EDS1-Dependent Cell Death and the Antioxidant System in Arabidopsis Leaves is Deregulated by the Mammalian Bax
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Induction of the Bax Gene
2.3. Biometric Measurements
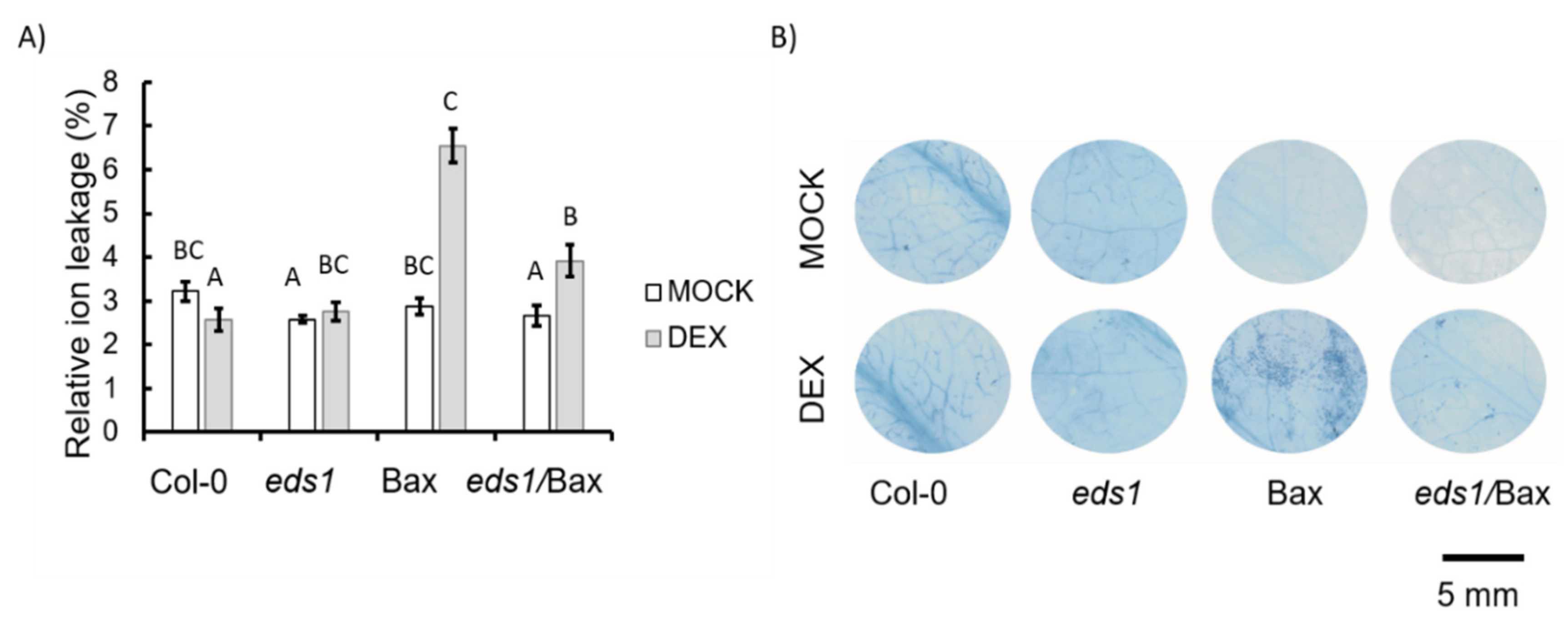
2.4. Relative Electrolyte Leakage
2.5. Trypan Blue Staining
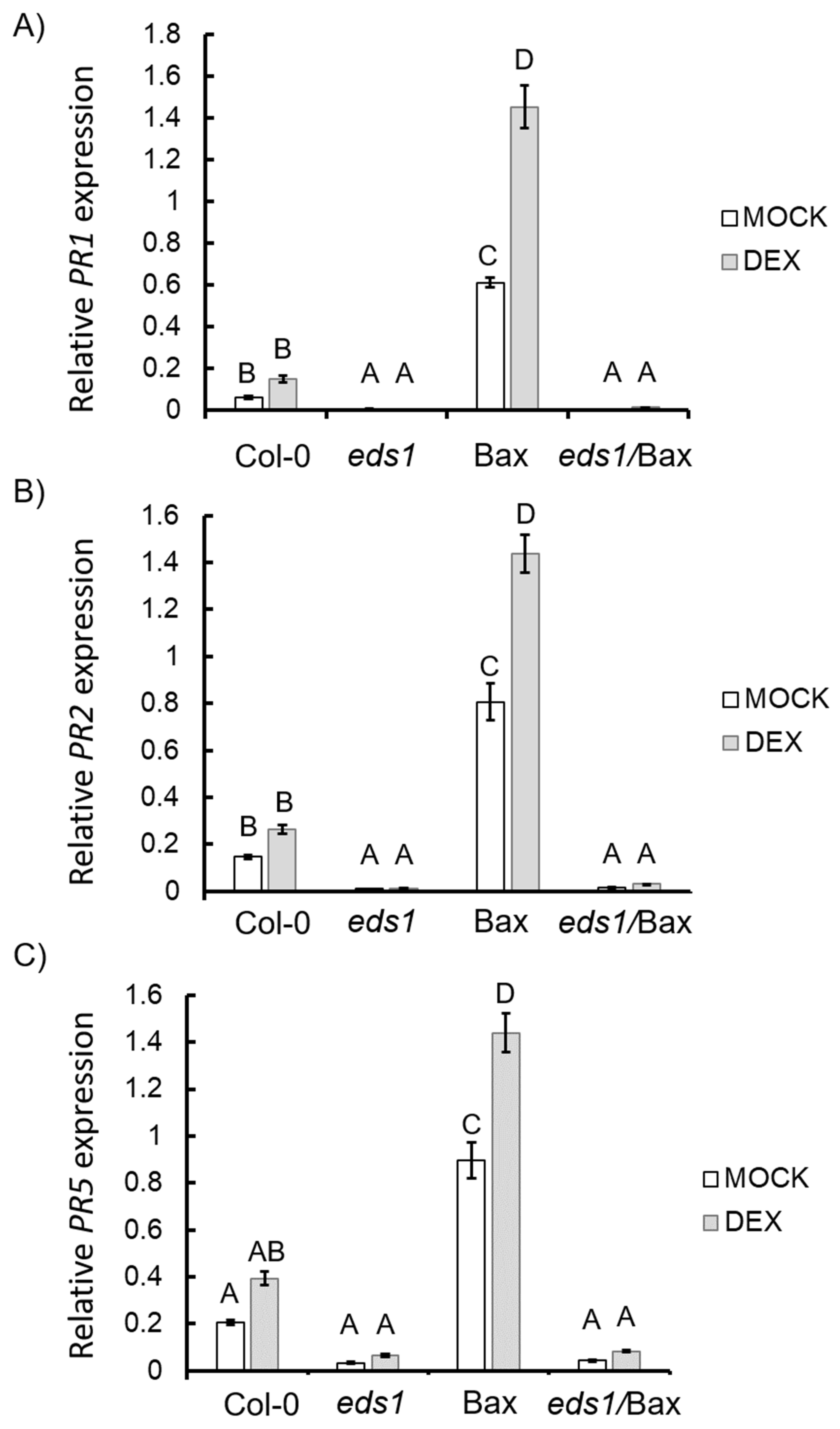
2.6. RNA Isolation, cDNA Synthesis, and Quantitative Real-Time PCR
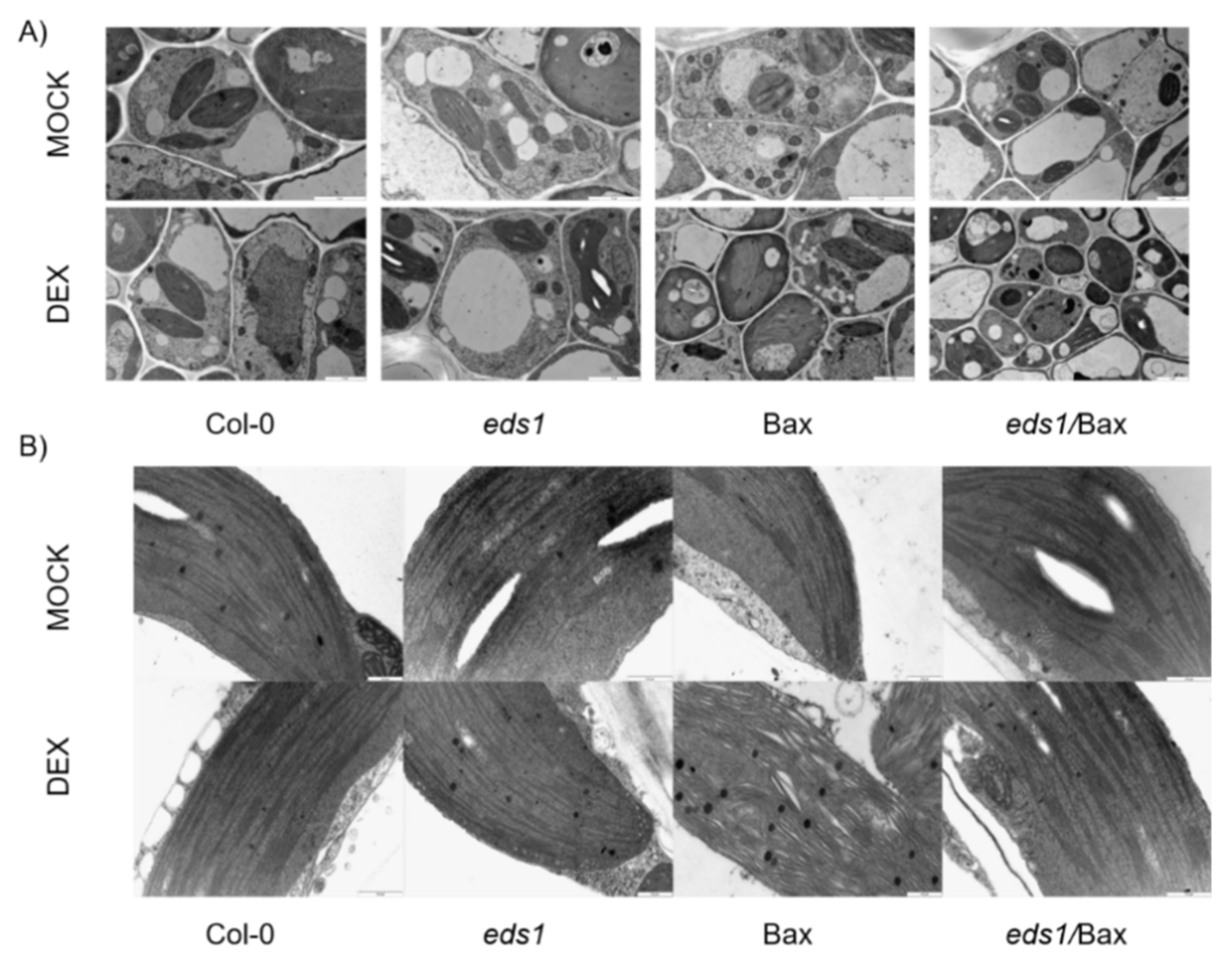
2.7. Anatomic and Ultrastructural Analysis
2.8. Protein Extraction
2.9. Total Soluble Protein Content
2.10. Measurements of Antioxidant Enzyme Activity
2.11. H2O2 Measurement and DAB Staining
2.12. Chlorophyll a Fluorescence Measurements
3. Results
3.1. Artificially Bax-Induced HR-Like Cell Death Propagation and Growth Inhibition Depend on EDS1
3.2. EDS1-Dependent Regulation of Antioxidant System and ROS Metabolism is Required for Bax-Induced HR-Like Cell Death
3.3. EDS1 is Essential for the Induction of Hypersensitive Response and Senescence During Bax-Induced Plant Cell Death
3.4. Arabidopsis thaliana Type I Metacaspases Seem to Be Involved in Bax-Induced Plant Cell Death
3.5. Improved Antioxidant System and Impaired HR Result in the Mitigation of Mammalian Bax-Induced Cellular Organelle Destruction
3.6. EDS1 Does Not Significantly Affects the Efficiency of PSII During Bax-Induced Plant Cell Death
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Domínguez, F.; Cejudo, F.J. Programmed cell death (PCD): An essential process of cereal seed development and germination. Front. Plant Sci. 2014, 5, 366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukuda, H. Programmed cell death of tracheary elements as a paradigm in plants. Plant Mol. Biol. 2000, 44, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Jabs, T.; Dietrich, R.A.; Dangl, J.L. Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science 1996, 273, 1853–1856. [Google Scholar] [CrossRef] [PubMed]
- Mühlenbock, P.; Plaszczyca, M.; Plaszczyca, M.; Mellerowicz, E.; Karpinski, S. Lysigenous Aerenchyma Formation in Arabidopsis Is Controlled by LESION SIMULATING DISEASE1. Plant Cell 2007, 19, 3819–3830. [Google Scholar] [CrossRef] [Green Version]
- Elmore, S. Apoptosis: A Review of Programmed Cell Death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef] [PubMed]
- Pennell, R.I.; Lamb, C. Programmed Cell Death in Plants. Plant Cell 1997, 9, 1157–1168. [Google Scholar] [CrossRef] [Green Version]
- Kawai-Yamada, M.; Jin, L.; Yoshinaga, K.; Hirata, A.; Uchimiya, H. Mammalian Bax-induced plant cell death can be down-regulated by overexpression of Arabidopsis Bax Inhibitor-1 (AtBI-1). Proc. Natl. Acad. Sci. USA 2001, 98, 12295–12300. [Google Scholar] [CrossRef] [Green Version]
