Mesenchymal MACF1 Facilitates SMAD7 Nuclear Translocation to Drive Bone Formation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Gene Expression Profile
2.2. Human Bone Specimen
2.3. Generation of the MACF1 Conditional Knockout (cKO) Mouse
2.4. Study Design and Animal Grouping
2.5. Cell Preparation
2.6. RNA and Real Time Quantitative PCR (qPCR)
2.7. Protein and Western Blot
2.8. Immunofluorescence
2.9. Co-Immunoprecipitation and iTRAQ-LC-MS/MS
2.10. Cleared Skeleton Preparation
2.11. Bone Densitometry and Micro-Computed Tomography
2.12. Three-Point Bending Test
2.13. Histochemical, Calcein Labeling, and Histomorphometric Analysis
2.14. Serological Factors
2.15. Cell Culture Staining
2.16. Electric Cell-Substrate Impedance Sensing
2.17. Plasmid Preparation
2.18. Data Processing and Statistics
3. Results
3.1. Mesenchymal Microtubule Actin Crosslinking Factor 1 (MACF1) Is Decreased in Osteoporosis Patients
3.2. Mesenchymal Deletion of Microtubule Actin Crosslinking Factor 1 (MACF1)
3.3. Loss of Microtubule Actin Crosslinking Factor 1 (MACF1) Inhibits Osteogenic Differentiation in Mesenchymal Stem Cells (MSCs)
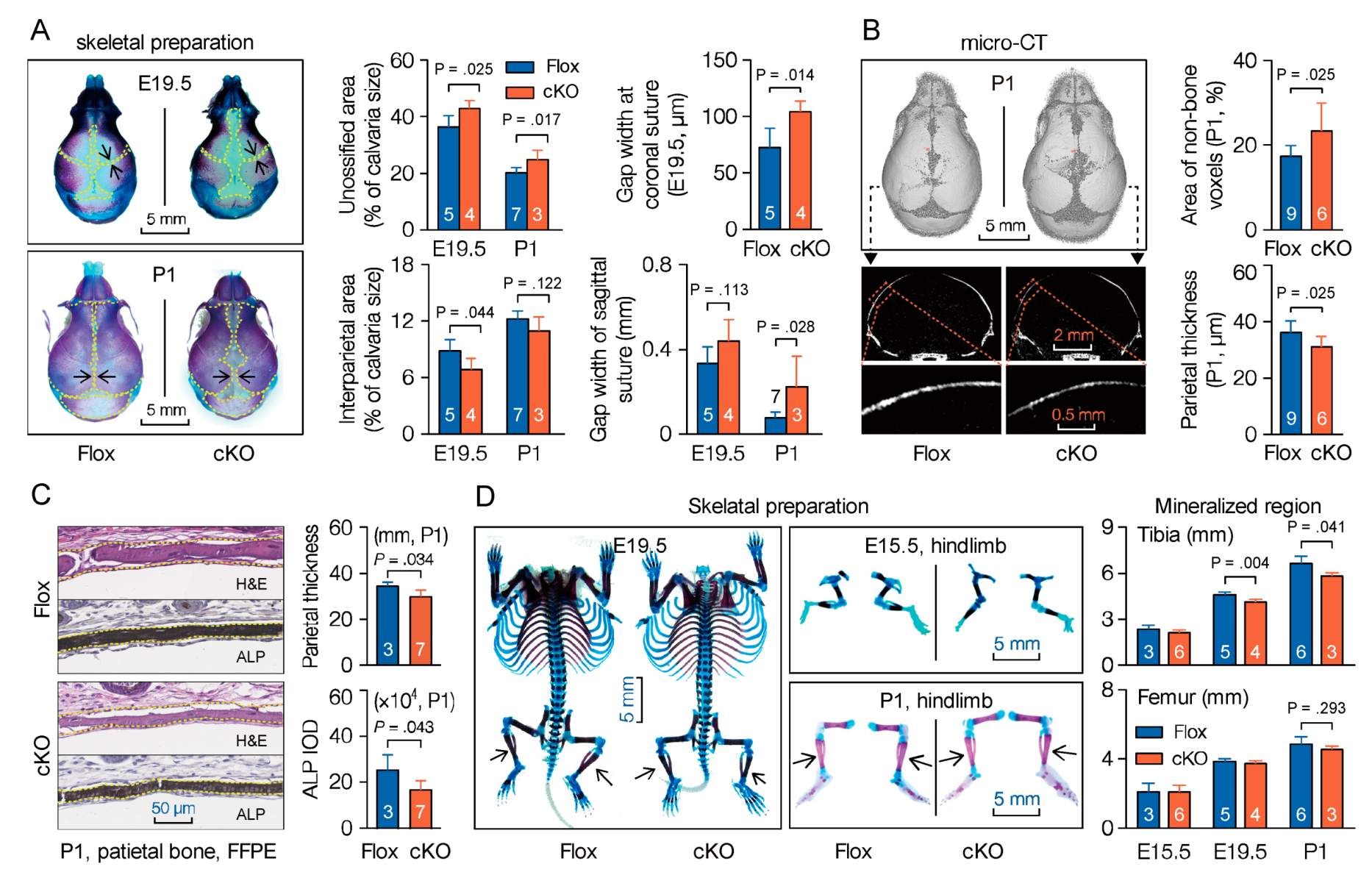
3.4. Mesenchymal Deletion of Microtubule Actin Crosslinking Factor 1 (MACF1) Impairs Early-Stage Bone Development in Mice
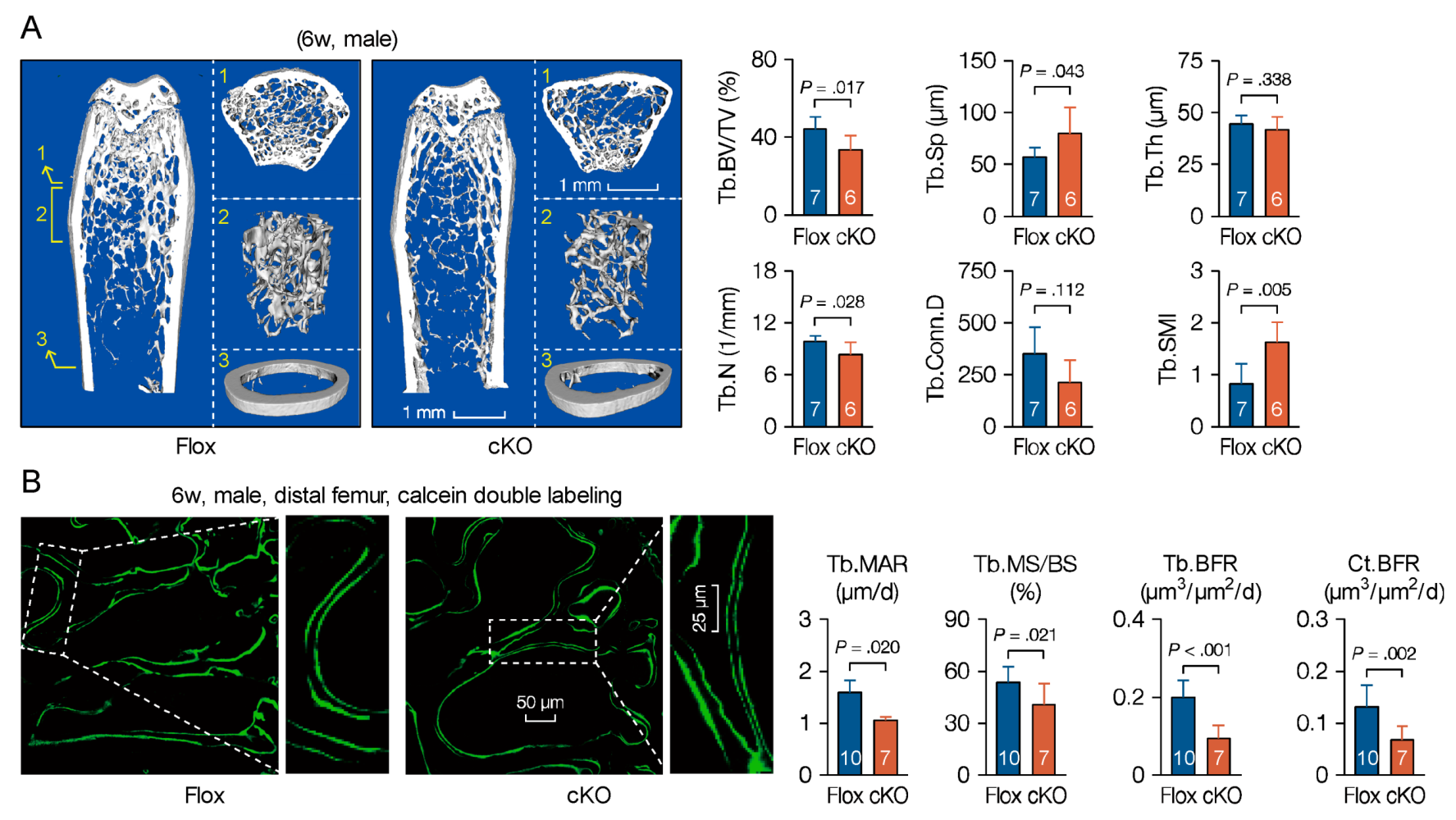
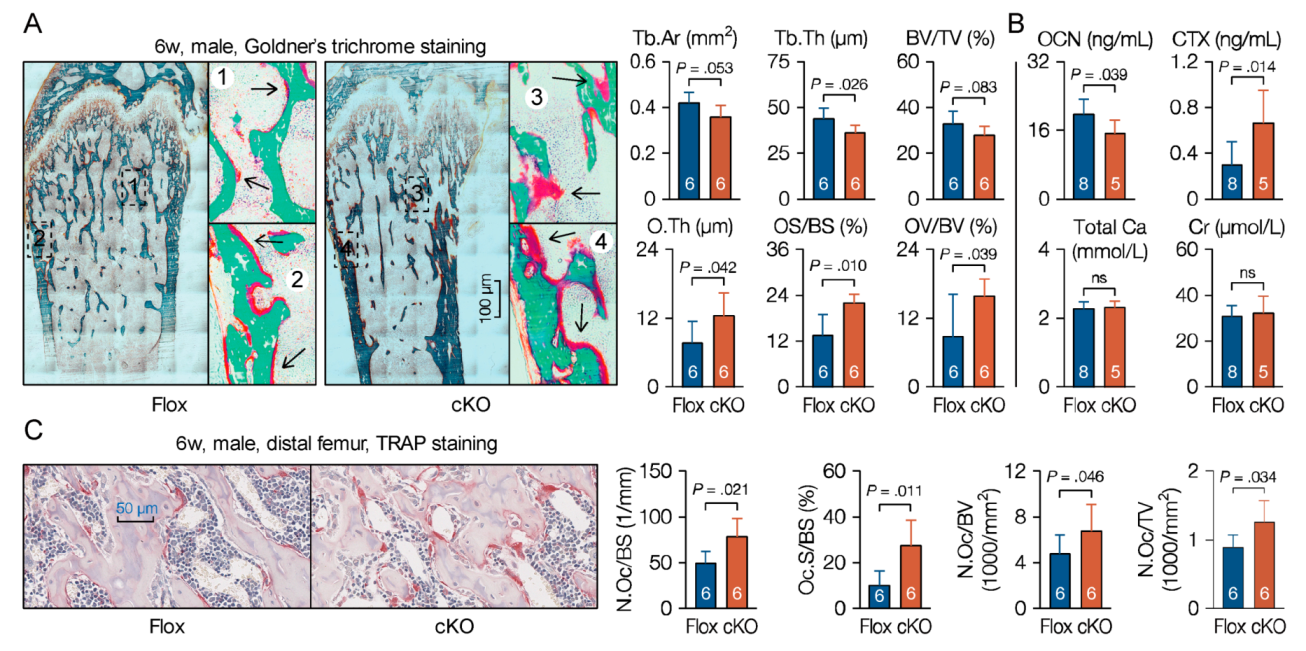
3.5. Microtubule Actin Crosslinking Factor 1 (MACF1) Is Required for Bone Formation
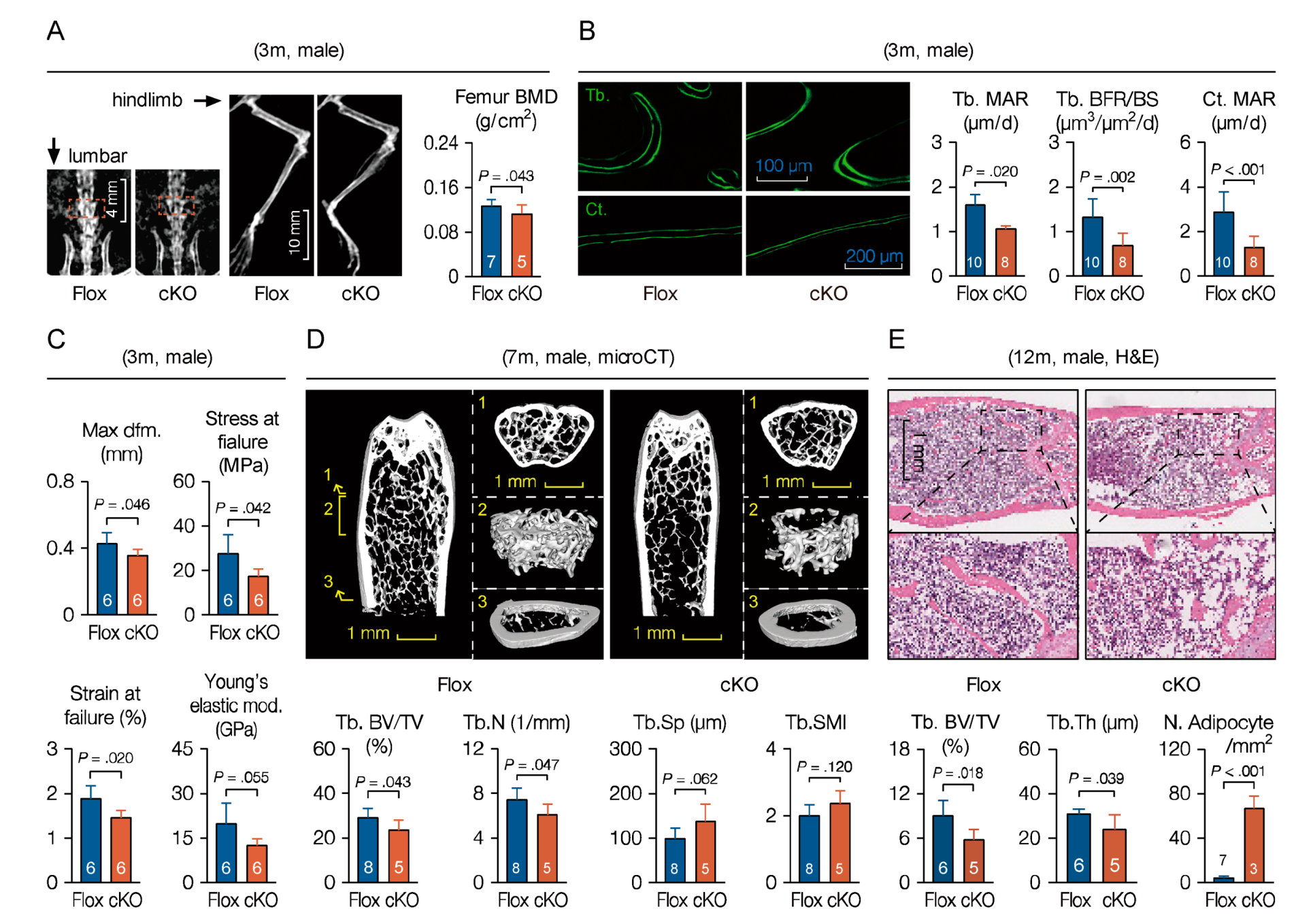
3.6. Loss of Microtubule Actin Crosslinking Factor 1 (MACF1) Weakens Bone Properties in Adult Mice
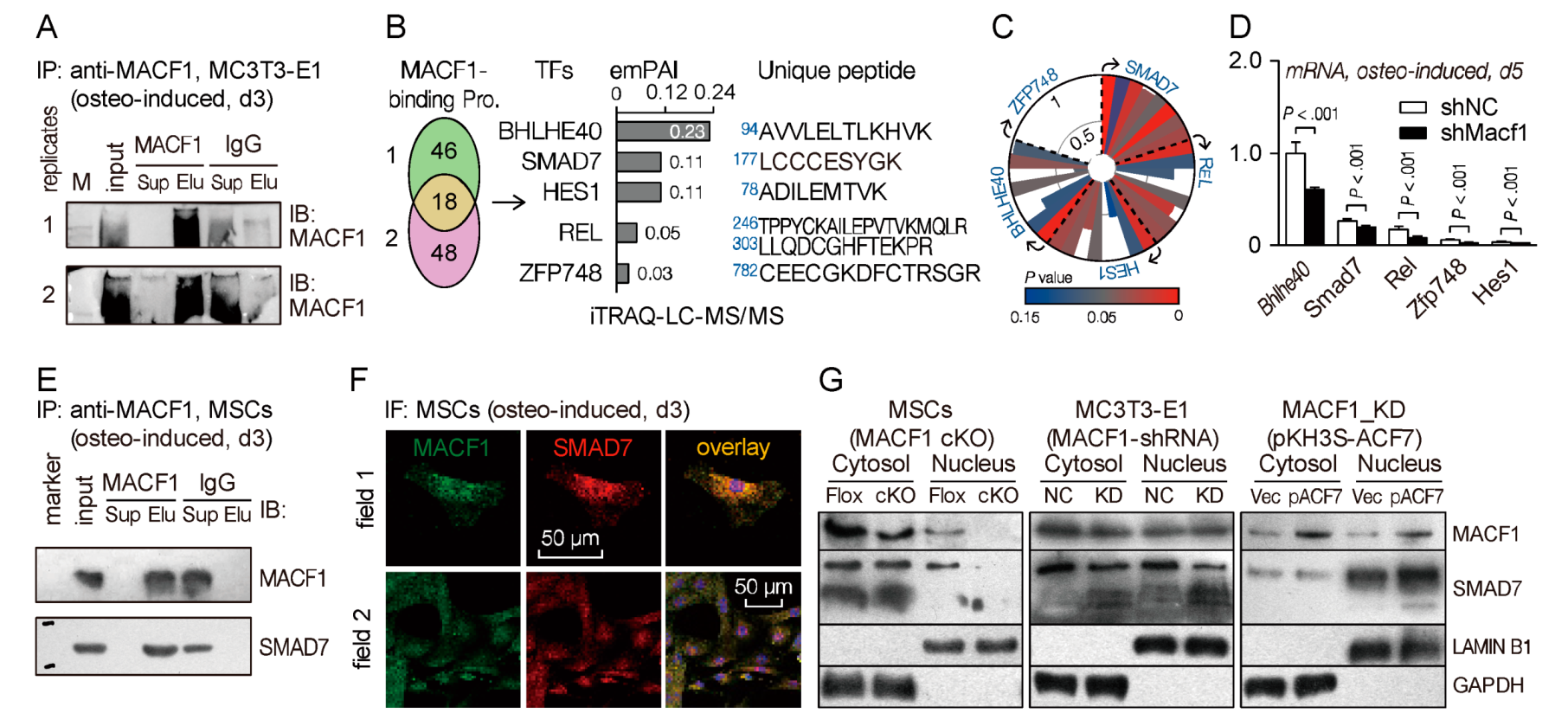
3.7. Microtubule Actin Crosslinking Factor 1 (MACF1) Interacts with SMAD Family Member 7 (SMAD7) in Mesenchymal Stem Cells (MSCs)
