The Emerging Roles of Exosomes as EMT Regulators in Cancer
Abstract
:1. Introduction
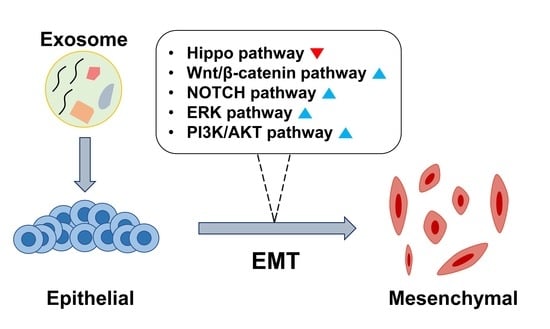
2. Exosome-Induced EMT
2.1. Hippo Pathway
2.2. β-Catenin Signaling Pathway
2.3. ERK Pathway
3. Therapeutic Candidates for Preventing Exosomal EMT Induction
4. Clinical Application
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ABCC1 | ATP binding cassette subfamily C member 1 |
| AKT | protein kinase B |
| AREG | amphiregulin |
| BCL9 | B-cell CLL/lymphoma 9 protein |
| BRG1 | brahma-related gene 1 |
| CAF | cancer associated fibroblast |
| COL10A1 | collagen type X alpha 1 chain |
| CSC | cancer stem cell |
| CTGF | connective tissue growth factor |
| DANCR | differentiation antagonizing non-protein coding RNA |
| DUSP1 | dual specificity protein phosphatase 1 |
| EGF | epidermal growth factor |
| EGFR | epidermal growth factor receptor |
| EMN | exosome-mimetic nanosystem |
| EMT | epithelial-mesenchymal-transition |
| EpCAM | epithelial cell adhesion molecule |
| ERK | extracellular signal regulated kinase |
| EV | extracellular vesicle |
| EZH2 | enhancer of zeste homolog 2 |
| FAK | focal adhesion kinase |
| FBXW7 | F-Box and WD repeat domain containing 7 |
| FGF | fibroblast growth factor |
| FOXF2 | forkhead box F2 |
| FOXO3a | forkhead box O3 |
| GBM | glioblastoma multiform |
| HCC | hepatocellular carcinoma |
| HGF | hepatocyte growth factor |
| HIF-1α | hypoxia-inducible factor 1-alpha |
| HLE | human lens epithelial cell |
| HMB1 | high mobility group box 1 |
| HNSCC | head and neck squamous cell carcinoma |
| ITGB1 | integrin subunit beta 1 |
| KIT | KIT proto-oncogene |
| LAMP3 | lysosome-associated membrane protein 3 |
| LEF1 | lymphoid enhancer-binding factor 1 |
| MAP3K2 | mitogen-activated protein kinase kinase kinase 2 |
| MAPK | mitogen-activated protein kinase |
| MEK | MAP kinase-ERK kinase |
| MOAP1 | modulator of apoptosis 1 |
| MYO6 | myosine 6 |
| NSCLC | non-small-cell lung carcinoma |
| OSCC | oral squamous cell carcinoma |
| PCP | planar cell polarity |
| PDAC | pancreatic ductal adenocarcinoma |
| PDCD4 | programmed cell death protein 4 |
| PDT | photodynamic therapy |
| PI3K | phosphoinositide 3-kinase |
| PRMT5 | protein arginine methyltransferase 5 |
| PTEN | phosphatase and tensin homolog |
| PTPRB | protein tyrosine phosphatase receptor type B |
| RAB27B | RAB27B, member RAS oncogene family |
| RECK | reversion-inducing-cysteine-rich protein with kazal motifs |
| ROCK1 | rho-associated, coiled-coil-containing protein kinase 1 |
| ROR | RNA regulator of reprogramming |
| SATB2 | special AT-rich sequence-binding protein 2 |
| SGK3 | serum and glucocorticoid kinase 3 |
| SLAI1 | snail family transcriptional repressor 1 |
| SLC31A1 | solute carrier family 31 member 1 |
| SOCS6 | suppressor of cytokine signaling 6 |
| Sox2ot | SOX2 overlapping transcript |
| SOX9 | SRY-Box transcription factor 9 |
| SRC | SRC proto-oncogene tyrosine kinase |
| STAT3 | signal transducer and activator of transcription 3 |
| TAZ | transcriptional coactivator with PDZ-binding motif |
| TCEAL7 | transcription elongation factor A like 7 |
| TEAD | transcriptional enhanced associate domain |
| TFF3 | trefoil factor 3 |
| TGF-β | transforming growth factor β |
| TIMP3 | metalloproteinase inhibitor 3 |
| TNC | tenascin C |
| TRIM44 | tripartite motif-containing protein 44 |
| TUSC3 | tumor suppressor candidate 3 |
| UBR2 | ubiquitin regulatory protein ligase E3 component n-recognin 2 |
| USP22 | ubiquitin specific peptidase 22 |
| YAP | yes-associated protein |
| YY1 | YY1 transcription factor |
| ZEB1/2 | zinc finger E-box binding homeobox 1 |
References
