Protein S100-A7 Derived from Digested Dentin Is a Critical Molecule for Dentin Pulp Regeneration
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of DMCs
2.2. Reverse-Phase High Performance Liquid Chromatography (RP-HPLC) Analysis and Fraction Collection
2.3. Animal Studies Using Fractions of u-DMCs or d-DMCs
2.3.1. Pulp Capping Experiment
2.3.2. Histological Evaluation
2.3.3. Tertiary Dentin Volume Quantification by µ-CT Analysis
2.4. Cell Studies
2.4.1. Cell Isolation and Culture
2.4.2. WST-1 Assay
2.4.3. Alkaline Phosphatase (ALP) and Alizarin Red Staining
2.5. Proteomic Analysis
2.6. Direct Pulp Capping Using Identified Proteins
2.7. In Vivo Damaged Pulp Model and Immunohistochemical Staining
2.8. Statistical Analysis
3. Results
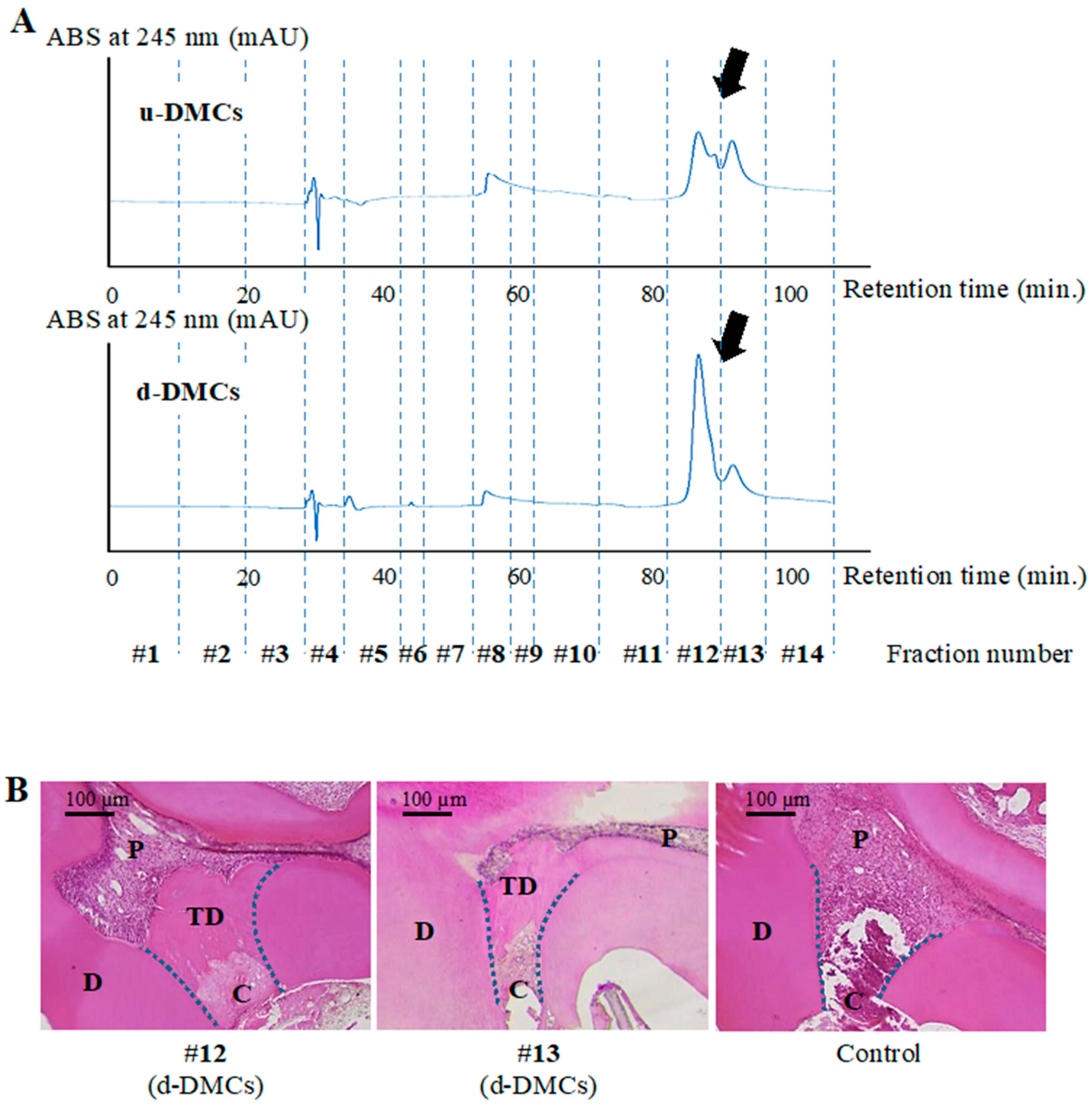
3.1. HPLC Analysis and Fraction Collection
3.2. Determination of Bioactive Fractions In Vivo
3.3. Effects of the Fractions on RPPCs In Vitro
3.4. Identification of the Bioactive Molecules
3.5. Effects of Proteins S100-A7 and S100-A8 on Tertiary Dentinogenesis
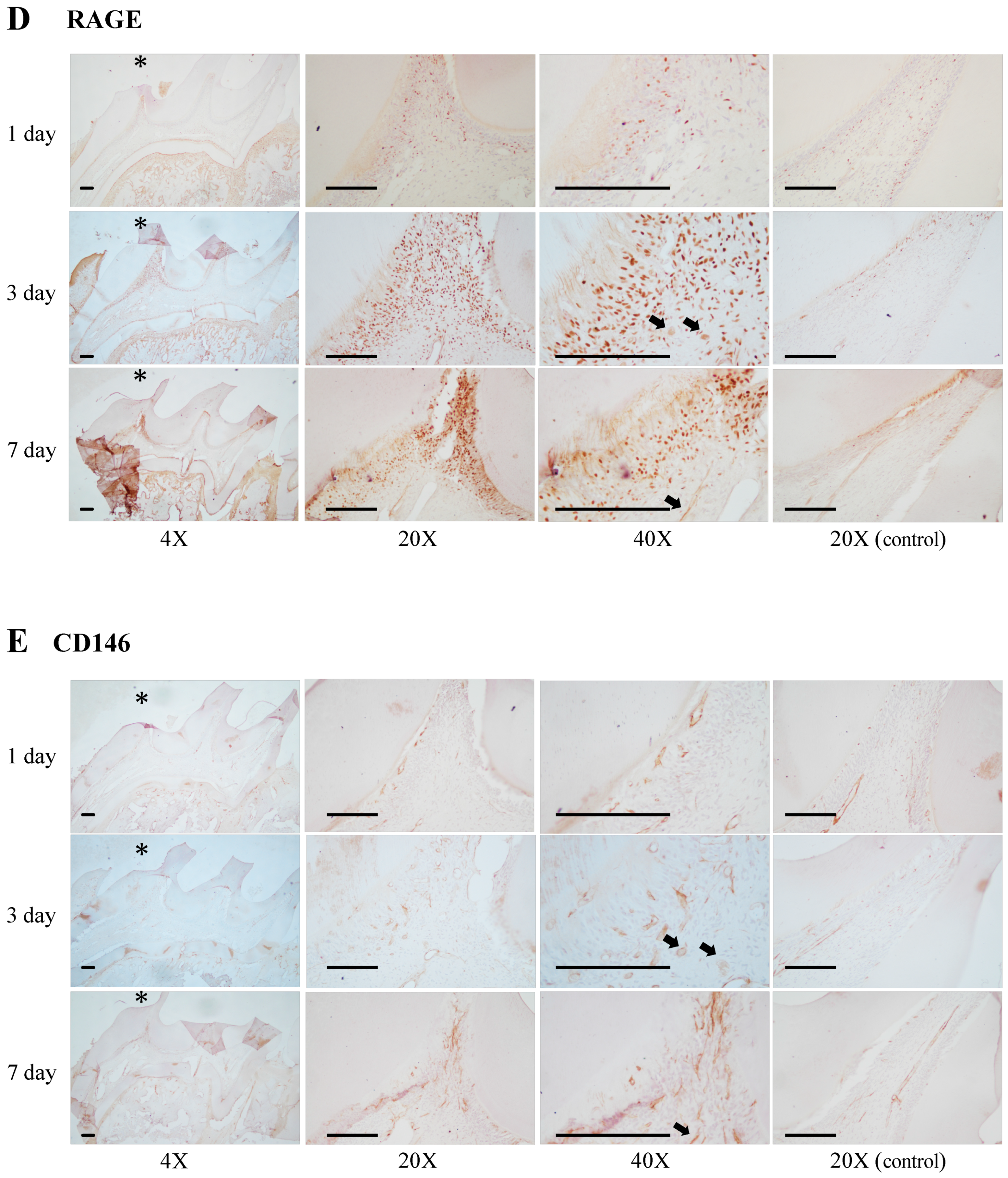
3.6. Expression of Receptor for Advanced Glycation end Products (RAGE) in Damaged Pulp
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Smith, A.J.; Scheven, B.A.; Takahashi, Y.; Ferracane, J.L.; Shelton, R.M.; Cooper, P.R. Dentine as a Bioactive Extracellular Matrix. Arch. Oral Biol. 2012, 57, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Gronthos, S.; Mankani, M.; Brahim, J.; Robey, P.G.; Shi, S. Postnatal Human Dental Pulp Stem Cells (DPSCs) in Vitro and In vivo. Proc. Natl. Acad. Sci. USA 2000, 97, 13625–13630. [Google Scholar] [CrossRef] [PubMed]
- Simon, S.; Smith, A.J.; Lumley, P.J.; Berdal, A.; Smith, G.; Finney, S.; Cooper, P.R. Molecular Characterization of Young and Mature Odontoblasts. Bone 2009, 45, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Miles, J.P.; Gluskin, A.H.; Chambers, D.; Peters, O.A. Pulp Capping with Mineral Trioxide Aggregate (MTA): A Retrospective Analysis of Carious Pulp Exposures Treated by Undergraduate Dental Students. Oper. Dent. 2010, 35, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Barthel, C.; Rosenkranz, B.; Leuenberg, A.; Roulet, J. Pulp Capping of Carious Exposures: Treatment Outcome after 5 and 10 Years: A Retrospective Study. J. Endod. 2000, 26, 525–528. [Google Scholar] [CrossRef] [PubMed]