3.8. Microtubule Actin Crosslinking Factor 1 (MACF1) Facilitates Nucleus Translocation of SMAD Family Member 7 (SMAD7) to Drive Osteogenic Differentiation
4. Discussion
4.1. Microtubule Actin Crosslinking Factor 1 (MACF1) Is Essential for Regulating Bone Development and Formation
4.2. Microtubule Actin Crosslinking Factor 1 (MACF1) Interacts Directly with SMAD Family Member 7 (SMAD7) to Initiate Osteogenic Differentiation
4.3. Microtubule Actin Crosslinking Factor 1 (MACF1) Is a Novel Potential Target for Primary Osteoporosis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ka, M.; Jung, E.M.; Mueller, U.; Kim, W.Y. MACF1 regulates the migration of pyramidal neurons via microtubule dynamics and GSK-3 signaling. Dev. Biol. 2014, 395, 4–18. [Google Scholar] [CrossRef] [Green Version]
- Leung, C.L.; Sun, D.; Zheng, M.; Knowles, D.R.; Liem, R.K. Microtubule actin cross-linking factor (MACF): A hybrid of dystonin and dystrophin that can interact with the actin and microtubule cytoskeletons. J. Cell Biol. 1999, 147, 1275–1286. [Google Scholar] [CrossRef]
- Suozzi, K.C.; Wu, X.; Fuchs, E. Spectraplakins: Master orchestrators of cytoskeletal dynamics. J. Cell Biol. 2012, 197, 465–475. [Google Scholar] [CrossRef] [Green Version]
- Lane, T.R.; Fuchs, E.; Slep, K.C. Structure of the ACF7 EF-Hand-GAR Module and Delineation of Microtubule Binding Determinants. Structure 2017, 25, 1130–1138. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Kodama, A.; Fuchs, E. ACF7 Regulates Cytoskeletal-Focal Adhesion Dynamics and Migration and Has ATPase Activity. Cell 2008, 135, 137–148. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.-J.; Lin, C.-M.; Lin, C.-S.; Perez-Olle, R.; Leung, C.L.; Liem, R.K.H. The role of microtubule actin cross-linking factor 1 (MACF1) in the Wnt signaling pathway. Genes Dev. 2006, 20, 1933–1945. [Google Scholar] [CrossRef] [Green Version]