- Pastushenko, I.; Blanpain, C. EMT Transition States during Tumor Progression and Metastasis. Trends Cell Biol. 2019, 29, 212–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Weinberg, R.A. Epithelial-to-mesenchymal transition in cancer: Complexity and opportunities. Front. Med. 2018, 12, 361–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, P.; Li, B.; Liu, F.; Zhang, M.; Wang, Q.; Liu, Y.; Yao, Y.; Li, D. The epithelial to mesenchymal transition (EMT) and cancer stem cells: Implication for treatment resistance in pancreatic cancer. Mol. Cancer 2017, 16, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.; Settleman, J. EMT, cancer stem cells and drug resistance: An emerging axis of evil in the war on cancer. Oncogene 2010, 29, 4741–4751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azmi, A.S. Unveiling the role of nuclear transport in epithelial-to-mesenchymal transition. Curr. Cancer Drug Targets 2013, 13, 906–914. [Google Scholar] [CrossRef]
- Xu, R.; Won, J.Y.; Kim, C.H.; Kim, D.E.; Yim, H. Roles of the Phosphorylation of Transcriptional Factors in Epithelial-Mesenchymal Transition. J. Oncol. 2019, 2019, 5810465. [Google Scholar] [CrossRef]
- Dave, N.; Guaita-Esteruelas, S.; Gutarra, S.; Frias, À.; Beltran, M.; Peiró, S.; de Herreros, A.G. Functional cooperation between Snail1 and twist in the regulation of ZEB1 expression during epithelial to mesenchymal transition. J. Biol. Chem. 2011, 286, 12024–12032. [Google Scholar] [CrossRef] [Green Version]
- Peinado, H.; Olmeda, D.; Cano, A. Snail, Zeb and bHLH factors in tumour progression: An alliance against the epithelial phenotype? Nat. Rev. Cancer 2007, 7, 415–428. [Google Scholar] [CrossRef]
- Blackwell, R.H.; Foreman, K.E.; Gupta, G.N. The Role of Cancer-Derived Exosomes in Tumorigenicity & Epithelial-to-Mesenchymal Transition. Cancers 2017, 9, 105. [Google Scholar] [CrossRef] [Green Version]
- Mashouri, L.; Yousefi, H.; Aref, A.R.; Ahadi, A.M.; Molaei, F.; Alahari, S.K. Exosomes: Composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol. Cancer 2019, 18, 75. [Google Scholar] [CrossRef]
- Huang, T.; Song, C.; Zheng, L.; Xia, L.; Li, Y.; Zhou, Y. The roles of extracellular vesicles in gastric cancer development, microenvironment, anti-cancer drug resistance, and therapy. Mol. Cancer 2019, 18, 62. [Google Scholar] [CrossRef]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genom. Proteom. Bioinf. 2015, 13, 17–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gruenberg, J.; van der Goot, F.G. Mechanisms of pathogen entry through the endosomal compartments. Nat. Rev. Mol. Cell Biol. 2006, 7, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Maia, J.; Caja, S.; Strano Moraes, M.C.; Couto, N.; Costa-Silva, B. Exosome-Based Cell-Cell Communication in the Tumor Microenvironment. Front. Cell Dev. Biol. 2018, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Vlassov, A.V.; Magdaleno, S.; Setterquist, R.; Conrad, R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta 2012, 1820, 940–948. [Google Scholar] [CrossRef]
- Yoshioka, Y.; Konishi, Y.; Kosaka, N.; Katsuda, T.; Kato, T.; Ochiya, T. Comparative marker analysis of extracellular vesicles in different human cancer types. J. Extracell. Vesicles 2013, 2, 20424. [Google Scholar] [CrossRef]
- Yu, F.-X.; Zhao, B.; Guan, K.-L. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell 2015, 163, 811–828. [Google Scholar] [CrossRef] [Green Version]
- Camargo, F.D.; Gokhale, S.; Johnnidis, J.B.; Fu, D.; Bell, G.W.; Jaenisch, R.; Brummelkamp, T.R. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 2007, 17, 2054–2060. [Google Scholar] [CrossRef] [Green Version]
- Boopathy, G.T.K.; Hong, W. Role of Hippo Pathway-YAP/TAZ Signaling in Angiogenesis. Front. Cell Dev. Biol. 2019, 7, 49. [Google Scholar] [CrossRef]
- Liu, H.; Du, S.; Lei, T.; Wang, H.; He, X.; Tong, R.; Wang, Y. Multifaceted regulation and functions of YAP/TAZ in tumors (Review). Oncol. Rep. 2018, 40, 16–28. [Google Scholar] [CrossRef] [PubMed]
- Huh, H.D.; Kim, D.H.; Jeong, H.-S.; Park, H.W. Regulation of TEAD Transcription Factors in Cancer Biology. Cells 2019, 8, 600. [Google Scholar] [CrossRef] [Green Version]