- Iohara, K.; Nakashima, M.; Ito, M.; Ishikawa, M.; Nakasima, A.; Akamine, A. Dentin Regeneration by Dental Pulp Stem Cell Therapy with Recombinant Human Bone Morphogenetic Protein 2. J. Dent. Res. 2004, 83, 590–595. [Google Scholar] [CrossRef] [PubMed]
- Yang, I.-S.; Lee, D.-S.; Park, J.-T.; Kim, H.-J.; Son, H.-H.; Park, J.-C. Tertiary Dentin Formation after Direct Pulp Capping with Odontogenic Ameloblast-Associated Protein in Rat Teeth. J. Endod. 2010, 36, 1956–1962. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Handa, K.; Koike, T.; Saito, T. The Possibility of Genistein as a New Direct Pulp Capping Agent. Dent. Mater. J. 2013, 32, 976–985. [Google Scholar] [CrossRef] [PubMed]
- Saarialho-Kere, U.; Kerkelä, E.; Suomela, S.; Jahkola, T.; Keski-Oja, J.; Lohi, J. Epilysin (MMP-28) Expression Is Associated with Cell Proliferation During Epithelial Repair. J. Investig. Dermatol. 2002, 119, 14–21. [Google Scholar] [CrossRef]
- Salmela, M.T.; Pender, S.L.F.; Karjalainen-Lindsberg, M.-L.; Puolakkainen, P.; MacDonald, T.T.; Saarialho-Kere, U. Collagenase-1 (MMP-1), Matrilysin-1 (MMP-7), and Stromelysin-2 (MMP-10) Are Expressed by Migrating Enterocytes during Intestinal Wound Healing. Scand. J. Gastroenterol. 2004, 39, 1095–1104. [Google Scholar] [CrossRef] [PubMed]
- Rohani, M.G.; Parks, W.C. Matrix Remodeling by MMPs during Wound Repair. Matrix Biol. 2015, 44–46, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Hattori, N.; Mochizuki, S.; Kishi, K.; Nakajima, T.; Takaishi, H.; D’Armiento, J.; Okada, Y. MMP-13 Plays a Role in Keratinocyte Migration, Angiogenesis, and Contraction in Mouse Skin Wound Healing. Am. J. Pathol. 2009, 175, 533–546. [Google Scholar] [CrossRef] [Green Version]
- Adair-Kirk, T.L.; Atkinson, J.J.; Broekelmann, T.J.; Doi, M.; Tryggvason, K.; Miner, J.H.; Mecham, R.P.; Senior, R.M. A Site on Laminin A5, AQARSAASKVKVSMKF, Induces Inflammatory Cell Production of Matrix Metalloproteinase-9 and Chemotaxis. J. Immunol. 2003, 171, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Brennan, E.P.; Tang, X.