- Lacomme, C.; Cruz, S.S. Bax-induced cell death in tobacco is similar to the hypersensitive response. Proc. Natl. Acad. Sci. USA 1999, 96, 7956–7961. [Google Scholar] [CrossRef] [Green Version]
- Aravind, L.; Dixit, V.M.; Koonin, E.V. The domains of death: Evolution of the apoptosis machinery. Trends Biochem. Sci. 1999, 24, 47–53. [Google Scholar] [CrossRef]
- Schinzel, A.; Kaufmann, T.; Borner, C. Bcl-2 family members: Integrators of survival and death signals in physiology and pathology [corrected]. Biochim. Biophys. Acta 2004, 1644, 95–105. [Google Scholar] [CrossRef]
- Youle, R.J.; Strasser, A. The BCL-2 protein family: Opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008, 9, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Kvansakul, M.; Yang, H.; Fairlie, W.D.; Czabotar, P.E.; Fischer, S.F.; Perugini, M.A.; Huang, D.C.S.; Colman, P.M. Vaccinia virus anti-apoptotic F1L is a novel Bcl-2-like domain-swapped dimer that binds a highly selective subset of BH3-containing death ligands. Cell Death Differ. 2008, 15, 1564–1571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinou, J.-C.; Youle, R.J. Mitochondria in Apoptosis: Bcl-2 Family Members and Mitochondrial Dynamics. Dev. Cell 2011, 21, 92–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaux, D.L. Apoptogenic factors released from mitochondria. Biochim. Biophys. Acta 2011, 1813, 546–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kroemer, G.; Galluzzi, L.; Brenner, C. Mitochondrial membrane permeabilization in cell death. Physiol. Rev. 2007, 87, 99–163. [Google Scholar] [CrossRef] [PubMed]
- Yoshinaga, K.; Arimura, S.; Hirata, A.; Niwa, Y.; Yun, D.-J.; Tsutsumi, N.; Uchimiya, H.; Kawai-Yamada, M. Mammalian Bax initiates plant cell death through organelle destruction. Plant Cell Rep. 2005, 24, 408–417. [Google Scholar] [CrossRef]
- Baek, D.; Nam, J.; Koo, Y.D.; Kim, D.H.; Lee, J.; Jeong, J.C.; Kwak, S.; Chung, W.S.; Lim, C.O.; Bahk, J.D.; et al. Bax-induced cell death of is meditated through reactive oxygen-dependent and -independent processes. Plant Mol. Biol. 2004, 56, 15–27. [Google Scholar] [CrossRef]
- Sanchez, P.; Zabala, M.D.T.; Grant, M. AtBI-1, a plant homologue of Bax Inhibitor-1, suppresses Bax-induced cell death in yeast and is rapidly upregulated during wounding and pathogen challenge. Plant J. 2000, 21, 393–399. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, N.; Lam, E. Arabidopsis Bax inhibitor-1 functions as an attenuator of biotic and abiotic types of cell death. Plant J. 2006, 45, 884–894. [Google Scholar] [CrossRef]