- Adhikary, A.; Chakraborty, S.; Mazumdar, M.; Ghosh, S.; Mukherjee, S.; Manna, A.; Mohanty, S.; Nakka, K.K.; Joshi, S.; De, A.; et al. Inhibition of epithelial to mesenchymal transition by E-cadherin up-regulation via repression of slug transcription and inhibition of E-cadherin degradation: Dual role of scaffold/matrix attachment region-binding protein 1 (SMAR1) in breast cancer cells. J. Biol. Chem. 2014, 289, 25431–25444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.T.; Zhong, H.T.; Li, G.W.; Shen, J.X.; Ye, Q.Q.; Zhang, M.L.; Liu, J. Oncogenic functions of the EMT-related transcription factor ZEB1 in breast cancer. J. Transl. Med. 2020, 18, 51. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, J.; Ying, X.; Lin, P.C.; Zhou, B.P. Twist-mediated Epithelial-mesenchymal Transition Promotes Breast Tumor Cell Invasion via Inhibition of Hippo Pathway. Sci. Rep. 2016, 6, 24606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loh, C.Y.; Chai, J.Y.; Tang, T.F.; Wong, W.F.; Sethi, G.; Shanmugam, M.K.; Chong, P.P.; Looi, C.Y. The E-Cadherin and N-Cadherin Switch in Epithelial-to-Mesenchymal Transition: Signaling, Therapeutic Implications, and Challenges. Cells 2019, 8, 1118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.H.; Shin, J.E.; Park, H.W. The Role of Hippo Pathway in Cancer Stem Cell Biology. Mol. Cells 2018, 41, 83–92. [Google Scholar] [CrossRef]
- Noguchi, S.; Saito, A.; Nagase, T. YAP/TAZ Signaling as a Molecular Link between Fibrosis and Cancer. Int. J. Mol. Sci. 2018, 19, 3674. [Google Scholar] [CrossRef] [Green Version]
- Lehmann, W.; Mossmann, D.; Kleemann, J.; Mock, K.; Meisinger, C.; Brummer, T.; Herr, R.; Brabletz, S.; Stemmler, M.P.; Brabletz, T. ZEB1 turns into a transcriptional activator by interacting with YAP1 in aggressive cancer types. Nat. Commun. 2016, 7, 10498. [Google Scholar] [CrossRef]
- Wang, S.; Su, X.; Xu, M.; Xiao, X.; Li, X.; Li, H.; Keating, A.; Zhao, R.C. Exosomes secreted by mesenchymal stromal/stem cell-derived adipocytes promote breast cancer cell growth via activation of Hippo signaling pathway. Stem Cell Res. Ther. 2019, 10, 117. [Google Scholar] [CrossRef] [Green Version]
- Qu, Z.; Wu, J.; Wu, J.; Ji, A.; Qiang, G.; Jiang, Y.; Jiang, C.; Ding, Y. Exosomal miR-665 as a novel minimally invasive biomarker for hepatocellular carcinoma diagnosis and prognosis. Oncotarget 2017, 8, 80666–80678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Yang, C.; Yang, S.; Cheng, F.; Rao, J.; Wang, X. miR-665 promotes hepatocellular carcinoma cell migration, invasion, and proliferation by decreasing Hippo signaling through targeting PTPRB. Cell Death Dis. 2018, 9, 954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corrado, C.; Saieva, L.; Raimondo, S.; Santoro, A.; De Leo, G.; Alessandro, R. Chronic myelogenous leukaemia exosomes modulate bone marrow microenvironment through activation of epidermal growth factor receptor. J. Cell Mol. Med. 2016, 20, 1829–1839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, N.; Morrison, C.D.; Liu, P.; Miecznikowski, J.; Bshara, W.; Han, S.; Zhu, Q.; Omilian, A.R.; Li, X.; Zhang, J. TAZ induces growth factor-independent proliferation through activation of EGFR ligand amphiregulin. Cell Cycle 2012, 11, 2922–2930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Yun, F.; Shi, L.; Li, Z.-H.; Luo, N.-R.; Jia, Y.-F. Roles of Signaling Pathways in the Epithelial-Mesenchymal Transition in Cancer. Asian Pac. J. Cancer Prev. 2015, 16, 6201–6206. [Google Scholar] [CrossRef] [Green Version]
- Wei, C.-Y.; Zhu, M.-X.; Yang, Y.-W.; Zhang, P.-F.; Yang, X.; Peng, R.; Gao, C.; Lu, J.-C.; Wang, L.; Deng, X.-Y.; et al. Downregulation of RNF128 activates Wnt/β-catenin signaling to induce cellular EMT and stemness via CD44 and CTTN ubiquitination in melanoma. J. Hematol. Oncol. 2019, 12, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, M.; Li, S.-N.; Anjum, K.M.; Gui, L.-X.; Zhu, S.-S.; Liu, J.; Chen, J.-K.; Liu, Q.-F.; Ye, G.-D.; Wang, W.-J.; et al. A double-negative feedback loop between Wnt-β-catenin signaling and HNF4α regulates epithelial-mesenchymal transition in hepatocellular carcinoma. J. Cell Sci. 2013, 126, 5692–5703. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Weinberg, R.A. Epithelial-mesenchymal transition: At the crossroads of development and tumor metastasis. Dev. Cell 2008, 14, 818–829. [Google Scholar] [CrossRef] [Green Version]
