LKB1 Differently Regulates Adipogenesis in Intramuscular and Subcutaneous Adipocytes through Metabolic and Cytokine-Related Signaling Pathways
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Isolation and Culture
2.2. Establishment of LKB1-Overexpressing Cells
2.3. Flow Cytometric Analysis
2.4. Oil-Red-O Staining
2.5. Protein Isolation and Western Blotting
2.6. Quantitative Real-Time PCR (qPCR)
2.7. RNA Isolation, Library Construction and RNA-Seq Analysis
2.8. Pathway Enrichment Assay
2.9. Statistical Analysis
3. Results
3.1. Overexpression of LKB1 Inhibits Pig IMF and SCF Preadipocytes Differentiation
3.2. LKB1 Overexpression Alters the Transcriptional Profile of Pig SCF Adipocytes
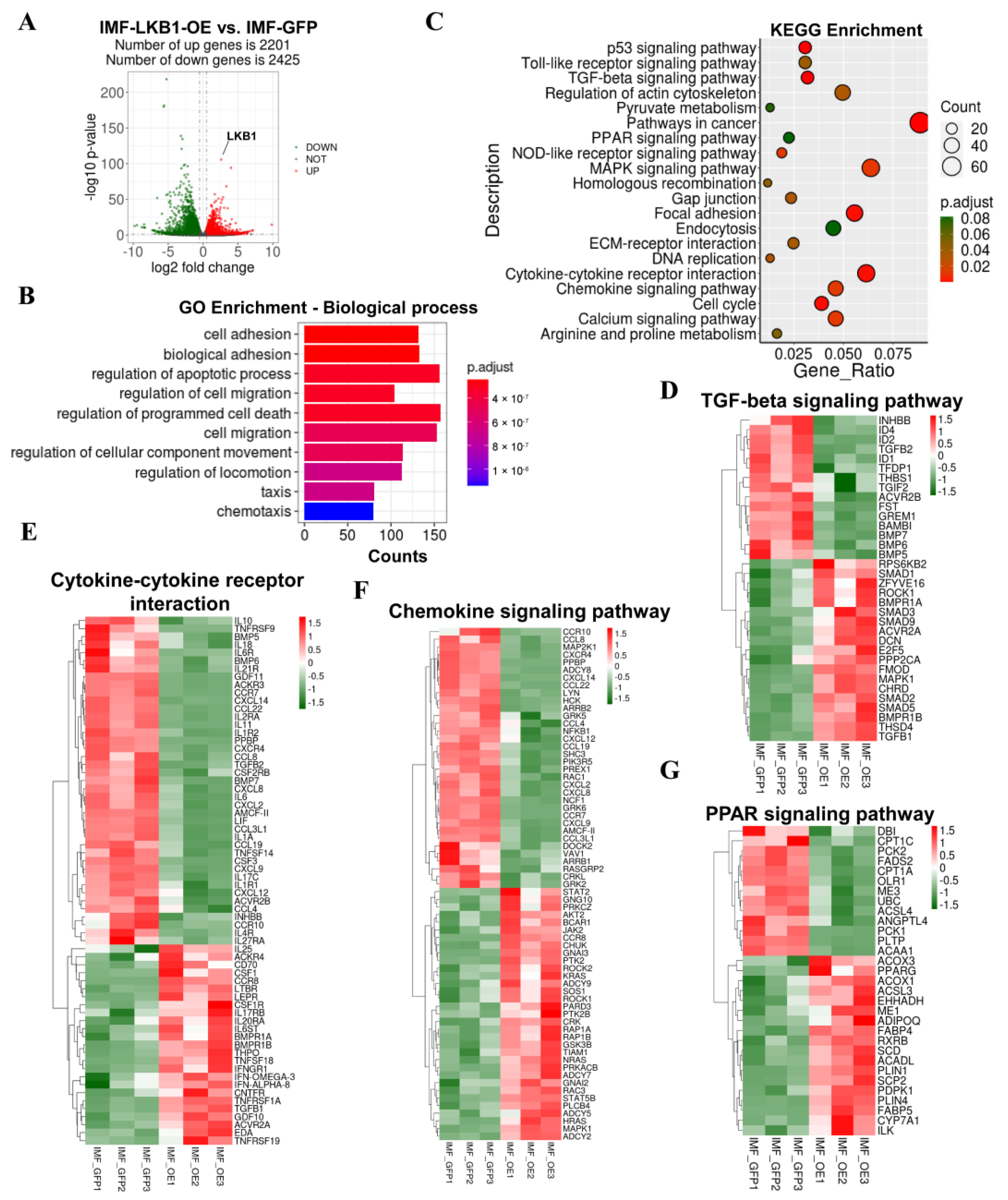
3.3. LKB1 Overexpression Alters the Transcriptional Profile of Pig IMF Adipocytes
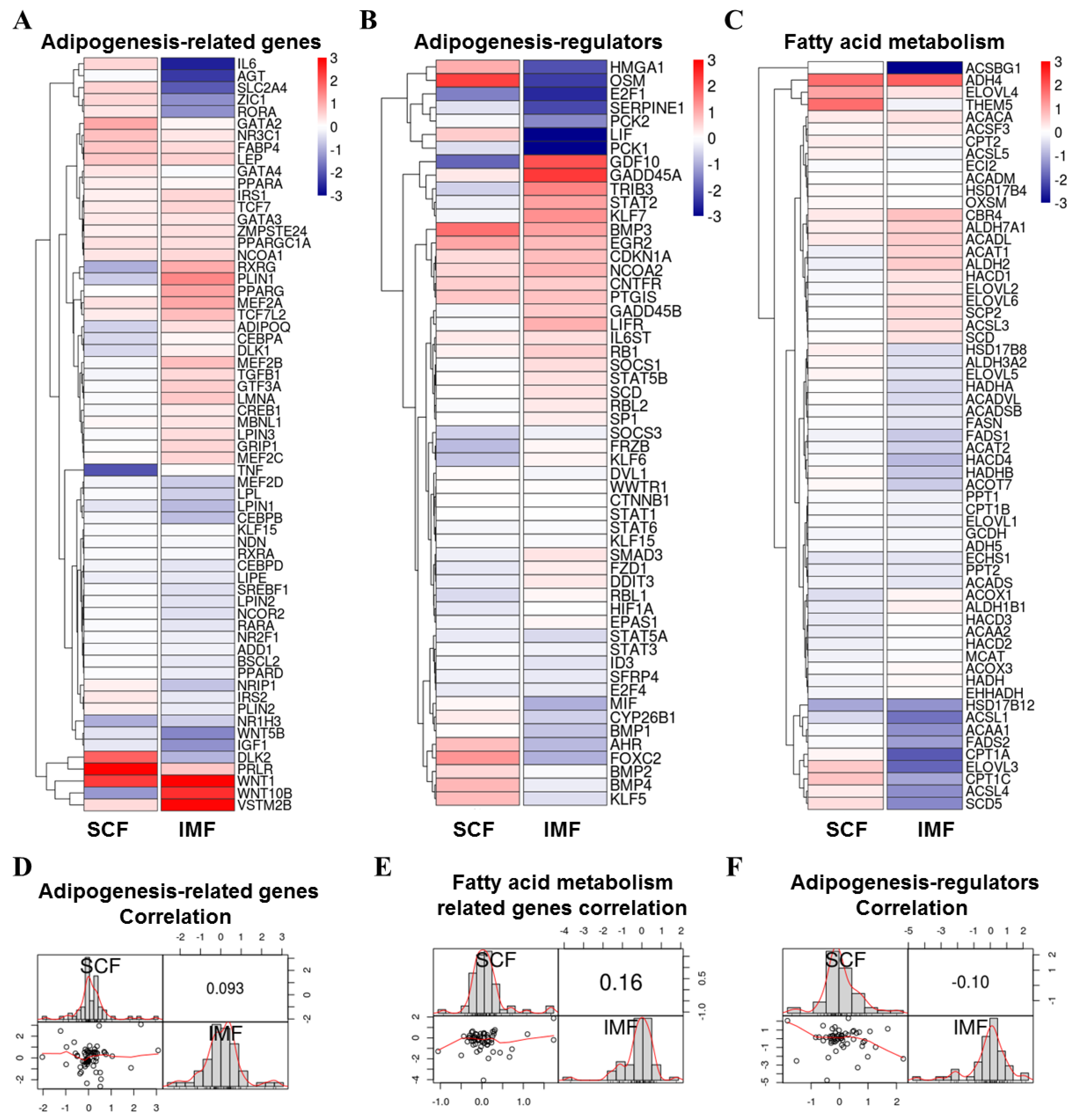
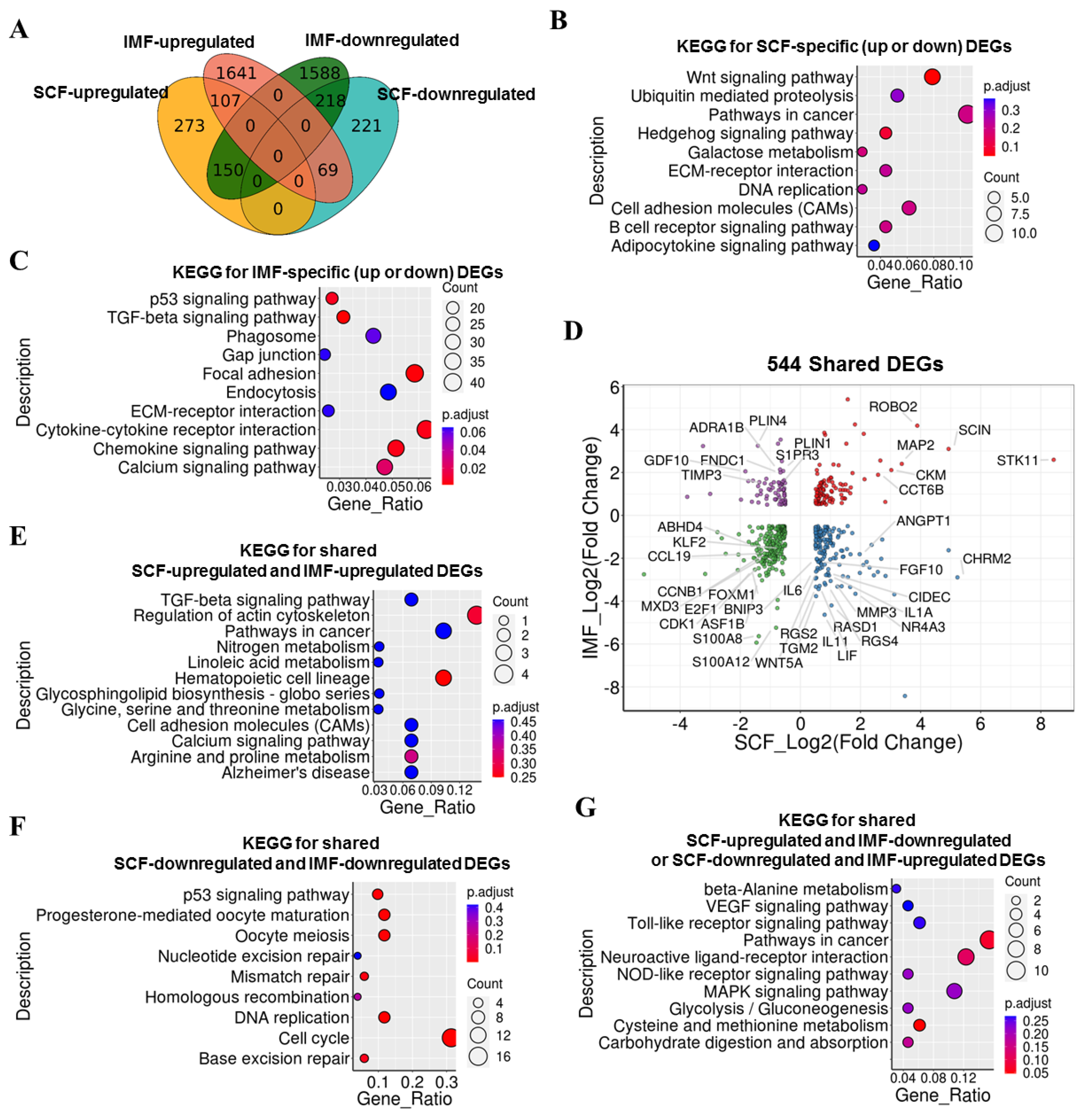
3.4. Comparison of Adipogenesis Genes of Pig IMF and SCF Adipocytes upon LKB1 Overexpression
3.5. Comparison of LKB1-Induced DEGs in Pig IMF and SCF Adipocytes
3.6. LKB1 Overexpression Inhibits Differentiation of Mouse IMF and iWAT Preadipocytes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Data Availability Statement
References
- Lim, S.; Son, K.R.; Song, I.C.; Park, H.S.; Jin, C.J.; Jang, H.C.; Park, K.S.; Kim, Y.B.; Lee, H.K. Fat in liver/muscle correlates more strongly with insulin sensitivity in rats than abdominal fat. Obesity (Silver Spring) 2009, 17, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Dodson, M.V.; Mir, P.S.; Hausman, G.J.; Guan, L.L.; Du, M.; Jiang, Z.; Fernyhough, M.E.; Bergen, W.G. Obesity, metabolic syndrome, and adipocytes. J. Lipids 2011, 2011, 721686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez, X.; Monin, G.; Talmant, A.; Mourot, J.; Lebret, B. Influence of intramuscular fat content on the quality of pig meat—1. Composition of the lipid fraction and sensory characteristics of m. longissimus lumborum. Meat Sci. 1999, 53, 59–65. [Google Scholar] [CrossRef]
