566. [Google Scholar] [CrossRef]
- Mallory, M.; Gogineni, E.; Jones, G.C.; Greer, L.; Simone, C.B. Therapeutic hyperthermia: The old, the new, and the upcoming. Crit. Rev. Oncol. Hematol. 2016, 97, 56–64. [Google Scholar] [CrossRef]
- Guri, Y.; Hall, M.N. mTOR Signaling Confers Resistance to Targeted Cancer Drugs. Trends Cancer 2016, 2, 688–697. [Google Scholar] [CrossRef] [PubMed]
- Frankowski, K.J.; Wang, C.; Patnaik, S.; Schoenen, F.J.; Southall, N.; Li, D.; Teper, Y.; Sun, W.; Kandela, I.; Hu, D.; et al. Metarrestin, a perinucleolar compartment inhibitor, effectively suppresses metastasis. Sci. Transl. Med. 2018, 10, eaap8307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dove, B.K.; You, J.H.; Reed, M.L.; Emmett, S.R.; Brooks, G.; Hiscox, J.A. Changes in nucleolar morphology and proteins during infection with the coronavirus infectious bronchitis virus. Cell. Microbiol. 2006, 8, 1147–1157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiscox, J.A. RNA viruses: Hijacking the dynamic nucleolus. Nat. Rev. Microbiol. 2007, 5, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Rawlinson, S.M.; Moseley, G.W. The nucleolar interface of RNA viruses. Cell. Microbiol. 2015, 17, 1108–1120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Latonen, L. Nucleolar aggresomes as counterparts of cytoplasmic aggresomes in proteotoxic stress. Proteasome inhibitors induce nuclear ribonucleoprotein inclusions that accumulate several key factors of neurodegenerative diseases and cancer. Bioessays 2011, 33, 386–395. [Google Scholar] [CrossRef] [PubMed]
- Latonen, L. Phase-to-Phase With Nucleoli—Stress Responses, Protein Aggregation and Novel Roles of RNA. Front. Cell. Neurosci. 2019, 13, 151. [Google Scholar] [CrossRef] [PubMed]
- Kastan, M.B. Wild-type p53: Tumors can’t stand it. Cell 2007, 128, 837–840. [Google Scholar] [CrossRef] [PubMed]
- Bourdon, J.C.; Fernandes, K.; Murray-Zmijewski, F.; Liu, G.; Diot, A.; Xirodimas, D.P.; Saville, M.K.; Lane, D.P. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2007, 19, 2122–2137. [Google Scholar] [CrossRef]
- Joruiz, S.M.; Bourdon, J.C. p53 Isoforms: Key Regulators of the Cell Fate Decision. Cold Spring Harb. Perspect. Med. 2016, 6, a026039. [Google Scholar] [CrossRef]
- Gadea, G.; Arsic, N.; Fernandes, K.; Diot, A.; Joruiz, S.M.; Abdallah, S.; Meuray, V.; Vinot, S.; Anguille, C.; Remenyi, J.; et al. TP53 drives invasion through expression of its Δ133p53β variant. Elife 2016, 5, e14734. [Google Scholar] [PubMed]
- Zhang, H.; Zhao, Y.; Sun, P.; Zhao, M.; Su, Z.; Jin, X.; Song, W. p53β: A new prognostic marker for patients with clear-cell renal cell carcinoma from 5.3 years of median follow-up. Carcinogenesis 2018, 39, 368–374. [Google Scholar] [CrossRef] [PubMed]
- Holmberg Olausson, K.; Nistér, M.; Lindström, M. p53 -Dependent and -Independent Nucleolar Stress Responses. Cells 2012, 1, 774–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deisenroth, C.; Zhang, Y. Ribosome biogenesis surveillance: Probing the ribosomal protein-Mdm2-p53 pathway. Oncogene 2010, 29, 4253–4260. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Kogo, R.; Kawahara, K.; Sasaki, M.; Nishio, M.; Maehama, T.; Sasaki, T.; Mimori, K.; Mori, M. A new PICTure of nucleolar stress. Cancer Sci. 2012, 103, 632–637. [Google Scholar] [CrossRef] [Green Version]
- Okahara, F.; Itoh, K.; Nakagawara, A.; Murakami, M.; Kanaho, Y.; Maehama, T. Critical Role of PICT-1, a Tumor Suppressor Candidate, in Phosphatidylinositol 3,4,5-Trisphosphate Signals and Tumorigenic Transformation. Mol. Biol. Cell 2006, 17, 4888–4895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stȩpiński, D. The nucleolus, an ally, and an enemy of cancer cells. Histochem. Cell Biol. 2018, 150, 607–629. [Google Scholar] [CrossRef]
- Moon, A.; Lim, S.J.; Jo, Y.H.; Lee, S.; Kim, J.Y.; Lee, J.; Park, J.H. Downregulation of GLTSCR2 expression is correlated with breast cancer progression. Pathol. Res. Pract. 2013, 209, 700–704. [Google Scholar] [CrossRef]