- Yue, X.; Lan, F.; Xia, T. Hypoxic Glioma Cell-Secreted Exosomal miR-301a Activates Wnt/β-catenin Signaling and Promotes Radiation Resistance by Targeting TCEAL7. Mol. Ther. 2019, 27, 1939–1949. [Google Scholar] [CrossRef]
- Nam, R.K.; Benatar, T.; Wallis, C.J.D.; Amemiya, Y.; Yang, W.; Garbens, A.; Naeim, M.; Sherman, C.; Sugar, L.; Seth, A. MiR-301a regulates E-cadherin expression and is predictive of prostate cancer recurrence. Prostate 2016, 76, 869–884. [Google Scholar] [CrossRef]
- Xia, Y.; Wei, K.; Hu, L.-Q.; Zhou, C.-R.; Lu, Z.-B.; Zhan, G.-S.; Pan, X.-L.; Pan, C.-F.; Wang, J.; Wen, W.; et al. Exosome-mediated transfer of miR-1260b promotes cell invasion through Wnt/β-catenin signaling pathway in lung adenocarcinoma. J. Cell Physiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Wei, K.; Yang, F.-M.; Hu, L.-Q.; Pan, C.-F.; Pan, X.-L.; Wu, W.-B.; Wang, J.; Wen, W.; He, Z.-C.; et al. miR-1260b, mediated by YY1, activates KIT signaling by targeting SOCS6 to regulate cell proliferation and apoptosis in NSCLC. Cell Death Dis. 2019, 10, 112. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.-L.; Fan, Y.-L.; Jiang, J.; Li, K.-D.; Zheng, M.; Chen, W.; Ma, X.-R.; Geng, N.; Chen, Q.-M.; Chen, Y.; et al. C-kit induces epithelial-mesenchymal transition and contributes to salivary adenoid cystic cancer progression. Oncotarget 2014, 5, 1491–1501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.L.; Wang, W.; Lan, X.L.; Zeng, Z.C.; Liang, Y.S.; Yan, Y.R.; Song, F.Y.; Wang, F.F.; Zhu, X.H.; Liao, W.J.; et al. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol. Cancer 2019, 18, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, C.; Shan, Z.; Hong, J.; Yang, L. MicroRNA-92a promotes epithelial-mesenchymal transition through activation of PTEN/PI3K/AKT signaling pathway in non-small cell lung cancer metastasis. Int. J. Oncol. 2017, 51, 235–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, J.C.; Lima, N.d.S.; Sarian, L.O.; Matheu, A.; Ribeiro, M.L.; Derchain, S.F.M. Exosome-mediated breast cancer chemoresistance via miR-155 transfer. Sci. Rep. 2018, 8, 829. [Google Scholar] [CrossRef] [Green Version]
- Shen, R.; Wang, Y.; Wang, C.-X.; Yin, M.; Liu, H.-L.; Chen, J.-P.; Han, J.-Q.; Wang, W.-B. MiRNA-155 mediates TAM resistance by modulating SOCS6-STAT3 signalling pathway in breast cancer. Am. J. Transl. Res. 2015, 7, 2115–2126. [Google Scholar]
- Ouyang, M.; Li, Y.; Ye, S.; Ma, J.; Lu, L.; Lv, W.; Chang, G.; Li, X.; Li, Q.; Wang, S.; et al. MicroRNA profiling implies new markers of chemoresistance of triple-negative breast cancer. PLoS ONE 2014, 9, e96228. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Kong, X.; Lv, L.; Gao, J. TGF-β1 acts through miR-155 to down-regulate TP53INP1 in promoting epithelial-mesenchymal transition and cancer stem cell phenotypes. Cancer Lett. 2015, 359, 288–298. [Google Scholar] [CrossRef]
- Kong, X.; Liu, F.; Gao, J. MiR-155 promotes epithelial-mesenchymal transition in hepatocellular carcinoma cells through the activation of PI3K/SGK3/β-catenin signaling pathways. Oncotarget 2016, 7, 66051–66060. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Lee, E.; Jung, J.; Lee, J.W.; Kim, H.J.; Kim, J.; Yoo, H.J.; Lee, H.J.; Chae, S.Y.; Jeon, S.M.; et al. microRNA-155 positively regulates glucose metabolism via PIK3R1-FOXO3a-cMYC axis in breast cancer. Oncogene 2018, 37, 2982–2991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Liu, J.; Ma, J.; Sun, T.; Zhou, Q.; Wang, W.; Wang, G.; Wu, P.; Wang, H.; Jiang, L.; et al. Exosomal circRNAs: Biogenesis, effect and application in human diseases. Mol. Cancer 2019, 18, 116. [Google Scholar] [CrossRef] [PubMed]
- Su, L.-L.; Chang, X.-J.; Zhou, H.-D.; Hou, L.-B.; Xue, X.-Y. Exosomes in esophageal cancer: A review on tumorigenesis, diagnosis and therapeutic potential. World J. Clin. Cases 2019, 7, 908–916. [Google Scholar] [CrossRef] [PubMed]