- Van Laack, R.L.; Stevens, S.G.; Stalder, K.J. The influence of ultimate pH and intramuscular fat content on pork tenderness and tenderization. J. Anim. Sci. 2001, 79, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Grunert, K.G.; Bredahl, L.; Brunsø, K. Consumer perception of meat quality and implications for product development in the meat sector-a review. Meat Sci. 2004, 66, 259–272. [Google Scholar] [CrossRef]
- Gardan, D.; Gondret, F.; Louveau, I. Lipid metabolism and secretory function of porcine intramuscular adipocytes compared with subcutaneous and perirenal adipocytes. Am. J. Physiol. Endocrinol. Metab. 2006, 291, E372–E380. [Google Scholar] [CrossRef]
- Gondret, F.; Guitton, N.; Guillerm-Regost, C.; Louveau, I. Regional differences in porcine adipocytes isolated from skeletal muscle and adipose tissues as identified by a proteomic approach. J. Anim. Sci. 2008, 86, 2115–2125. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.H.; Lu, J.X.; Chen, Y.; Zhao, Y.Q.; Guo, P.H.; Yang, J.T.; Zang, R.X. Comparison of the adipogenesis in intramuscular and subcutaneous adipocytes from Bamei and Landrace pigs. Biochem. Cell Biol. 2014, 92, 259–267. [Google Scholar] [CrossRef]
- Wang, S.; Zhou, G.; Shu, G.; Wang, L.; Zhu, X.; Gao, P.; Xi, Q.; Zhang, Y.; Yuan, L.; Jiang, Q. Glucose utilization, lipid metabolism and BMP-Smad signaling pathway of porcine intramuscular preadipocytes compared with subcutaneous preadipocytes. Cell. Physiol. Biochem. 2013, 31, 981–996. [Google Scholar] [CrossRef]