- Wituszyńska, W.; Szechyńska-Hebda, M.; Sobczak, M.; Rusaczonek, A.; Kozłowska-Makulska, A.; Witoń, D.; Karpiński, S. Lesion simulating disease 1 and enhanced disease susceptibility 1 differentially regulate UV-C-induced photooxidative stress signalling and programmed cell death in Arabidopsis thaliana. Plant Cell Environ. 2015, 38, 315–330. [Google Scholar] [CrossRef]
- Gilroy, S.; Białasek, M.; Suzuki, N.; Górecka, M.; Devireddy, A.R.; Karpiński, S.; Mittler, R. ROS, Calcium, and Electric Signals: Key Mediators of Rapid Systemic Signaling in Plants. Plant Physiol. 2016, 171, 1606–1615. [Google Scholar] [CrossRef] [PubMed]
- Karpiński, S.; Szechyńska-Hebda, M.; Wituszyńska, W.; Burdiak, P. Light acclimation, retrograde signalling, cell death and immune defences in plants. Plant Cell Environ. 2013, 36, 736–744. [Google Scholar] [CrossRef] [PubMed]
- Aoyama, T.; Chua, N.H. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. Cell Mol. Biol. 1997, 11, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Glazebrook, J.; Rogers, E.E.; Ausubel, F.M. Isolation of Arabidopsis Mutants with Enhanced Disease Susceptibility by Direct Screening. Genetics 1996, 143, 973–982. [Google Scholar]
- Parker, J.E.; Holub, E.B.; Frost, L.N.; Falk, A.; Gunn, N.D.; Daniels, M.J. Characterization of EDS1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 1996, 8, 2033–2046. [Google Scholar]
- Falk, A.; Feys, B.J.; Frost, L.N.; Jones, J.D.G.; Daniels, M.J.; Parker, J.E. EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 1999, 96, 3292–3297. [Google Scholar] [CrossRef] [Green Version]
- Bernacki, M.J.; Czarnocka, W.; Rusaczonek, A.; Witoń, D.; Kęska, S.; Czyż, J.; Szechyńska-Hebda, M.; Karpiński, S. LSD1, EDS1 and PAD4-dependent conditional correlation among salicylic acid, hydrogen peroxide, water use efficiency, and seed yield in Arabidopsis thaliana. Physiol. Plant. 2018, 165, 369–382. [Google Scholar] [CrossRef]
- Mateo, A.; Mühlenbock, P.; Rustérucci, C.; Chang, C.C.-C.; Miszalski, Z.; Karpinska, B.; Parker, J.E.; Mullineaux, P.M.; Karpinski, S. LESION SIMULATING DISEASE 1 is required for acclimation to conditions that promote excess excitation energy. Plant Physiol. 2004, 136, 2818–2830. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.-F.; Xu, L.; Tan, W.-J.; Chen, L.; Qi, H.; Xie, L.-J.; Chen, M.-X.; Liu, B.-Y.; Yu, L.-J.; Yao, N.; et al. Disruption of the Arabidopsis Defense Regulator Genes SAG101, EDS1, and PAD4 Confers Enhanced Freezing Tolerance. Mol. Plant 2015, 8, 1536–1549. [Google Scholar] [CrossRef] [Green Version]
- Jirage, D.; Tootle, T.L.; Reuber, T.L.; Frost, L.N.; Feys, B.J.; Parker, J.E.; Ausubel, F.M.; Glazebrook, J. Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA 1999, 96, 13583–13588. [Google Scholar] [CrossRef] [Green Version]