-H.; Stewart-Akers, A.M.; Gudas, L.J.; Badylak, S.F. Chemoattractant Activity of Degradation Products of Fetal and Adult Skin Extracellular Matrix for Keratinocyte Progenitor Cells. J. Tissue Eng. Regen. Med. 2008, 2, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, S.; Takahashi, Y.; Abe, M.; Michikami, I.; Imazato, S.; Wakisaka, S.; Hayashi, M.; Ebisu, S. Activation of the Wnt/-Catenin Pathway and Tissue Inhibitor of Metalloprotease 1 during Tertiary Dentinogenesis. J. Biochem. (Tokyo) 2013, 153, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Chaussain-Miller, C.; Fioretti, F.; Goldberg, M.; Menashi, S. The Role of Matrix Metalloproteinases (MMPs) in Human Caries. J. Dent. Res. 2006, 85, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.T.; Hannas, A.R.; Leite, A.L.; Bolanho, A.; Zarella, B.L.; Santos, J.; Carrilho, M.; Tjäderhane, L.; Buzalaf, M.A.R. Activity of Matrix Metalloproteinases in Bovine versus Human Dentine. Caries Res. 2011, 45, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Wu, X.; Lei, W.; Pang, L.; Wan, C.; Shi, Z.; Zhao, L.; Nagy, T.R.; Peng, X.; Hu, J.; et al. TGF-Β1–Induced Migration of Bone Mesenchymal Stem Cells Couples Bone Resorption with Formation. Nat. Med. 2009, 15, 757–765. [Google Scholar] [CrossRef] [PubMed]
- Mundy, G.R.; Bonewald, L.F. Role of TGF? In Bone Remodeling. Ann. N. Y. Acad. Sci. 1990, 593, 91–97. [Google Scholar] [CrossRef]
- Dallas, S.L.; Rosser, J.L.; Mundy, G.R.; Bonewald, L.F. Proteolysis of Latent Transforming Growth Factor-β (TGF-β)-Binding Protein-1 by Osteoclasts: A CELLULAR MECHANISM FOR RELEASE OF TGF-β FROM BONE MATRIX. J. Biol. Chem. 2002, 277, 21352–21360. [Google Scholar] [CrossRef]
- Tatti, O.; Vehvilainen, P.; Lehti, K.; Keskioja, J. MT1-MMP Releases Latent TGF-Β1 from Endothelial Cell Extracellular Matrix via Proteolytic Processing of LTBP-1. Exp. Cell Res. 2008, 314, 2501–2514. [Google Scholar] [CrossRef] [PubMed]
- Gajjeraman, S.; Narayanan, K.; Hao, J.; Qin, C.; George, A. Matrix Macromolecules in Hard Tissues Control the Nucleation and Hierarchical Assembly of Hydroxyapatite. J. Biol. Chem. 2007, 282, 1193–1204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gericke, A.; Qin, C.; Sun, Y.; Redfern, R.; Redfern, D.; Fujimoto, Y.; Taleb, H.; Butler, W.T.; Boskey, A.L. Different Forms of DMP1 Play Distinct Roles in Mineralization. J. Dent. Res. 2010, 89, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Sreenath, T.; Haruyama, N.; Honeycutt, C.; Terse, A.; Cho, A.; Kohler, T.; Müller, R.; Goldberg, M.; Kulkarni, A.B. Dentin Sialoprotein and Dentin Phosphoprotein Have Distinct Roles in Dentin Mineralization. Matrix Biol. 2009, 28, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Gibson, M.P.; Liu, Q.; Zhu, Q.; Lu, Y.; Jani, P.; Wang, X.; Liu, Y.; Paine, M.L.; Snead, M.L.; Feng, J.Q.; et al. Role of the NH2-Terminal Fragment of Dentin Sialophosphoprotein in Dentinogenesis. Eur. J. Oral Sci. 2013, 121, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Gibson, M.P.; Liu, Q.; Liu, Y.; Lu, Y.; Wang, X.; Feng, J.Q.; Qin, C. Proteolytic Processing of Dentin Sialophosphoprotein (DSPP) Is Essential to Dentinogenesis. J. Biol. Chem. 2012, 287, 30426–30435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, J.G.; Smith, A.J.; Shelton, R.M.; Cooper, P.R. Recruitment of Dental Pulp Cells by Dentine and Pulp Extracellular Matrix Components. Exp. Cell Res. 2012, 318, 2397–2406. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, M.; Takahashi, Y.; Komichi, S.; Cooper, P.R.; Hayashi, M. Dentinogenic Effects of Extracted Dentin Matrix Components Digested with Matrix Metalloproteinases. Sci. Rep. 2018, 8, 10690. [Google Scholar] [CrossRef]
- Tomson, P.L.; Grover, L.M.; Lumley, P.J.; Sloan, A.J.; Smith, A.J.; Cooper, P.R. Dissolution of Bio-Active Dentine Matrix Components by Mineral Trioxide Aggregate. J. Dent. 2007, 35, 636–642. [Google Scholar] [CrossRef]
- Yoneda, N.; Noiri, Y.; Matsui, S.; Kuremoto, K.; Maezono, H.; Ishimoto, T.; Nakano, T.; Ebisu, S.; Hayashi, M. Development of a Root Canal Treatment Model in the Rat. Sci. Rep. 2017, 7, 3315. [Google Scholar] [CrossRef]
- Kuremoto, K.; Noiri, Y.; Ishimoto, T.; Yoneda, N.; Yamamoto, R.; Maezono, H.; Nakano, T.; Hayashi, M.; Ebisu, S. Promotion of Endodontic Lesions in Rats by a Novel Extraradicular Biofilm Model Using Obturation Materials. Appl. Environ. Microbiol. 2014, 80, 3804–3810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant Enables High Peptide Identification Rates, Individualized p.p.b.-Range Mass Accuracies and Proteome-Wide Protein Quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Maeda, O.; Usami, N.; Kondo, M.; Takahashi, M.; Goto, H.; Shimokata, K.; Kusugami, K.; Sekido, Y. Plakoglobin (γ-Catenin) Has TCF/LEF Family-Dependent Transcriptional Activity in β-Catenin-Deficient Cell Line. Oncogene 2004, 23, 964–972. [Google Scholar] [CrossRef] [PubMed]