- Kakinuma, T.; Ichikawa, H.; Tsukada, Y.; Nakamura, T.; Toh, B.-H. Interaction between p230 and MACF1 is associated with transport of a glycosyl phosphatidyl inositol-anchored protein from the Golgi to the cell periphery. Exp. Cell Res. 2004, 298, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Antonellis, P.J.; Pollock, L.M.; Chou, S.-W.; Hassan, A.; Geng, R.; Chen, X.; Fuchs, E.; Alagramam, K.N.; Auer, M.; McDermott, B.M. ACF7 Is a Hair-Bundle Antecedent, Positioned to Integrate Cuticular Plate Actin and Somatic Tubulin. J. Neurosci. 2014, 34, 305–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ka, M.; Kim, W.-Y. Microtubule-Actin Crosslinking Factor 1 Is Required for Dendritic Arborization and Axon Outgrowth in the Developing Brain. Mol. Neurobiol. 2016, 53, 6018–6032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- May-Simera, H.L.; Gumerson, J.D.; Gao, C.; Campos, M.; Cologna, S.M.; Beyer, T.; Boldt, K.; Kaya, K.D.; Patel, N.; Kretschmer, F.; et al. Loss of MACF1 Abolishes Ciliogenesis and Disrupts Apicobasal Polarity Establishment in the Retina. Cell Rep. 2016, 17, 1399–1413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duhamel, S.; Goyette, M.-A.; Thibault, M.-P.; Filion, D.; Gaboury, L.; Côté, J.-F. The E3 Ubiquitin Ligase HectD1 Suppresses EMT and Metastasis by Targeting the +TIP ACF7 for Degradation. Cell Rep. 2018, 22, 1016–1030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Escobar-Aguirre, M.; Zhang, H.; Jamieson-Lucy, A.; Mullins, M.C. Microtubule-actin crosslinking factor 1 (Macf1) domain function in Balbiani body dissociation and nuclear positioning. PLoS Genet. 