SRSF3 Is a Critical Requirement for Inclusion of Exon 3 of BIS Pre-mRNA
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Transfection
2.2. Reverse Transcription PCR (RT-PCR) and Quantitative Real-Time PCR (qRT-PCR)
2.3. Western Blot Assay and Subcellular Fractionation
2.4. Construction of Expression Vectors
2.5. Biotin Pull-Down Assay
2.6. Statistics
3. Results
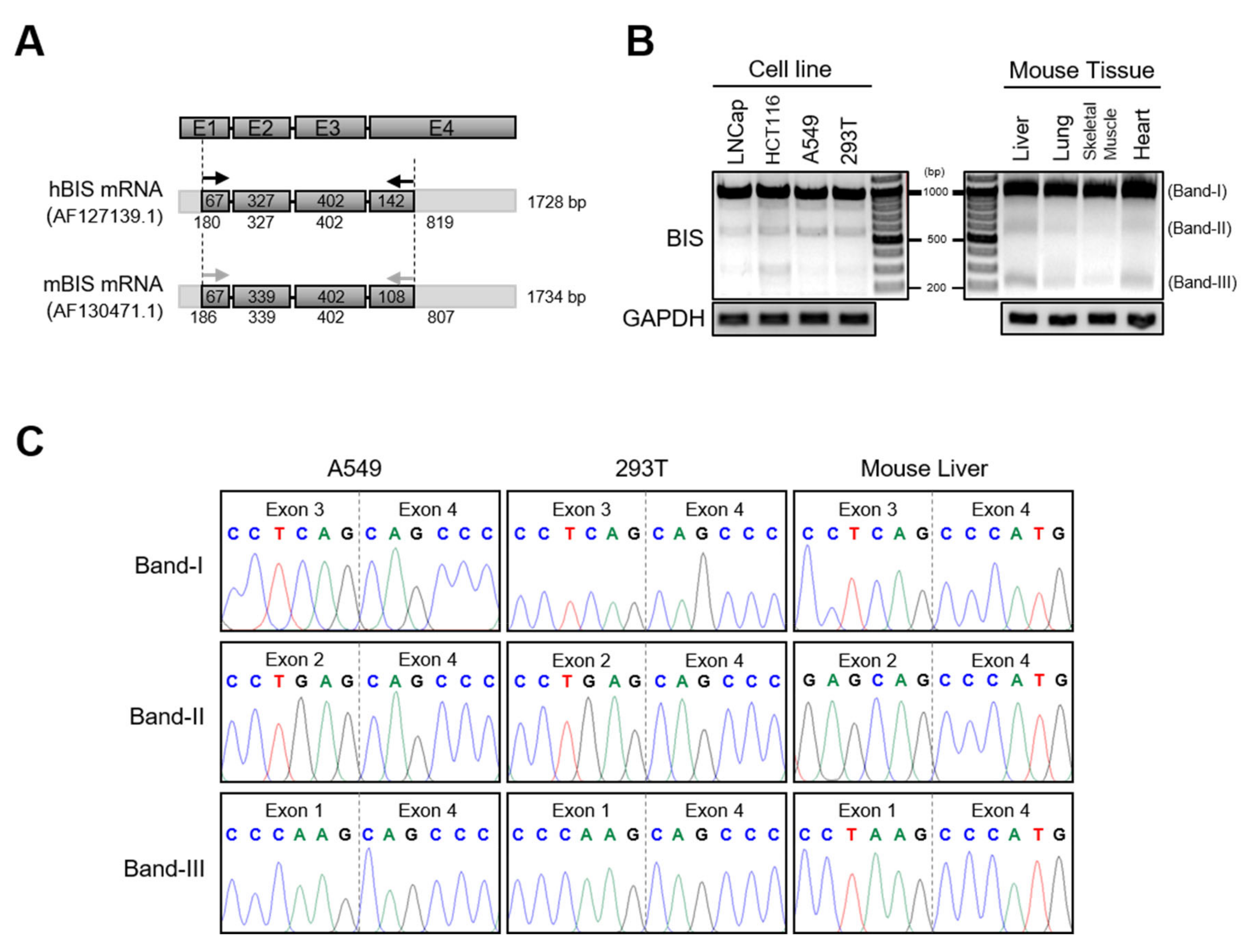
3.1. Identification of BIS mRNA Isoforms in Human Cell Lines and Mouse Tissues
3.2. SRSF3 Is Involved in the Inclusion of Exon 3 of BIS Pre-mRNA
3.3. Interaction of SRSF3 with Exon 3 Is Essential for the Inclusion of Exon 3 of BIS Pre-mRNA
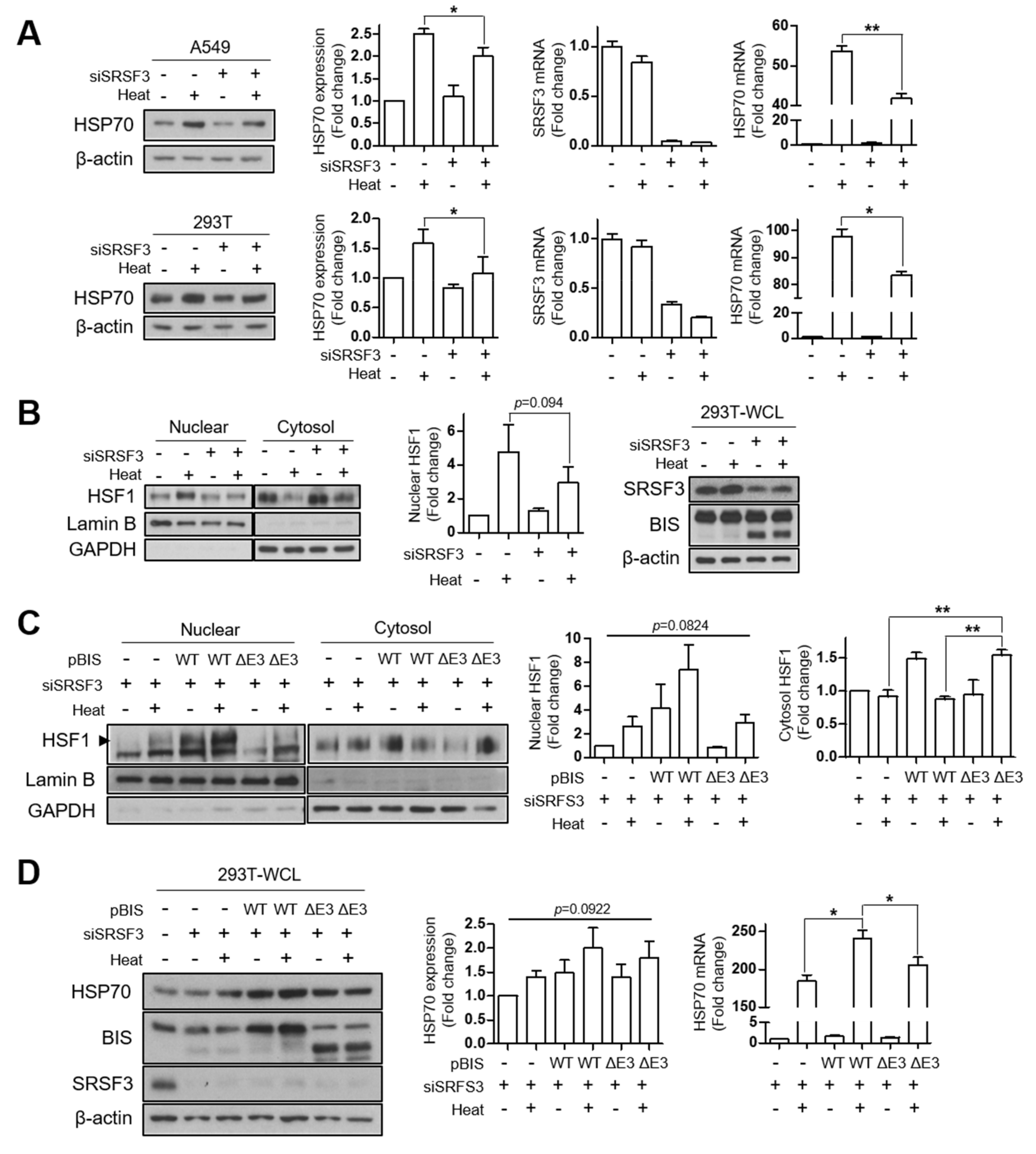
3.4. Depletion of SRSF3 Inhibited HSF1 Translocation by Heat Shock Stress
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sturner, E.; Behl, C. The Role of the Multifunctional BAG3 Protein in Cellular Protein Quality Control and in Disease. Front. Mol. Neurosci. 2017, 10, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.-H.; Takahashi, T.; Yasuhara, N.; Inazawa, J.; Kamada, S.; Tsujimoto, Y. Bis, a Bcl-2-binding protein that synergizes with Bcl-2 in preventing cell death. Oncogene 1999, 18, 6183–6190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosati, A.; Graziano, V.; De Laurenzi, V.; Pascale, M.; Turco, M.C. BAG3: A multifaceted protein that regulates major cell pathways. Cell Death Dis. 2011, 2, e141. [Google Scholar] [CrossRef]
- Myers, V.D.; McClung, J.M.; Wang, J.; Tahrir, F.G.; Gupta, M.K.; Gordon, J.; Kontos, C.H.; Khalili, K.; Cheung, J.Y.; Feldman, A.M. The Multifunctional Protein BAG3: A Novel Therapeutic Target in Cardiovascular Disease. JACC Basic Transl. Sci. 2018, 3, 122–131. [Google Scholar] [CrossRef] [PubMed]
- De Marco, M.; Basile, A.; Iorio, V.; Festa, M.; Falco, A.; Ranieri, B.; Pascale, M.; Sala, G.; Remondelli, P.; Capunzo, M.; et al. Role of BAG3 in cancer progression: A therapeutic opportunity. Semin. Cell Dev. Biol. 2018, 78, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Homma, S.; Iwasaki, M.; Shelton, G.D.; Engvall, E.; Reed, J.C.; Takayama, S. BAG3 deficiency results in fulminant myopathy and early lethality. Am. J. Pathol. 2006, 169, 761–773. [Google Scholar] [CrossRef] [Green Version]