- Ling, X.-H.; Chen, Z.-Y.; Luo, H.-W.; Liu, Z.-Z.; Liang, Y.-K.; Chen, G.-X.; Jiang, F.-N.; Zhong, W.-D.E. BCL9, a coactivator for Wnt/β-catenin transcription, is targeted by miR-30c and is associated with prostate cancer progression. Oncol. Lett. 2016, 11, 2001–2008. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Chen, S.; Fu, Q. Exosomes from CD133(+) cells carrying circ-ABCC1 mediate cell stemness and metastasis in colorectal cancer. J. Cell Biochem. 2020. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.-W.; Cao, C.-H.; Han, K.; Zhao, Y.-X.; Cai, M.-Y.; Xiang, Z.-C.; Zhang, J.-X.; Chen, J.-W.; Zhong, L.-P.; Huang, Y.; et al. APC-activated long noncoding RNA inhibits colorectal carcinoma pathogenesis through reduction of exosome production. J. Clin. Investig. 2019, 129, 727–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.B.; Yan, C.; Mu, L.; Mi, Y.L.; Zhao, H.; Hu, H.; Li, X.L.; Tao, D.D.; Wu, Y.Q.; Gong, J.P.; et al. Exosomal Wnt-induced dedifferentiation of colorectal cancer cells contributes to chemotherapy resistance. Oncogene 2019, 38, 1951–1965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, K.; Shamskhou, E.A.; Orcholski, M.E.; Nathan, A.; Reddy, S.; Honda, H.; Mani, V.; Zeng, Y.; Ozen, M.O.; Wang, L.; et al. Loss of Endothelium-Derived Wnt5a Is Associated With Reduced Pericyte Recruitment and Small Vessel Loss in Pulmonary Arterial Hypertension. Circulation 2019, 139, 1710–1724. [Google Scholar] [CrossRef]
- Chen, Y.; Zeng, C.; Zhan, Y.; Wang, H.; Jiang, X.; Li, W. Aberrant low expression of p85α in stromal fibroblasts promotes breast cancer cell metastasis through exosome-mediated paracrine Wnt10b. Oncogene 2017, 36, 4692–4705. [Google Scholar] [CrossRef] [Green Version]
- Luga, V.; Zhang, L.; Viloria-Petit, A.M.; Ogunjimi, A.A.; Inanlou, M.R.; Chiu, E.; Buchanan, M.; Hosein, A.N.; Basik, M.; Wrana, J.L. Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell 2012, 151, 1542–1556. [Google Scholar] [CrossRef] [Green Version]
- Luga, V.; Wrana, J.L. Tumor-stroma interaction: Revealing fibroblast-secreted exosomes as potent regulators of Wnt-planar cell polarity signaling in cancer metastasis. Cancer Res. 2013, 73, 6843–6847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Z.; Feng, Y. Exosomes Derived From Hypoxic Colorectal Cancer Cells Promote Angiogenesis Through Wnt4-Induced β-Catenin Signaling in Endothelial Cells. Oncol. Res. 2017, 25, 651–661. [Google Scholar] [CrossRef] [PubMed]
- Koch, R.; Demant, M.; Aung, T.; Diering, N.; Cicholas, A.; Chapuy, B.; Wenzel, D.; Lahmann, M.; Güntsch, A.; Kiecke, C.; et al. Populational equilibrium through exosome-mediated Wnt signaling in tumor progression of diffuse large B-cell lymphoma. Blood 2014, 123, 2189–2198. [Google Scholar] [CrossRef] [PubMed]
- Kalra, H.; Gangoda, L.; Fonseka, P.; Chitti, S.V.; Liem, M.; Keerthikumar, S.; Samuel, M.; Boukouris, S.; Al Saffar, H.; Collins, C.; et al. Extracellular vesicles containing oncogenic mutant β-catenin activate Wnt signalling pathway in the recipient cells. J. Extracell. Vesicles 2019, 8, 1690217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menck, K.; Klemm, F.; Gross, J.C.; Pukrop, T.; Wenzel, D.; Binder, C. Induction and transport of Wnt 5a during macrophage-induced malignant invasion is mediated by two types of extracellular vesicles. Oncotarget 2013, 4, 2057–2066. [Google Scholar] [CrossRef] [Green Version]
- Ekström, E.J.; Bergenfelz, C.; von Bülow, V.; Serifler, F.; Carlemalm, E.; Jönsson, G.; Andersson, T.; Leandersson, K. WNT5A induces release of exosomes containing pro-angiogenic and immunosuppressive factors from malignant melanoma cells. Mol. Cancer 2014, 13, 88. [Google Scholar] [CrossRef] [Green Version]
- You, J.; Li, M.; Cao, L.M.; Gu, Q.H.; Deng, P.B.; Tan, Y.; Hu, C.P. Snail1-dependent cancer-associated fibroblasts induce epithelial-mesenchymal transition in lung cancer cells via exosomes. QJM 2019, 112, 581–590. [Google Scholar] [CrossRef]
- Mao, J.; Liang, Z.; Zhang, B.; Yang, H.; Li, X.; Fu, H.; Zhang, X.; Yan, Y.; Xu, W.; Qian, H. UBR2 Enriched in p53 Deficient Mouse Bone Marrow Mesenchymal Stem Cell-Exosome Promoted Gastric Cancer Progression via Wnt/β-Catenin Pathway. Stem Cells 2017, 35, 2267–2279. [Google Scholar] [CrossRef] [Green Version]