2017, 13, e1006983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, J.; Zhang, Y.; Liang, W.G.; Gou, X.; Lee, P.; Liu, H.; Lyu, W.; Tang, W.-J.; Chen, S.-Y.; Yang, F.; et al. In vivo epidermal migration requires focal adhesion targeting of ACF7. Nat. Commun. 2016, 7, 11692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Li, N.; Xiong, N.; You, Q.; Li, J.; Yu, J.; Qing, H.; Wang, T.; Cordell, H.J.; Isacson, O.; et al. Genetic Variants of Microtubule Actin Cross-linking Factor 1 (MACF1) Confer Risk for Parkinson’s Disease. Mol. Neurobiol. 2017, 54, 2878–2888. [Google Scholar] [CrossRef]
- Ma, Y.; Yue, J.; Zhang, Y.; Shi, C.; Odenwald, M.; Liang, W.G.; Wei, Q.; Goel, A.; Gou, X.; Zhang, J.; et al. ACF7 regulates inflammatory colitis and intestinal wound response by orchestrating tight junction dynamics. Nat. Commun. 2017, 8, 15375. [Google Scholar] [CrossRef]
- Ka, M.; Moffat, J.J.; Kim, W.-Y. MACF1 Controls Migration and Positioning of Cortical GABAergic Interneurons in Mice. Cereb. Cortex 2017, 27, 5525–5538. [Google Scholar] [CrossRef] [Green Version]
- Kodama, A.; Karakesisoglou, I.; Wong, E.; Vaezi, A.; Fuchs, E. ACF7: An essential integrator of microtubule dynamics. Cell 2003, 115, 343–354. [Google Scholar] [CrossRef] [Green Version]
- Yin, C.; Zhang, Y.; Hu, L.; Tian, Y.; Chen, Z.; Li, D.; Zhao, F.; Su, P.; Ma, X.; Zhang, G.; et al. Mechanical unloading reduces microtubule actin crosslinking factor 1 expression to inhibit β-catenin signaling and osteoblast proliferation. J. Cell. Physiol. 2018, 233, 5405–5419. [Google Scholar] [CrossRef]