- Yoo, H.J.; Im, C.N.; Youn, D.Y.; Yun, H.H.; Lee, J.H. Bis is Induced by Oxidative Stress via Activation of HSF1. Korean J. Physiol. Pharmacol. 2014, 18, 403–409. [Google Scholar] [CrossRef] [Green Version]
- Franceschelli, S.; Rosati, A.; Lerose, R.; De Nicola, S.; Turco, M.C.; Pascale, M. bag3 gene expression is regulated by heat shock factor 1. J. Cell Physiol. 2008, 215, 575–577. [Google Scholar] [CrossRef]
- Song, S.; Kole, S.; Precht, P.; Pazin, M.J.; Bernier, M. Activation of heat shock factor 1 plays a role in pyrrolidine dithiocarbamate-mediated expression of the co-chaperone BAG3. Int J. Biochem. Cell Biol. 2010, 42, 1856–1863. [Google Scholar] [CrossRef] [Green Version]
- Cesaro, E.; Montano, G.; Rosati, A.; Crescitelli, R.; Izzo, P.; Turco, M.C.; Costanzo, P. WT1 protein is a transcriptional activator of the antiapoptotic bag3 gene. Leukemia 2010, 24, 1204–1206. [Google Scholar] [CrossRef]
- Gentilella, A.; Passiatore, G.; Deshmane, S.; Turco, M.C.; Khalili, K. Activation of BAG3 by Egr-1 in response to FGF-2 in neuroblastoma cells. Oncogene 2008, 27, 5011–5018. [Google Scholar] [CrossRef] [Green Version]
- Ben Aicha, S.; Lessard, J.; Pelletier, M.; Fournier, A.; Calvo, E.; Labrie, C. Transcriptional profiling of genes that are regulated by the endoplasmic reticulum-bound transcription factor AIbZIP/CREB3L4 in prostate cells. Physiol. Genom. 2007, 31, 295–305. [Google Scholar] [CrossRef] [Green Version]
- Bruno, A.P.; Festa, M.; Dal Piaz, F.; Rosati, A.; Turco, M.C.; Giuditta, A.; Marzullo, L. Identification of a synaptosome-associated form of BAG3 protein. Cell Cycle 2008, 7, 3104–3105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, Z.X.; Meng, X.; Zhang, H.Y.; Guan, Y.; Wang, H.Q. Caspase-dependent cleavage of BAG3 in proteasome inhibitors-induced apoptosis in thyroid cancer cells. Biochem. Biophys. Res. Commun. 2008, 369, 894–898. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Q.; Meng, X.; Gao, Y.Y.; Liu, B.Q.; Niu, X.F.; Zhang, H.Y.; Du, Z.X. Characterization of BAG3 cleavage during apoptosis of pancreatic cancer cells. J. Cell Physiol. 