- Rietz, S.; Stamm, A.; Malonek, S.; Wagner, S.; Becker, D.; Medina-Escobar, N.; Vlot, A.C.; Feys, B.J.; Niefind, K.; Parker, J.E. Different roles of Enhanced Disease Susceptibility1 (EDS1) bound to and dissociated from Phytoalexin Deficient4 (PAD4) in Arabidopsis immunity. New Phytol. 2011, 191, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Feys, B.J.; Moisan, L.J.; Newman, M.-A.; Parker, J.E. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 2001, 20, 5400–5411. [Google Scholar] [CrossRef] [PubMed]
- Feys, B.J.; Wiermer, M.; Bhat, R.A.; Moisan, L.J.; Medina-Escobar, N.; Neu, C.; Cabral, A.; Parker, J.E. Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 2005, 17, 2601–2613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, S.; Jeong, R.-D.; Venugopal, S.C.; Lapchyk, L.; Navarre, D.; Kachroo, A.; Kachroo, P. SAG101 Forms a Ternary Complex with EDS1 and PAD4 and Is Required for Resistance Signaling against Turnip Crinkle Virus. PLOS Pathog. 2011, 7, e1002318. [Google Scholar] [CrossRef] [Green Version]
- Czarnocka, W.; Van Der Kelen, K.; Willems, P.; Szechynska-Hebda, M.; Shahnejat-Bushehri, S.; Balazadeh, S.; Rusaczonek, A.; Mueller-Roeber, B.; Van Breusegem, F.; Karpinski, S. The dual role of LESION SIMULATING DISEASE 1 as a condition-dependent scaffold protein and transcription regulator. Plant Cell Environ. 2017, 40, 2644–2662. [Google Scholar] [CrossRef] [Green Version]
- Oracz, K.; Karpiński, S. Phytohormones Signaling Pathways and ROS Involvement in Seed Germination. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Hruz, T.; Laule, O.; Szabo, G.; Wessendorp, F.; Bleuler, S.; Oertle, L.; Widmayer, P.; Gruissem, W.; Zimmermann, P. Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinform. 2008, 2008, 420747. [Google Scholar] [CrossRef]
- Ramakers, C.; Ruijter, J.M.; Deprez, R.H.L.; Moorman, A.F.M. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef]
- Golinowski, W.; Grundler, F.M.W.; Sobczak, M. Changes in the structure ofArabidopsis thaliana during female development of the plant-parasitic nematode Heterodera schachtii. Protoplasma 1996, 194, 103–116. [Google Scholar] [CrossRef]
- Labudda, M.; Różańska, E.; Czarnocka, W.; Sobczak, M.; Dzik, J.M. Systemic changes in photosynthesis and reactive oxygen species homeostasis in shoots of Arabidopsis thaliana infected with the beet cyst nematode Heterodera schachtii. Mol. Plant Pathol. 2018, 19, 1690–1704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spector, T. Refinement of the coomassie blue method of protein quantitation. A simple and linear spectrophotometric assay for less than or equal to 0.5 to 50 microgram of protein. Anal. Biochem. 1978, 86, 142–146. [Google Scholar] [CrossRef]