- Wu, W.; Zhang, J.; Zhao, C.; Sun, Y.; Pang, W.; Yang, G. CTRP6 Regulates Porcine Adipocyte Proliferation and Differentiation by the AdipoR1/MAPK Signaling Pathway. J. Agric. Food Chem. 2017, 65, 5512–5522. [Google Scholar] [CrossRef]
- Giardiello, F.M.; Trimbath, J.D. Peutz-Jeghers syndrome and management recommendations. Clin. Gastroenterol. Hepatol. 2006, 4, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, Q.; Song, P.; Zou, M.H. Liver kinase b1 is required for white adipose tissue growth and differentiation. Diabetes 2013, 62, 2347–2358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shan, T.; Xiong, Y.; Zhang, P.; Li, Z.; Jiang, Q.; Bi, P.; Yue, F.; Yang, G.; Wang, Y.; Liu, X.; et al. Lkb1 controls brown adipose tissue growth and thermogenesis by regulating the intracellular localization of CRTC3. Nat. Commun. 2016, 7, 12205. [Google Scholar] [CrossRef] [PubMed]
- Gormand, A.; Berggreen, C.; Amar, L.; Henriksson, E.; Lund, I.; Albinsson, S.; Göransson, O. LKB1 signalling attenuates early events of adipogenesis and responds to adipogenic cues. J. Mol. Endocrinol. 2014, 53, 117–130. [Google Scholar] [CrossRef] [Green Version]
- Xi, P.; Xue, J.; Wu, Z.; Wang, H.; Han, J.; Liang, H.; Tian, D. Liver kinase B1 induces browning phenotype in 3T3-L1 adipocytes. Gene 2019, 682, 33–41. [Google Scholar] [CrossRef]
- Bunnell, B.A.; Flaat, M.; Gagliardi, C.; Patel, B.; Ripoll, C. Adipose-derived stem cells: Isolation, expansion and differentiation. Methods 2008, 45, 115–120. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.; Yu, W.; Wei, W.; Zhang, X.; Tian, Y.; Sherif, M.; Liu, X.; Dong, C.; Wu, W.; Zhang, L.; et al. Melatonin reduces intramuscular fat deposition by promoting lipolysis and increasing mitochondrial function. J. Lipid Res. 2019, 60, 767–782. [Google Scholar] [CrossRef]