- Wolf, R.; Howard, O.M.Z.; Dong, H.-F.; Voscopoulos, C.; Boeshans, K.; Winston, J.; Divi, R.; Gunsior, M.; Goldsmith, P.; Ahvazi, B.; et al. Chemotactic Activity of S100A7 (Psoriasin) Is Mediated by the Receptor for Advanced Glycation End Products and Potentiates Inflammation with Highly Homologous but Functionally Distinct S100A15. J. Immunol. Baltim. Md 1950 2008, 181, 1499–1506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zackular, J.P.; Chazin, W.J.; Skaar, E.P. Nutritional Immunity: S100 Proteins at the Host-Pathogen Interface. J. Biol. Chem. 2015, 290, 18991–18998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cassoni, P.; Sapino, A.; Haagensen, D.E.; Naldoni, C.; Bussolati, G. Mitogenic Effect of the 15-KDa Gross Cystic Disease Fluid Protein (GCDFP-15) on Breast-Cancer Cell Lines and on Immortal Mammary Cells. Int. J. Cancer 2006, 60, 216–220. [Google Scholar] [CrossRef] [PubMed]
- Eckhardt, A.; Jágr, M.; Pataridis, S.; Mikšík, I. Proteomic Analysis of Human Tooth Pulp: Proteomics of Human Tooth. J. Endod. 2014, 40, 1961–1966. [Google Scholar] [CrossRef]
- Jágr, M.; Eckhardt, A.; Pataridis, S.; Mikšík, I. Comprehensive Proteomic Analysis of Human Dentin. Eur. J. Oral Sci. 2012, 120, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.J.; Smith, G. Proteolytic Activity of Rabbit Incisor Dentine. Arch. Oral Biol. 1984, 29, 1049–1050. [Google Scholar] [CrossRef]
- Yamakoshi, Y.; Kinoshita, S.; Izuhara, L.; Karakida, T.; Fukae, M.; Oida, S. DPP and DSP Are Necessary for Maintaining TGF-Β1 Activity in Dentin. J. Dent. Res. 2014, 93, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Yamakoshi, Y.; Hu, J.C.-C.; Iwata, T.; Kobayashi, K.; Fukae, M.; Simmer, J.P. Dentin Sialophosphoprotein Is Processed by MMP-2 and MMP-20 in Vitro and in Vivo. J. Biol. Chem. 2006, 281, 38235–38243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishimoto, K.; Hayano, S.; Yanagita, T.; Kurosaka, H.; Kawanabe, N.; Itoh, S.; Ono, M.; Kuboki, T.; Kamioka, H.; Yamashiro, T. Topical Application of Lithium Chloride on the Pulp Induces Dentin Regeneration. PLoS ONE 2015, 10, e0121938. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Chen, W.; Xia, Z.; Liu, Y.; Peggrem, S.; Geng, T.; Yang, Z.; Li, H.; Xu, B.; Zhang, C.; et al. Local Application of Ibandronate/Gelatin Sponge Improves Osteotomy Healing in Rabbits. PLoS ONE 2015, 10, e0125807. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wang, L.; Yu, J.; Lei, G.; Yan, M.; Smith, G.; Cooper, P.R.; Tang, C.; Zhang, G.; Smith, A.J. Dentin Matrix Proteins (DMPs) Enhance Differentiation of BMMSCs via ERK and P38 MAPK Pathways. Cell Tissue Res. 2014, 356, 171–182. [Google Scholar] [CrossRef]