- Kostyuk, V.A.; Potapovich, A.I. Superoxide—Driven oxidation of quercetin and a simple sensitive assay for determination of superoxide dismutase. Biochem. Int. 1989, 19, 1117–1124. [Google Scholar] [PubMed]
- Rusaczonek, A.; Czarnocka, W.; Kacprzak, S.; Witoń, D.; Ślesak, I.; Szechyńska-Hebda, M.; Gawroński, P.; Karpiński, S. Role of phytochromes A and B in the regulation of cell death and acclimatory responses to UV stress in Arabidopsis thaliana. J. Exp. Bot. 2015, 66, 6679–6695. [Google Scholar] [CrossRef] [Green Version]
- Czarnocka, W.; Karpiński, S. Friend or foe? Reactive oxygen species production, scavenging and signaling in plant response to environmental stresses. Free Radic. Biol. Med. 2018, 122, 4–20. [Google Scholar] [CrossRef]
- Zurbriggen, M.D.; Carrillo, N.; Hajirezaei, M.-R. ROS signaling in the hypersensitive response. Plant Signal. Behav. 2010, 5, 393–396. [Google Scholar] [CrossRef] [Green Version]
- Rustérucci, C.; Aviv, D.H.; Holt, B.F.; Dangl, J.L.; Parker, J.E. The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell 2001, 13, 2211–2224. [Google Scholar] [CrossRef] [Green Version]
- Gadjev, I.; Vanderauwera, S.; Gechev, T.S.; Laloi, C.; Minkov, I.N.; Shulaev, V.; Apel, K.; Inzé, D.; Mittler, R.; Breusegem, F.V. Transcriptomic Footprints Disclose Specificity of Reactive Oxygen Species Signaling in Arabidopsis. Plant Physiol. 2006, 141, 436–445. [Google Scholar] [CrossRef] [Green Version]
- Czarnocka, W.; Fichman, Y.; Bernacki, M.; Różańska, E.; Sańko-Sawczenko, I.; Mittler, R.; Karpiński, S. FMO1 Is Involved in Excess Light Stress-Induced Signal Transduction and Cell Death Signaling. Cells 2020, 9, 2163. [Google Scholar] [CrossRef]
- Op den Camp, R.G.L.; Przybyla, D.; Ochsenbein, C.; Laloi, C.; Kim, C.; Danon, A.; Wagner, D.; Hideg, E.; Göbel, C.; Feussner, I.; et al. Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 2003, 15, 2320–2332. [Google Scholar] [CrossRef] [Green Version]
- Carmody, M.; Crisp, P.A.; d’Alessandro, S.; Ganguly, D.; Gordon, M.; Havaux, M.; Albrecht-Borth, V.; Pogson, B.J. Uncoupling High Light Responses from Singlet Oxygen Retrograde Signaling and Spatial-Temporal Systemic Acquired Acclimation1[OPEN]. Plant Physiol. 2016, 171, 1734–1749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pontier, D.; Gan, S.; Amasino, R.M.; Roby, D.; Lam, E. Markers for hypersensitive response and senescence show distinct patterns of expression. Plant Mol. Biol. 1999, 39, 1243–1255. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.D.; Aarts, N.; Feys, B.J.; Dong, X.; Parker, J.E. Constitutive disease resistance requires EDS1 in the Arabidopsis mutants cpr1 and cpr6 and is partially EDS1-dependent in cpr5. Plant J. Cell Mol. Biol. 2001, 26, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Xing, D.; Chen, Z. Effects of mutations and constitutive overexpression of EDS1 and PAD4 on plant resistance to different types of microbial pathogens. Plant Sci. 2006, 171, 251–262. [Google Scholar] [CrossRef]
- Lohman, K.N.; Gan, S.; John, M.C.; Amasino, R.M. Molecular analysis of natural leaf senescence in Arabidopsis thaliana. Physiol. Plant. 1994, 92, 322–328. [Google Scholar] [CrossRef]
- Lord, C.E.N.; Gunawardena, A.H.L.A.N. Environmentally induced programmed cell death in leaf protoplasts of Aponogeton madagascariensis. Planta 2011, 233, 407–421. [Google Scholar] [CrossRef]
- Fagundes, D.; Bohn, B.; Cabreira, C.; Leipelt, F.; Dias, N.; Bodanese-Zanettini, M.H.; Cagliari, A. Caspases in plants: Metacaspase gene family in plant stress responses. Funct. Integr. Genomics 2015, 15, 639–649. [Google Scholar] [CrossRef]
- Yu, L.-H.; Kawai-Yamada, M.; Naito, M.; Watanabe, K.; Reed, J.C.; Uchimiya, H. Induction of mammalian cell death by a plant Bax inhibitor. FEBS Lett. 2002, 512, 308–312. [Google Scholar] [CrossRef] [Green Version]
- Alvarez, M.E.; Pennell, R.I.; Meijer, P.J.; Ishikawa, A.; Dixon, R.A.; Lamb, C. Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 1998, 92, 773–784. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.-C.; Du, Y.-Y.; An, G.-Y.; Zhou, Y.; Miao, C.; Song, C.-P. Analysis of Global Expression Profiles of Arabidopsis Genes Under Abscisic Acid and H2O2 Applications. J. Integr. Plant Biol. 2006, 48, 62–74. [Google Scholar] [CrossRef]
- Kliebenstein, D.J.; Monde, R.A.; Last, R.L. Superoxide dismutase in Arabidopsis: An eclectic enzyme family with disparate regulation and protein localization. Plant Physiol. 1998, 118, 637–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Starkov, A.A.; Polster, B.M.; Fiskum, G. Regulation of hydrogen peroxide production by brain mitochondria by calcium and Bax. J. Neurochem. 2002, 83, 220–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melis, A. Photosystem-II damage and repair cycle in chloroplasts: What modulates the rate of photodamage in vivo? Trends Plant Sci. 1999, 4, 130–135. [Google Scholar] [CrossRef]
- Pospíšil, P. Production of reactive oxygen species by photosystem II. Biochim. Biophys. Acta BBA—Bioenerg. 2009, 1787, 1151–1160. [Google Scholar] [CrossRef] [Green Version]
- Asada, K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006, 141, 391–396. [Google Scholar] [CrossRef] [Green Version]