- Wu, W.; Zhang, D.; Yin, Y.; Ji, M.; Xu, K.; Huang, X.; Peng, Y.; Zhang, J. Comprehensive transcriptomic view of the role of the LGALS12 gene in porcine subcutaneous and intramuscular adipocytes. BMC Genom. 2019, 20, 509. [Google Scholar] [CrossRef]
- Berry, R.; Rodeheffer, M.S. Characterization of the adipocyte cellular lineage in vivo. Nat. Cell Biol. 2013, 15, 302–308. [Google Scholar] [CrossRef]
- Zhao, C.; Yao, X.; Chen, X.; Wu, W.; Xi, F.; Yang, G.; Yu, T. Knockdown of ubiquitin D inhibits adipogenesis during the differentiation of porcine intramuscular and subcutaneous preadipocytes. Cell Prolif. 2018, 51, e12401. [Google Scholar] [CrossRef] [Green Version]
- Zhou, G.; Wang, S.; Wang, Z.; Zhu, X.; Shu, G.; Liao, W.; Yu, K.; Gao, P.; Xi, Q.; Wang, X.; et al. Global comparison of gene expression profiles between intramuscular and subcutaneous adipocytes of neonatal landrace pig using microarray. Meat Sci. 2010, 86, 440–450. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.Y.; You, W.J.; Zhou, Y.B.; Chen, W.T.; Wang, Y.Z.; Shan, T.Z. Cold-induced lipid dynamics and transcriptional programs in white adipose tissue. BMC Biol. 2019, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Minoru, K.; Susumu, G.; Yoko, S.; Masayuki, K.; Miho, F.; Mao, T.J.N.A.R. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014, 42, 199–205. [Google Scholar]
- Xie, X.; Song, J.; Li, G. MiR-21a-5p suppresses bisphenol A-induced pre-adipocyte differentiation by targeting map2k3 through MKK3/p38/MAPK. Biochem. Biophys. Res. Commun. 2016, 473, 140–146. [Google Scholar] [CrossRef]
- Cohen, D.M.; Won, K.J.; Nguyen, N.; Lazar, M.A.; Chen, C.S.; Steger, D.J. ATF4 licenses C/EBPβ activity in human mesenchymal stem cells primed for adipogenesis. Elife 2015, 4, e06821. [Google Scholar] [CrossRef]