- Hu, L.; Su, P.; Yin, C.; Zhang, Y.; Li, R.; Yan, K.; Chen, Z.; Li, D.; Zhang, G.; Wang, L.; et al. Microtubule actin crosslinking factor 1 promotes osteoblast differentiation by promoting β-catenin/TCF1/Runx2 signaling axis. J. Cell. Physiol. 2018, 233, 1574–1584. [Google Scholar] [CrossRef]
- Hu, L.; Su, P.; Li, R.; Yan, K.; Chen, Z.; Shang, P.; Qian, A. Knockdown of microtubule actin crosslinking factor 1 inhibits cell proliferation in MC3T3-E1 osteoblastic cells. BMB Rep. 2015, 48, 583–588. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Yin, C.; Hu, L.; Chen, Z.; Zhao, F.; Li, D.; Ma, J.; Ma, X.; Su, P.; Qiu, W.; et al. MACF1 Overexpression by Transfecting the 21 kbp Large Plasmid PEGFP-C1A-ACF7 Promotes Osteoblast Differentiation and Bone Formation. Hum. Gene Ther. 2018, 29, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Benisch, P.; Schilling, T.; Klein-Hitpass, L.; Frey, S.P.; Seefried, L.; Raaijmakers, N.; Krug, M.; Regensburger, M.; Zeck, S.; Schinke, T.; et al. The transcriptional profile of mesenchymal stem cell populations in primary osteoporosis is distinct and shows overexpression of osteogenic inhibitors. PLoS ONE 2012, 7, e45142. [Google Scholar] [CrossRef] [PubMed]
- Wagner, W.; Bork, S.; Horn, P.; Krunic, D.; Walenda, T.; Diehlmann, A.; Benes, V.; Blake, J.; Huber, F.-X.; Eckstein, V.; et al. Aging and replicative senescence have related effects on human stem and progenitor cells. PLoS ONE 2009, 4, e5846. [Google Scholar] [CrossRef]
- Zhu, H.; Guo, Z.-K.; Jiang, X.-X.; Li, H.; Wang, X.-Y.; Yao, H.-Y.; Zhang, Y.; Mao, N. A protocol for isolation and culture of mesenchymal stem cells from mouse compact bone. Nat. Protoc. 2010, 5, 550–560. [Google Scholar] [CrossRef] [PubMed]