- Qian, S.; Tan, X.; Liu, X.; Liu, P.; Wu, Y. Exosomal Tenascin-c induces proliferation and invasion of pancreatic cancer cells by WNT signaling. Onco Targets Ther. 2019, 12, 3197–3205. [Google Scholar] [CrossRef] [Green Version]
- Nagaharu, K.; Zhang, X.; Yoshida, T.; Katoh, D.; Hanamura, N.; Kozuka, Y.; Ogawa, T.; Shiraishi, T.; Imanaka-Yoshida, K. Tenascin C induces epithelial-mesenchymal transition-like change accompanied by SRC activation and focal adhesion kinase phosphorylation in human breast cancer cells. Am. J. Pathol. 2011, 178, 754–763. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Jiang, P.; Li, J.; Peng, M.; Zhao, X.; Zhang, X.; Chen, K.; Zhang, Y.; Liu, H.; Gan, L.; et al. Tumor-derived exosomal lnc-Sox2ot promotes EMT and stemness by acting as a ceRNA in pancreatic ductal adenocarcinoma. Oncogene 2018, 37, 3822–3838. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Shi, J.; Zhang, K.; Xue, J.; Li, J.; Yang, J.; Chen, J.; Wei, J.; Ren, H.; Liu, X. Sox2 inhibits Wnt-β-catenin signaling and metastatic potency of cisplatin-resistant lung adenocarcinoma cells. Mol. Med. Rep. 2017, 15, 1693–1701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardin, H.; Helein, H.; Meyer, K.; Robertson, S.; Zhang, R.; Zhong, W.; Lloyd, R.V. Thyroid cancer stem-like cell exosomes: Regulation of EMT via transfer of lncRNAs. Lab. Investig. 2018, 98, 1133–1142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, P.; Zhao, Y.; Li, Z.; Yao, R.; Ma, M.; Gao, Y.; Zhao, L.; Zhang, Y.; Huang, B.; Lu, J. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014, 5, e1287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lou, Y.; Jiang, H.; Cui, Z.; Wang, L.; Wang, X.; Tian, T. Linc-ROR induces epithelial-to-mesenchymal transition in ovarian cancer by increasing Wnt/β-catenin signaling. Oncotarget 2017, 8, 69983–69994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Li, C.; Wang, S.; Wang, Z.; Jiang, J.; Wang, W.; Li, X.; Chen, J.; Liu, K.; Li, C.; et al. Exosomes Derived from Hypoxic Oral Squamous Cell Carcinoma Cells Deliver miR-21 to Normoxic Cells to Elicit a Prometastatic Phenotype. Cancer Res. 2016, 76, 1770–1780. [Google Scholar] [CrossRef] [Green Version]
- Han, M.; Liu, M.; Wang, Y.; Chen, X.; Xu, J.; Sun, Y.; Zhao, L.; Qu, H.; Fan, Y.; Wu, C. Antagonism of miR-21 reverses epithelial-mesenchymal transition and cancer stem cell phenotype through AKT/ERK1/2 inactivation by targeting PTEN. PLoS ONE 2012, 7, e39520. [Google Scholar] [CrossRef] [Green Version]
- Zerial, M.; McBride, H. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2001, 2, 107–117. [Google Scholar] [CrossRef]
- Qu, J.L.; Qu, X.J.; Zhao, M.F.; Teng, Y.E.; Zhang, Y.; Hou, K.Z.; Jiang, Y.H.; Yang, X.H.; Liu, Y.P. Gastric cancer exosomes promote tumour cell proliferation through PI3K/Akt and MAPK/ERK activation. Dig. Liver Dis. 2009, 41, 875–880. [Google Scholar] [CrossRef]
- He, M.; Qin, H.; Poon, T.C.W.; Sze, S.-C.; Ding, X.; Co, N.N.; Ngai, S.-M.; Chan, T.-F.; Wong, N. Hepatocellular carcinoma-derived exosomes promote motility of immortalized hepatocyte through transfer of oncogenic proteins and RNAs. Carcinogenesis 2015, 36, 1008–1018. [Google Scholar] [CrossRef] [Green Version]
- Berrondo, C.; Flax, J.; Kucherov, V.; Siebert, A.; Osinski, T.; Rosenberg, A.; Fucile, C.; Richheimer, S.; Beckham, C.J. Expression of the Long Non-Coding RNA HOTAIR Correlates with Disease Progression in Bladder Cancer and Is Contained in Bladder Cancer Patient Urinary Exosomes. PLoS ONE 2016, 11, e0147236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conigliaro, A.; Cicchini, C. Exosome-Mediated Signaling in Epithelial to Mesenchymal Transition and Tumor Progression. J. Clin. Med. 2018, 8, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amicone, L.; Citarella, F.; Cicchini, C. Epigenetic regulation in hepatocellular carcinoma requires long noncoding RNAs. BioMed Res. Int. 2015, 2015, 473942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Battistelli, C.; Cicchini, C.; Santangelo, L.; Tramontano, A.; Grassi, L.; Gonzalez, F.J.; de Nonno, V.; Grassi, G.; Amicone, L.; Tripodi, M. The Snail repressor recruits EZH2 to specific genomic sites through the enrollment of the lncRNA HOTAIR in epithelial-to-mesenchymal transition. Oncogene 2017, 36, 942–955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, F.; Chen, B.; Dong, P.; Zheng, J. HOTAIR Epigenetically Modulates PTEN Expression via MicroRNA-29b: A Novel Mechanism in Regulation of Liver Fibrosis. Mol. Ther. 2017, 25, 205–217. [Google Scholar] [CrossRef] [Green Version]