- Donato, R.; Cannon, B.R.; Sorci, G.; Riuzzi, F.; Hsu, K.; Weber, D.J.; Geczy, C.L. Functions of S100 Proteins. Curr. Mol. Med. 2013, 13, 24–57. [Google Scholar] [CrossRef] [Green Version]
- Foell, D.; Wittkowski, H.; Vogl, T.; Roth, J. S100 Proteins Expressed in Phagocytes: A Novel Group of Damage-Associated Molecular Pattern Molecules. J. Leukoc. Biol. 2007, 81, 28–37. [Google Scholar] [CrossRef]
- McLachlan, J.L.; Sloan, A.J.; Smith, A.J.; Landini, G.; Cooper, P.R. S100 and Cytokine Expression in Caries. Infect. Immun. 2004, 72, 4102–4108. [Google Scholar] [CrossRef] [Green Version]
- Stern, D.; Du Yan, S.; Fang Yan, S.; Marie Schmidt, A. Receptor for Advanced Glycation Endproducts: A Multiligand Receptor Magnifying Cell Stress in Diverse Pathologic Settings. Adv. Drug Deliv. Rev. 2002, 54, 1615–1625. [Google Scholar] [CrossRef]
- Sugars, R.; Karlström, E.; Christersson, C.; Olsson, M.-L.; Wendel, M.; Fried, K. Expression of HMGB1 during Tooth Development. Cell Tissue Res. 2007, 327, 511–519. [Google Scholar] [CrossRef]
- Qi, S.C.; Cui, C.; Yan, Y.H.; Sun, G.H.; Zhu, S.R. Effects of High-Mobility Group Box 1 on the Proliferation and Odontoblastic Differentiation of Human Dental Pulp Cells. Int. Endod. J. 2013, 46, 1153–1163. [Google Scholar] [CrossRef] [PubMed]
- Gläser, R.; Meyer-Hoffert, U.; Harder, J.; Cordes, J.; Wittersheim, M.; Kobliakova, J.; Fölster-Holst, R.; Proksch, E.; Schröder, J.-M.; Schwarz, T. The Antimicrobial Protein Psoriasin (S100A7) Is Upregulated in Atopic Dermatitis and after Experimental Skin Barrier Disruption. J. Investig. Dermatol. 2009, 129, 641–649. [Google Scholar] [Green Version]
- Meyer, J.E.; Harder, J.; Sipos, B.; Maune, S.; Klöppel, G.; Bartels, J.; Schröder, J.-M.; Gläser, R. Psoriasin (S100A7) Is a Principal Antimicrobial Peptide of the Human Tongue. Mucosal Immunol. 2008, 1, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Nakatomi, M.; Ida-Yonemochi, H.; Ohshima, H. Osteopontin Is Essential for Type I Collagen Secretion in Reparative Dentin. J. Dent. Res. 2016, 95, 1034–1041. [Google Scholar] [CrossRef] [PubMed]