- Bernacki, M.J.; Czarnocka, W.; Witoń, D.; Rusaczonek, A.; Szechyńska-Hebda, M.; Ślesak, I.; Dąbrowska-Bronk, J.; Karpiński, S. ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1) affects development, photosynthesis, and hormonal homeostasis in hybrid aspen (Populus tremula L. × P. tremuloides). J. Plant Physiol. 2018, 226, 91–102. [Google Scholar] [CrossRef]
- Morel, J.-B.; Dangl, J.L. The hypersensitive response and the induction of cell death in plants. Cell Death Differ. 1997, 4, 671–683. [Google Scholar] [CrossRef] [Green Version]
- Lam, E.; Kato, N.; Lawton, M. Programmed cell death, mitochondria and the plant hypersensitive response. Nature 2001, 411, 848–853. [Google Scholar] [CrossRef]
- Green, R.; Fluhr, R. UV-B-Induced PR-1 Accumulation Is Mediated by Active Oxygen Species. Plant Cell 1995, 7, 203–212. [Google Scholar] [CrossRef] [Green Version]
- Mur, L.A.J.; Kenton, P.; Lloyd, A.J.; Ougham, H.; Prats, E. The hypersensitive response; the centenary is upon us but how much do we know? J. Exp. Bot. 2008, 59, 501–520. [Google Scholar] [CrossRef] [Green Version]
- McLellan, H.; Gilroy, E.M.; Yun, B.-W.; Birch, P.R.J.; Loake, G.J. Functional redundancy in the Arabidopsis Cathepsin B gene family contributes to basal defence, the hypersensitive response and senescence. New Phytol. 2009, 183, 408–418. [Google Scholar] [CrossRef]
- El Oirdi, M.; Bouarab, K. Plant signalling components EDS1 and SGT1 enhance disease caused by the necrotrophic pathogen Botrytis cinerea. New Phytol. 2007, 175, 131–139. [Google Scholar] [CrossRef]
- Peng, J.-L.; Dong, H.-S.; Dong, H.-P.; Delaney, T.P.; Bonasera, J.M.; Beer, S.V. Harpin-elicited hypersensitive cell death and pathogen resistance require the NDR1 and EDS1 genes. Physiol. Mol. Plant Pathol. 2003, 62, 317–326. [Google Scholar] [CrossRef]
- Hu, G.; de Hart, A.K.A.; Li, Y.; Ustach, C.; Handley, V.; Navarre, R.; Hwang, C.-F.; Aegerter, B.J.; Williamson, V.M.; Baker, B. EDS1 in tomato is required for resistance mediated by TIR-class R genes and the receptor-like R gene Ve. Plant J. Cell Mol. Biol. 2005, 42, 376–391. [Google Scholar] [CrossRef]
- Clarke, J.D.; Liu, Y.; Klessig, D.F.; Dong, X. Uncoupling PR gene expression from NPR1 and bacterial resistance: Characterization of the dominant Arabidopsis cpr6-1 mutant. Plant Cell 1998, 10, 557–569. [Google Scholar] [CrossRef]
- Aarts, N.; Metz, M.; Holub, E.; Staskawicz, B.J.; Daniels, M.J.; Parker, J.E. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 1998, 95, 10306–10311. [Google Scholar] [CrossRef] [Green Version]
- Coll, N.S.; Vercammen, D.; Smidler, A.; Clover, C.; Van Breusegem, F.; Dangl, J.L.; Epple, P. Arabidopsis type I metacaspases control cell death. Science 2010, 330, 1393–1397. [Google Scholar] [CrossRef]