- Okamura, K.; Takayama, K.; Kawahara, K.; Harada, T.; Nishio, M.; Otsubo, K.; Ijichi, K.; Kohno, M.; Iwama, E.; Fujii, A.; et al. PICT1 expression is a poor prognostic factor in non-small cell lung cancer. Oncoscience 2015, 1, 375. [Google Scholar] [CrossRef]
- Kim, J.Y.; Park, J.H.; Lee, S. GLTSCR2 contributes to the death resistance and invasiveness of hypoxia-selected cancer cells. FEBS Lett. 2012, 586, 3435–3440. [Google Scholar] [CrossRef]
- Fontana, L.; Partridge, L.; Longo, V.D. Extending healthy life span-from yeast to humans. Science 2010, 328, 321–326. [Google Scholar] [CrossRef] [PubMed]
- De Cabo, R.; Carmona-Gutierrez, D.; Bernier, M.; Hall, M.N.; Madeo, F. The search for antiaging interventions: From elixirs to fasting regimens. Cell 2014, 157, 1515–1526. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Partridge, L. Promoting health and longevity through diet: From model organisms to humans. Cell 2015, 161, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Blagosklonny, M.V. Aging and immortality: Quasi-programmed senescence and its pharmacologic inhibition. Cell Cycle 2006, 5, 2087–2102. [Google Scholar] [CrossRef] [PubMed]
- Di Francesco, A.; di Germanio, C.; Bernier, M.; de Cabo, R. A time to fast. Science 2018, 362, 770–775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Longo, V.D. Programmed longevity, youthspan, and juventology. Aging Cell 2019, 18, e12843. [Google Scholar] [CrossRef]
- Deprez, M.A.; Eskes, E.; Winderickx, J.; Wilms, T. The TORC1-Sch9 pathway as a crucial mediator of chronological lifespan in the yeast Saccharomyces cerevisiae. FEMS Yeast Res. 2018, 18, foy048. [Google Scholar] [CrossRef]
- Tsang, C.K.; Chen, M.; Cheng, X.; Qi, Y.; Chen, Y.; Das, I.; Li, X.; Vallat, B.; Fu, L.W.; Qian, C.N.; et al. SOD1 Phosphorylation by mTORC1 Couples Nutrient Sensing and Redox Regulation. Mol. Cell 2018, 70, 502–515.e8. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, T. A new role of the rDNA and nucleolus in the nucleus—RDNA instability maintains genome integrity. BioEssays 2008, 30, 267–272. [Google Scholar] [CrossRef]
- Ganley, A.R.; Kobayashi, T. Ribosomal DNA and cellular senescence: New evidence supporting the connection between rDNA and aging. FEMS Yeast Res. 2014, 14, 49–59. [Google Scholar] [CrossRef]
- Wang, M.; Lemos, B. Ribosomal DNA harbors an evolutionarily conserved clock of biological aging. Genome Res. 2019, 29, 325–333. [Google Scholar] [CrossRef] [PubMed]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed]



| Nucleolar and rDNA Features | |
|---|---|
| Yeast | Humans |
| Nucleolus and nucleus are not disassembled in mitosis | Nucleolus is disassembled in mitosis along with the nucleus |
| Bipartite composition | Tripartite composition |
| Absence of a perinucleolar domain | Presence of a perinucleolar domain |
| All rRNA genes together | 5S and 45S genes in different genome loci |
| The rDNA in a single array at chromosome XII right arm | The rDNA 45S array in the five acrocentric chromosomes (13, 14, 15, 21, 22); 5S in chromosome 1 |
| 100–200 copies (haploid) of a 9.1 Kb unit | 300–400 copies (haploid) of the 43 Kb unit (45S) |
| The rDNA is attached to the nuclear envelope | The rDNA is not always attached to the nuclear envelope |
| Presence of a cryptic RNApol II promoter at the rDNA | Absence of a cryptic RNApol II promoter at the rDNA |
| Silencing complexes: RENT (Net1, Sir2, Cdc14); Tof2-Lrs4/Csm1 | Silencing complexes NoRC (TIP5, SNF2h); eNoSC (SIRT1, NML, SUV39H1) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matos-Perdomo, E.; Machín, F. Nucleolar and Ribosomal DNA Structure under Stress: Yeast Lessons for Aging and Cancer. Cells 2019, 8, 779. https://doi.org/10.3390/cells8080779
Matos-Perdomo E, Machín F. Nucleolar and Ribosomal DNA Structure under Stress: Yeast Lessons for Aging and Cancer. Cells. 2019; 8(8):779. https://doi.org/10.3390/cells8080779
Chicago/Turabian StyleMatos-Perdomo, Emiliano, and Félix Machín. 2019. "Nucleolar and Ribosomal DNA Structure under Stress: Yeast Lessons for Aging and Cancer" Cells 8, no. 8: 779. https://doi.org/10.3390/cells8080779