- Bouxsein, M.L.; Boyd, S.K.; Christiansen, B.A.; Guldberg, R.E.; Jepsen, K.J.; Müller, R. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J. Bone Miner. Res. 2010, 25, 1468–1486. [Google Scholar] [CrossRef] [PubMed]
- Sharir, A.; Barak, M.M.; Shahar, R. Whole bone mechanics and mechanical testing. Vet. J. 2008, 177, 8–17. [Google Scholar] [CrossRef]
- Gruber, H.E. Adaptations of Goldner’s Masson trichrome stain for the study of undecalcified plastic embedded bone. Biotech. Histochem. 1992, 67, 30–34. [Google Scholar] [CrossRef]
- van’t Hof, R.J.; Rose, L.; Bassonga, E.; Daroszewska, A. Open source software for semi-automated histomorphometry of bone resorption and formation parameters. Bone 2017, 99, 69–79. [Google Scholar] [CrossRef] [Green Version]
- Bagnaninchi, P.O.; Drummond, N. Real-time label-free monitoring of adipose-derived stem cell differentiation with electric cell-substrate impedance sensing. Proc. Natl. Acad. Sci. USA 2011, 108, 6462–6467. [Google Scholar] [CrossRef] [Green Version]
- Bernier, G.; Mathieu, M.; De Repentigny, Y.; Vidal, S.M.; Kothary, R. Cloning and characterization of mouse ACF7, a novel member of the dystonin subfamily of actin binding proteins. Genomics 1996, 38, 19–29. [Google Scholar] [CrossRef]
- Goryunov, D.; He, C.Z.; Lin, C.S.; Leung, C.L.; Liem, R.K.H. Nervous-tissue-specific elimination of microtubule-actin crosslinking factor 1a results in multiple developmental defects in the mouse brain. Mol. Cell. Neurosci. 2010, 44, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Logan, M.; Martin, J.F.; Nagy, A.; Lobe, C.; Olson, E.N.; Tabin, C.J. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis 2002, 33, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Elefteriou, F.; Yang, X. Genetic mouse models for bone studies—Strengths and limitations. Bone 2011, 49, 1242–1254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q.; Sinha, K.; Deng, J.M.; Yasuda, H.; Krahe, R.; Behringer, R.R.; De Crombrugghe, B. Mesenchymal deletion of histone demethylase NO66 in mice promotes bone formation. J. Bone Miner. Res. 2015, 30, 1608–1617. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Niu, N.; Li, L.; Shao, R.; Ouyang, H.; Zou, W. H3K36 trimethylation mediated by SETD2 regulates the fate of bone marrow mesenchymal stem cells. PLoS Biol. 2018, 16, e2006522. [Google Scholar] [CrossRef] [Green Version]
- Hilton, M.J. Skeletal Development and Repair: Methods and Protocols; Springer: London, UK, 2014. [Google Scholar]
- Wu, M.; Chen, G.; Li, Y.-P. TGF-β and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. 2016, 4, 16009. [Google Scholar] [CrossRef]
- Fassett, J.T.; Xu, X.; Kwak, D.; Wang, H.; Liu, X.; Hu, X.; Bache, R.J.; Chen, Y. Microtubule Actin Cross-linking Factor 1 regulates cardiomyocyte microtubule distribution and adaptation to hemodynamic overload. PLoS ONE 2013, 8, e73887. [Google Scholar] [CrossRef] [Green Version]
- Liang, Y.; Shi, C.; Yang, J.; Chen, H.; Xia, Y.; Zhang, P.; Wang, F.; Han, H.; Qin, H. ACF7 regulates colonic permeability. Int. J. Mol. Med. 2013, 31, 861–866. [Google Scholar] [CrossRef] [Green Version]
- Qiu, W.-X.; Ma, X.-L.; Lin, X.; Zhao, F.; Li, D.-J.; Chen, Z.-H.; Zhang, K.-W.; Zhang, R.; Wang, P.; Xiao, Y.-Y.; et al. Deficiency of Macf1 in osterix expressing cells decreases bone formation by Bmp2/Smad/Runx2 pathway. J. Cell. Mol. Med. 2020, 24, 317–327. [Google Scholar] [CrossRef] [Green Version]