2010, 224, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, M.; Homma, S.; Hishiya, A.; Dolezal, S.J.; Reed, J.C.; Takayama, S. BAG3 regulates motility and adhesion of epithelial cancer cells. Cancer Res. 2007, 67, 10252–10259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.J.; Lee, J.S.; Cui, M.N.; Yun, H.H.; Kim, H.Y.; Lee, S.H.; Lee, J.H. BIS targeting induces cellular senescence through the regulation of 14-3-3 zeta/STAT3/SKP2/p27 in glioblastoma cells. Cell Death Dis. 2014, 5, e1537. [Google Scholar] [CrossRef]
- Kim, H.Y.; Kim, Y.S.; Yun, H.H.; Im, C.N.; Ko, J.H.; Lee, J.H. ERK-mediated phosphorylation of BIS regulates nuclear translocation of HSF1 under oxidative stress. Exp. Mol. Med. 2016, 48, e260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baek, J.Y.; Yun, H.H.; Im, C.N.; Ko, J.H.; Jeong, S.M.; Lee, J.H. BIS overexpression does not affect the sensitivity of HEK 293T cells against apoptosis. Mol. Cell Toxicol. 2017, 13, 95–103. [Google Scholar] [CrossRef]
- Busch, A.; Hertel, K.J. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip. Rev. RNA 2012, 3, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Jeong, S. SR Proteins: Binders, Regulators, and Connectors of RNA. Mol. Cells 2017, 40, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Kornblihtt, A.R.; Schor, I.E.; Allo, M.; Dujardin, G.; Petrillo, E.; Munoz, M.J. Alternative splicing: A pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Bio. 2013, 14, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, L.-N.; Cheng, L.; Tu, S.; Guo, S.-J.; Le, H.-Y.; Xiong, Q.; Mo, R.; Li, C.-Y.; Jeong, J.-S.; et al. Bcl2-associated Athanogene 3 Interactome Analysis Reveals a New Role in Modulating Proteasome Activity. Mol. Cell. Proteom. 2013, 12, 2804. [Google Scholar] [CrossRef] [Green Version]
- Guisbert, K.S.K.; Guisbert, E. SF3B1 is a stress-sensitive splicing factor that regulates both HSF1 concentration and activity. PLoS ONE 2017, 12, e0176382. [Google Scholar] [CrossRef] [Green Version]
- Corbo, C.; Orru, S.; Salvatore, F. SRp20: An overview of its role in human diseases. Biochem. Bioph. Res. Commun. 2013, 436, 1–5. [Google Scholar] [CrossRef]