- Ota, Y.; Takahashi, K.; Otake, S.; Tamaki, Y.; Okada, M.; Aso, K.; Makino, Y.; Fujii, S.; Ota, T.; Haneda, M. Extracellular vesicle-encapsulated miR-30e suppresses cholangiocarcinoma cell invasion and migration via inhibiting epithelial-mesenchymal transition. Oncotarget 2018, 9, 16400–16417. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.-M.; Li, Z.; Han, X.-D.; Shi, J.-H.; Tu, D.-Y.; Song, W.; Zhang, J.; Qiu, X.-L.; Ren, Y.; Zhen, L.-L. MiR-30e inhibits tumor growth and chemoresistance via targeting IRS1 in Breast Cancer. Sci. Rep. 2017, 7, 15929. [Google Scholar] [CrossRef]
- Croset, M.; Pantano, F.; Kan, C.W.S.; Bonnelye, E.; Descotes, F.; Alix-Panabières, C.; Lecellier, C.-H.; Bachelier, R.; Allioli, N.; Hong, S.-S.; et al. miRNA-30 Family Members Inhibit Breast Cancer Invasion, Osteomimicry, and Bone Destruction by Directly Targeting Multiple Bone Metastasis-Associated Genes. Cancer Res. 2018, 78, 5259–5273. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.; Zhao, L.; Zhao, S.; Guo, T.; Li, F.; Li, Z.; Fang, L.; Wu, T.; Gu, C. MicroRNA-30e inhibits proliferation and invasion of non-small cell lung cancer via targeting SOX9. Hum. Cell 2019, 32, 326–333. [Google Scholar] [CrossRef]
- Xu, G.; Cai, J.; Wang, L.; Jiang, L.; Huang, J.; Hu, R.; Ding, F. MicroRNA-30e-5p suppresses non-small cell lung cancer tumorigenesis by regulating USP22-mediated Sirt1/JAK/STAT3 signaling. Exp. Cell Res. 2018, 362, 268–278. [Google Scholar] [CrossRef]
- Liu, K.; Xie, F.; Gao, A.; Zhang, R.; Zhang, L.; Xiao, Z.; Hu, Q.; Huang, W.; Huang, Q.; Lin, B.; et al. SOX2 regulates multiple malignant processes of breast cancer development through the SOX2/miR-181a-5p, miR-30e-5p/TUSC3 axis. Mol. Cancer 2017, 16, 62. [Google Scholar] [CrossRef] [PubMed]
- Laudato, S.; Patil, N.; Abba, M.L.; Leupold, J.H.; Benner, A.; Gaiser, T.; Marx, A.; Allgayer, H. P53-induced miR-30e-5p inhibits colorectal cancer invasion and metastasis by targeting ITGA6 and ITGB1. Int. J. Cancer 2017, 141, 1879–1890. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wang, X.; Zhu, Z.; Li, W.; Yu, G.; Jia, Z.; Wang, X. Prostate carcinoma cell-derived exosomal MicroRNA-26a modulates the metastasis and tumor growth of prostate carcinoma. Biomed. Pharmacother. 2019, 117, 109109. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Duan, W.; Sun, W. LncRNA SNHG6 promotes the migration, invasion, and epithelial-mesenchymal transition of colorectal cancer cells by miR-26a/EZH2 axis. Onco Targets Ther. 2019, 12, 3349–3360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.C.; Li, S.Y.; Jia, Y.F. MicroRNA-26a suppresses the malignant biological behaviors of papillary thyroid carcinoma by targeting ROCK1 and regulating PI3K/AKT signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8940–8949. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Guan, M.; Jia, Y.; Wang, D.; Pang, R.; Lv, F.; Xiao, Z.; Wang, L.; Zhang, H.; Xue, Y. The coordinated roles of miR-26a and miR-30c in regulating TGFβ1-induced epithelial-to-mesenchymal transition in diabetic nephropathy. Sci. Rep. 2016, 6, 37492. [Google Scholar] [CrossRef]
- Chen, X.; Xiao, W.; Chen, W.; Liu, X.; Wu, M.; Bo, Q.; Luo, Y.; Ye, S.; Cao, Y.; Liu, Y. MicroRNA-26a and -26b inhibit lens fibrosis and cataract by negatively regulating Jagged-1/Notch signaling pathway. Cell Death Differ. 2017, 24, 1431–1442. [Google Scholar] [CrossRef] [Green Version]
- Fan, B.; Li, L.; Xiao, D.J.; Xu, T. Study of miR-26a inhibiting epithelial-mesenchymal transition and invasion of Tu686 cell line through SMAD1. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 217–224. [Google Scholar] [CrossRef]