- Miyazawa, K.; Miyazono, K. Regulation of TGF-β Family Signaling by Inhibitory Smads. Cold Spring Harb. Perspect. Biol. 2017, 9, a022095. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Lee, W.Y.-W.; Lin, S.-E.; Ni, M.; Zhang, T.; Huang, X.-R.; Lan, H.-Y.; Li, G. Partial loss of Smad7 function impairs bone remodeling, osteogenesis and enhances osteoclastogenesis in mice. Bone 2014, 67, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Miyake, T.; Alli, N.S.; McDermott, J.C. Nuclear function of Smad7 promotes myogenesis. Mol. Cell. Biol. 2010, 30, 722–735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slep, K.C.; Rogers, S.L.; Elliott, S.L.; Ohkura, H.; Kolodziej, P.A.; Vale, R.D. Structural determinants for EB1-mediated recruitment of APC and spectraplakins to the microtubule plus end. J. Cell Biol. 2005, 168, 587–598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015, 43, D512–D520. [Google Scholar] [CrossRef] [PubMed] [Green Version]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, F.; Ma, X.; Qiu, W.; Wang, P.; Zhang, R.; Chen, Z.; Su, P.; Zhang, Y.; Li, D.; Ma, J.; et al. Mesenchymal MACF1 Facilitates SMAD7 Nuclear Translocation to Drive Bone Formation. Cells 2020, 9, 616. https://doi.org/10.3390/cells9030616
Zhao F, Ma X, Qiu W, Wang P, Zhang R, Chen Z, Su P, Zhang Y, Li D, Ma J, et al. Mesenchymal MACF1 Facilitates SMAD7 Nuclear Translocation to Drive Bone Formation. Cells. 2020; 9(3):616. https://doi.org/10.3390/cells9030616
Chicago/Turabian StyleZhao, Fan, Xiaoli Ma, Wuxia Qiu, Pai Wang, Ru Zhang, Zhihao Chen, Peihong Su, Yan Zhang, Dijie Li, Jianhua Ma, and et al. 2020. "Mesenchymal MACF1 Facilitates SMAD7 Nuclear Translocation to Drive Bone Formation" Cells 9, no. 3: 616. https://doi.org/10.3390/cells9030616