- Tang, Y.; Horikawa, I.; Ajiro, M.; Robles, A.I.; Fujita, K.; Mondal, A.M.; Stauffer, J.K.; Zheng, Z.M.; Harris, C.C. Downregulation of splicing factor SRSF3 induces p53 beta, an alternatively spliced isoform of p53 that promotes cellular senescence. Oncogene 2013, 32, 2792–2798. [Google Scholar] [CrossRef] [Green Version]
- Loh, T.J.; Moon, H.; Jang, H.N.; Liu, Y.; Choi, N.; Shen, S.F.; Williams, D.R.; Jung, D.W.; Zheng, X.; Shen, H. SR proteins regulate V-6 exon splicing of CD44 pre-mRNA. BMB Rep. 2016, 49, 612–616. [Google Scholar] [CrossRef] [Green Version]
- Jia, R.; Li, C.L.; Mccoy, J.P.; Deng, C.X.; Zheng, Z.M. SRp20 is a proto-oncogene critical for cell proliferation and tumor induction and maintenance. Int. J. Biol. Sci. 2010, 6, 806–826. [Google Scholar] [CrossRef]
- Sen, S.; Talukdar, I.; Webster, N.J.G. SRp20 and CUG-BP1 Modulate Insulin Receptor Exon 11 Alternative Splicing. Mol. Cell. Biol. 2009, 29, 871–880. [Google Scholar] [CrossRef] [Green Version]
- Munoz, U.; Puche, J.E.; Hannivoort, R.; Lang, U.E.; Cohen-Naftaly, M.; Friedman, S.L. Hepatocyte Growth Factor Enhances Alternative Splicing of the Kruppel-like Factor 6 (KLF6) Tumor Suppressor to Promote Growth through SRSF1. Mol. Cancer Res. 2012, 10, 1216–1227. [Google Scholar] [CrossRef] [Green Version]
- Jumaa, H.; Nielsen, P.J. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 1997, 16, 5077–5085. [Google Scholar] [CrossRef]
- Galiana-Arnoux, D.; Lejeune, F.; Gesnel, M.C.; Stevenin, J.; Breathnach, R.; Del Gatto-Konczak, F. The CD44 alternative v9 exon contains a splicing enhancer responsive to the SR proteins 9G8, ASF/SF2, and SRp20. J. Biol. Chem. 2003, 278, 32943–32953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, J.H.; Jia, J.; Jia, R. PTBP1 and PTBP2 impaired autoregulation of SRSF3 in cancer cells. Sci. Rep. 2015, 5, 14548. [Google Scholar] [CrossRef] [Green Version]
- Che, Y.Y.; Fu, L. Aberrant expression and regulatory network of splicing factor-SRSF3 in tumors. J. Cancer 2020, 11, 3502–3511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Judge, L.M.; Perez-Bermejo, J.A.; Truong, A.; Ribeiro, A.J.S.; Yoo, J.C.; Jensen, C.L.; Mandegar, M.A.; Huebsch, N.; Kaake, R.M.; So, P.L.; et al. A BAG3 chaperone complex maintains cardiomyocyte function during proteotoxic stress. JCI Insight. 2017, 2, e94623. [Google Scholar] [CrossRef] [PubMed]