- Watanabe, N.; Lam, E. BAX Inhibitor-1 Modulates Endoplasmic Reticulum Stress-mediated Programmed Cell Death in Arabidopsis. J. Biol. Chem. 2008, 283, 3200–3210. [Google Scholar] [CrossRef] [Green Version]
- Kołodziejek, I.; Kozioł, J.; Wałęza, M.; Mostowska, A. Ultrastructure of Mesophyll Cells and Pigment Content in Senescing Leaves of Maize and Barley. J. Plant Growth Regul. 2003, 22, 217–227. [Google Scholar] [CrossRef]
- Kawai-Yamada, M.; Ohori, Y.; Uchimiya, H. Dissection of Arabidopsis Bax Inhibitor-1 Suppressing Bax–, Hydrogen Peroxide–, and Salicylic Acid–Induced Cell Death. Plant Cell 2004, 16, 21–32. [Google Scholar] [CrossRef] [Green Version]
- Greenhalf, W.; Stephan, C.; Chaudhuri, B. Role of mitochondria and C-terminal membrane anchor of Bcl-2 in Bax induced growth arrest and mortality in Saccharomyces cerevisiae. FEBS Lett. 1996, 380, 169–175. [Google Scholar] [CrossRef] [Green Version]
- Scherz-Shouval, R.; Elazar, Z. Regulation of autophagy by ROS: Physiology and pathology. Trends Biochem. Sci. 2011, 36, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Naderi, J.; Somayajulu-Nitu, M.; Mukerji, A.; Sharda, P.; Sikorska, M.; Borowy-Borowski, H.; Antonsson, B.; Pandey, S. Water-soluble formulation of Coenzyme Q10 inhibits Bax-induced destabilization of mitochondria in mammalian cells. Apoptosis 2006, 11, 1359–1369. [Google Scholar] [CrossRef] [PubMed]
- Møller, I.M.; Sweetlove, L.J. ROS signalling—Specificity is required. Trends Plant Sci. 2010, 15, 370–374. [Google Scholar] [CrossRef]
- Galvez-Valdivieso, G.; Mullineaux, P.M. The role of reactive oxygen species in signalling from chloroplasts to the nucleus. Physiol. Plant. 2010, 138, 430–439. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bernacki, M.J.; Czarnocka, W.; Zaborowska, M.; Różańska, E.; Labudda, M.; Rusaczonek, A.; Witoń, D.; Karpiński, S. EDS1-Dependent Cell Death and the Antioxidant System in Arabidopsis Leaves is Deregulated by the Mammalian Bax. Cells 2020, 9, 2454. https://doi.org/10.3390/cells9112454
Bernacki MJ, Czarnocka W, Zaborowska M, Różańska E, Labudda M, Rusaczonek A, Witoń D, Karpiński S. EDS1-Dependent Cell Death and the Antioxidant System in Arabidopsis Leaves is Deregulated by the Mammalian Bax. Cells. 2020; 9(11):2454. https://doi.org/10.3390/cells9112454
Chicago/Turabian StyleBernacki, Maciej Jerzy, Weronika Czarnocka, Magdalena Zaborowska, Elżbieta Różańska, Mateusz Labudda, Anna Rusaczonek, Damian Witoń, and Stanisław Karpiński. 2020. "EDS1-Dependent Cell Death and the Antioxidant System in Arabidopsis Leaves is Deregulated by the Mammalian Bax" Cells 9, no. 11: 2454. https://doi.org/10.3390/cells9112454