- Yang, L.; Liu, Z.; Chen, C.; Cong, X.; Li, Z.; Zhao, S.; Ren, M. Low-dose radiation modulates human mesenchymal stem cell proliferation through regulating CDK and Rb. Am. J. Transl. Res. 2017, 9, 1914–1921. [Google Scholar]
- Jonker, J.W.; Suh, J.M.; Atkins, A.R.; Ahmadian, M.; Li, P.; Whyte, J.; He, M.; Juguilon, H.; Yin, Y.Q.; Phillips, C.T.; et al. A PPARγ-FGF1 axis is required for adaptive adipose remodelling and metabolic homeostasis. Nature 2012, 485, 391–394. [Google Scholar] [CrossRef]
- Xu, Z.; Liu, J.; Shan, T. New Roles of Lkb1 in Regulating Adipose Tissue Development and Thermogenesis. J. Cell Physiol. 2017, 232, 2296–2298. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, Y.; Chen, H.; Zhang, X.; Liu, X.; He, Z.; Cong, P.; Chen, Y.; Mo, D. Comparative Transcriptome Analysis Reveals a More Complicated Adipogenic Process in Intramuscular Stem Cells than That of Subcutaneous Vascular Stem Cells. J. Agric. Food Chem. 2019, 67, 4700–4708. [Google Scholar] [CrossRef]
- Hausman, G.J. Meat Science and Muscle Biology Symposium: The influence of extracellular matrix on intramuscular and extramuscular adipogenesis. J. Anim. Sci. 2012, 90, 942–949. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Ke, S.; Wang, Y.; Irwin, D.M.; Zhang, S.; Wang, Z. Prolonged treatment with 3-isobutyl-1-methylxanthine improves the efficiency of differentiating 3T3-L1 cells into adipocytes. Anal. Biochem. 2016, 507, 18–20. [Google Scholar] [CrossRef] [PubMed]