- Gentilella, A.; Khalili, K. Autoregulation of Co-Chaperone BAG3 Gene Transcription. J. Cell. Biochem. 2009, 108, 1117–1124. [Google Scholar] [CrossRef] [Green Version]
- Cui, M.N.; Yun, H.H.; Lee, N.E.; Kim, H.Y.; Im, C.N.; Kim, Y.S.; Lee, J.H. Depletion of BIS sensitizes A549 cells to treatment with cisplatin. Mol. Cell. Toxicol. 2016, 12, 63–71. [Google Scholar] [CrossRef]
- Jin, Y.H.; Ahn, S.G.; Kim, S.A. BAG3 affects the nucleocytoplasmic shuttling of HSF1 upon heat stress. Biochem. Bioph. Res. Commun. 2015, 464, 561–567. [Google Scholar] [CrossRef]
- Kano, S.; Nishida, K.; Kurebe, H.; Nishiyama, C.; Kita, K.; Akaike, Y.; Kajita, K.; Kurokawa, K.; Masuda, K.; Kuwano, Y.; et al. Oxidative stress-inducible truncated serine/arginine-rich splicing factor 3 regulates interleukin-8 production in human colon cancer cells. Am. J. Physiol. Cell Physiol. 2014, 306, C250–C262. [Google Scholar] [CrossRef] [Green Version]
- Kano, S.; Nishida, K.; Nishiyama, C.; Akaike, Y.; Kajita, K.; Kurokawa, K.; Masuda, K.; Kuwano, Y.; Tanahashi, T.; Rokutan, K. Truncated serine/arginine-rich splicing factor 3 accelerates cell growth through up-regulating c-Jun expression. J. Med. Invest. 2013, 60, 228–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sariyer, R.; De-Simone, F.I.; Donadoni, M.; Hoek, J.B.; Chang, S.L.; Sariyer, I.K. Alcohol-Mediated Missplicing of Mcl-1 Pre-mRNA is Involved in Neurotoxicity. Alcohol. Clin. Exp. Res. 2017, 41, 1715–1724. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baek, J.-Y.; Yun, H.-H.; Jung, S.-Y.; Lee, J.; Yoo, K.; Lee, J.-H. SRSF3 Is a Critical Requirement for Inclusion of Exon 3 of BIS Pre-mRNA. Cells 2020, 9, 2325. https://doi.org/10.3390/cells9102325
Baek J-Y, Yun H-H, Jung S-Y, Lee J, Yoo K, Lee J-H. SRSF3 Is a Critical Requirement for Inclusion of Exon 3 of BIS Pre-mRNA. Cells. 2020; 9(10):2325. https://doi.org/10.3390/cells9102325
Chicago/Turabian StyleBaek, Ji-Ye, Hye-Hyeon Yun, Soon-Young Jung, Jeehan Lee, Kyunghyun Yoo, and Jeong-Hwa Lee. 2020. "SRSF3 Is a Critical Requirement for Inclusion of Exon 3 of BIS Pre-mRNA" Cells 9, no. 10: 2325. https://doi.org/10.3390/cells9102325