- Horibe, K.; Hosoya, A.; Hiraga, T.; Nakamura, H. Expression and Localization of CRAMP in Rat Tooth Germ and during Reparative Dentin Formation. Clin. Oral Investig. 2018, 22, 2559–2566. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Jiang, H.; Gong, Q.; Fan, C.; Huang, Y.; Ling, J. Expression of High Mobility Group Box 1 in Inflamed Dental Pulp and Its Chemotactic Effect on Dental Pulp Cells. Biochem. Biophys. Res. Commun. 2014, 450, 1547–1552. [Google Scholar] [CrossRef] [PubMed]
- Rombouts, C.; Jeanneau, C.; Bakopoulou, A.; About, I. Dental Pulp Stem Cell Recruitment Signals within Injured Dental Pulp Tissue. Dent. J. 2016, 4, 8. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Zhang, Q.Z.; Karabucak, B.; Le, A.D. DPSCs from Inflamed Pulp Modulate Macrophage Function via the TNF-α/IDO Axis. J. Dent. Res. 2016, 95, 1274–1281. [Google Scholar] [CrossRef] [PubMed]





| Protein ID | Protein Description | Protein Ration | Peptide PEP * |
|---|---|---|---|
| tr|A0A024R1X 8|A0A024R1X8_HUMAN | Junction plakoglobin | 9999 | 0.005 |
| sp|P31151|S10A7_HUMAN | Protein S100-A7 | 9999 | 1.095 × 10−6 |
| sp|P05109|S10A8_HUMAN | Protein S100-A8 | in d-DMCs only | 1.270 × 10−27 |
| sp|P12273|PIP_HUMAN | Prolactin-inducible protein | in d-DMCs only | 0.036 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Komichi, S.; Takahashi, Y.; Okamoto, M.; Ali, M.; Watanabe, M.; Huang, H.; Nakai, T.; Cooper, P.; Hayashi, M. Protein S100-A7 Derived from Digested Dentin Is a Critical Molecule for Dentin Pulp Regeneration. Cells 2019, 8, 1002. https://doi.org/10.3390/cells8091002
Komichi S, Takahashi Y, Okamoto M, Ali M, Watanabe M, Huang H, Nakai T, Cooper P, Hayashi M. Protein S100-A7 Derived from Digested Dentin Is a Critical Molecule for Dentin Pulp Regeneration. Cells. 2019; 8(9):1002. https://doi.org/10.3390/cells8091002
Chicago/Turabian StyleKomichi, Shungo, Yusuke Takahashi, Motoki Okamoto, Manahil Ali, Masakatsu Watanabe, Hailing Huang, Takeo Nakai, Paul Cooper, and Mikako Hayashi. 2019. "Protein S100-A7 Derived from Digested Dentin Is a Critical Molecule for Dentin Pulp Regeneration" Cells 8, no. 9: 1002. https://doi.org/10.3390/cells8091002
APA StyleKomichi, S., Takahashi, Y., Okamoto, M., Ali, M., Watanabe, M., Huang, H., Nakai, T., Cooper, P., & Hayashi, M. (2019). Protein S100-A7 Derived from Digested Dentin Is a Critical Molecule for Dentin Pulp Regeneration. Cells, 8(9), 1002. https://doi.org/10.3390/cells8091002