- Contador, D.; Ezquer, F.; Espinosa, M.; Arango-Rodriguez, M.; Puebla, C.; Sobrevia, L.; Conget, P. Dexamethasone and rosiglitazone are sufficient and necessary for producing functional adipocytes from mesenchymal stem cells. Exp. Biol. Med. (Maywood) 2015, 240, 1235–1246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luan, G.; Li, G.; Ma, X.; Jin, Y.; Hu, N.; Li, J.; Wang, Z.; Wang, H. Dexamethasone-Induced Mitochondrial Dysfunction and Insulin Resistance-Study in 3T3-L1 Adipocytes and Mitochondria Isolated from Mouse Liver. Molecules 2019, 24, 1982. [Google Scholar] [CrossRef] [Green Version]
- Krycer, J.R.; Yugi, K.; Hirayama, A.; Fazakerley, D.J.; Quek, L.E.; Scalzo, R.; Ohno, S.; Hodson, M.P.; Ikeda, S.; Shoji, F.; et al. Dynamic Metabolomics Reveals that Insulin Primes the Adipocyte for Glucose Metabolism. Cell Rep. 2017, 21, 3536–3547. [Google Scholar] [CrossRef] [Green Version]
- Czech, M.P. Insulin action and resistance in obesity and type 2 diabetes. Nat. Med. 2017, 23, 804–814. [Google Scholar] [CrossRef]
- Sachs, S.; Zarini, S.; Kahn, D.E.; Harrison, K.A.; Perreault, L.; Phang, T.; Newsom, S.A.; Strauss, A.; Kerege, A.; Schoen, J.A.; et al. Intermuscular adipose tissue directly modulates skeletal muscle insulin sensitivity in humans. Am. J. Physiol. Endocrinol. Metab. 2019, 316, E866–E879. [Google Scholar] [CrossRef] [Green Version]
- Delarue, J.; Magnan, C. Free fatty acids and insulin resistance. Curr. Opin. Clin. Nutr. Metab. Care 2007, 10, 142–148. [Google Scholar] [CrossRef]
- Frühbeck, G.; Méndez-Giménez, L.; Fernández-Formoso, J.A.; Fernández, S.; Rodríguez, A. Regulation of adipocyte lipolysis. Nutr. Res. Rev. 2014, 27, 63–93. [Google Scholar] [CrossRef] [Green Version]
- Jiang, N.; Li, Y.; Shu, T.; Wang, J. Cytokines and inflammation in adipogenesis: An updated review. Front. Med. 2019, 13, 314–329. [Google Scholar] [CrossRef]
- Chang, E.J.; Lee, S.K.; Song, Y.S.; Jang, Y.J.; Park, H.S.; Hong, J.P.; Ko, A.R.; Kim, D.Y.; Kim, J.H.; Lee, Y.J.; et al. IL-34 is associated with obesity, chronic inflammation, and insulin resistance. J. Clin. Endocrinol. Metab. 2014, 99, E1263–E1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bost, F.; Aouadi, M.; Caron, L.; Binétruy, B. The role of MAPKs in adipocyte differentiation and obesity. Biochimie 2005, 87, 51–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Liu, F.; Yuan, Y.; Wu, J.; Wang, H.; Zhang, L.; Hu, P.; Li, Z.; Li, Q.; Ye, J. Metformin suppresses lipid accumulation in skeletal muscle by promoting fatty acid oxidation. Clin. Lab. 2014, 60, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.K.; Lee, S.H.; Jhun, J.Y.; Byun, J.K.; Jeong, J.H.; Lee, S.Y.; Kim, J.K.; Choi, J.Y.; Cho, M.L. Metformin Prevents Fatty Liver and Improves Balance of White/Brown Adipose in an Obesity Mouse Model by Inducing FGF21. Mediat. Inflamm. 2016, 2016, 5813030. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Z.; Zhou, Y.; Nong, Q.; You, W.; Wang, L.; Wang, Y.; Shan, T. LKB1 Differently Regulates Adipogenesis in Intramuscular and Subcutaneous Adipocytes through Metabolic and Cytokine-Related Signaling Pathways. Cells 2020, 9, 2599. https://doi.org/10.3390/cells9122599
Xu Z, Zhou Y, Nong Q, You W, Wang L, Wang Y, Shan T. LKB1 Differently Regulates Adipogenesis in Intramuscular and Subcutaneous Adipocytes through Metabolic and Cytokine-Related Signaling Pathways. Cells. 2020; 9(12):2599. https://doi.org/10.3390/cells9122599
Chicago/Turabian StyleXu, Ziye, Yanbing Zhou, Qiuyun Nong, Wenjing You, Liyi Wang, Yizhen Wang, and Tizhong Shan. 2020. "LKB1 Differently Regulates Adipogenesis in Intramuscular and Subcutaneous Adipocytes through Metabolic and Cytokine-Related Signaling Pathways" Cells 9, no. 12: 2599. https://doi.org/10.3390/cells9122599