- Geng, C.; Wei, J.; Wu, C. Yap-Hippo pathway regulates cerebral hypoxia-reoxygenation injury in neuroblastoma N2a cells via inhibiting ROCK1/F-actin/mitochondrial fission pathways. Acta Neurol. Belg. 2018, 1–14. [Google Scholar] [CrossRef]
- Slabáková, E.; Culig, Z.; Remšík, J.; Souček, K. Alternative mechanisms of miR-34a regulation in cancer. Cell Death Dis. 2017, 8, e3100. [Google Scholar] [CrossRef]
- Li, Y.-Y.; Tao, Y.-W.; Gao, S.; Li, P.; Zheng, J.-M.; Zhang, S.-E.; Liang, J.; Zhang, Y. Cancer-associated fibroblasts contribute to oral cancer cells proliferation and metastasis via exosome-mediated paracrine miR-34a-5p. EBioMedicine 2018, 36, 209–220. [Google Scholar] [CrossRef] [Green Version]
- Rokavec, M.; Öner, M.G.; Li, H.; Jackstadt, R.; Jiang, L.; Lodygin, D.; Kaller, M.; Horst, D.; Ziegler, P.K.; Schwitalla, S.; et al. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J. Clin. Investig. 2014, 124, 1853–1867. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Tian, X.; Li, Y.; Wang, Z.; Li, X.; Zhu, C. miR-27b and miR-34a enhance docetaxel sensitivity of prostate cancer cells through inhibiting epithelial-to-mesenchymal transition by targeting ZEB1. Biomed. Pharmacother. 2018, 97, 736–744. [Google Scholar] [CrossRef]
- Liang, J.; Li, Y.; Daniels, G.; Sfanos, K.; De Marzo, A.; Wei, J.; Li, X.; Chen, W.; Wang, J.; Zhong, X.; et al. LEF1 Targeting EMT in Prostate Cancer Invasion Is Regulated by miR-34a. Mol. Cancer Res. 2015, 13, 681–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, P.; Xiong, Y.; Watari, H.; Hanley, S.J.B.; Konno, Y.; Ihira, K.; Yamada, T.; Kudo, M.; Yue, J.; Sakuragi, N. MiR-137 and miR-34a directly target Snail and inhibit EMT, invasion and sphere-forming ability of ovarian cancer cells. J. Exp. Clin. Cancer Res. 2016, 35, 132. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Wang, G.; Liu, H.; Xiong, C. SATB2 targeted by methylated miR-34c-5p suppresses proliferation and metastasis attenuating the epithelial-mesenchymal transition in colorectal cancer. Cell Prolif. 2018, 51, e12455. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Li, Y.; Pan, Q.; Ye, M.; He, S.; Tian, Q.; Xue, M. MiR-34c/SOX9 axis regulates the chemoresistance of ovarian cancer cell to cisplatin-based chemotherapy. J. Cell Biochem. 2019, 120, 2940–2953. [Google Scholar] [CrossRef]
- Russo, V.; Paciocco, A.; Affinito, A.; Roscigno, G.; Fiore, D.; Palma, F.; Galasso, M.; Volinia, S.; Fiorelli, A.; Esposito, C.L.; et al. Aptamer-miR-34c Conjugate Affects Cell Proliferation of Non-Small-Cell Lung Cancer Cells. Mol. Ther. Nucleic Acids 2018, 13, 334–346. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Li, W.-Z.; Huang, M.-L.; Wei, H.-L.; Wang, T.; Fan, J.; Li, N.-L.; Ling, R. Regulation of cancerous progression and epithelial-mesenchymal transition by miR-34c-3p via modulation of MAP3K2 signaling in triple-negative breast cancer cells. Biochem. Biophys. Res. Commun. 2017, 483, 10–16. [Google Scholar] [CrossRef]
- Wang, J.-X.; Yang, Y.; Li, K. Long noncoding RNA DANCR aggravates retinoblastoma through miR-34c and miR-613 by targeting MMP-9. J. Cell Physiol. 2018, 233, 6986–6995. [Google Scholar] [CrossRef]
- Peng, D.; Wang, H.; Li, L.; Ma, X.; Chen, Y.; Zhou, H.; Luo, Y.; Xiao, Y.; Liu, L. miR-34c-5p promotes eradication of acute myeloid leukemia stem cells by inducing senescence through selective RAB27B targeting to inhibit exosome shedding. Leukemia 2018, 32, 1180–1188. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Du, L.-Y.; Guo, F.; Li, X.; Cheng, B. Exosomes derived from microRNA-101-3p-overexpressing human bone marrow mesenchymal stem cells suppress oral cancer cell proliferation, invasion, and migration. Mol. Cell Biochem. 2019, 458, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Cogdell, D.; Hu, L.; Yang, D.; Sood, A.K.; Xue, F.; Zhang, W. MiR-101 suppresses the epithelial-to-mesenchymal transition by targeting ZEB1 and ZEB2 in ovarian carcinoma. Oncol. Rep. 2014, 31, 2021–2028. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Zhao, Y.; Li, X.; Tao, Z.; Hou, M.; Ma, H. Overexpression of HIF-2α-Dependent NEAT1 Promotes the Progression of Non-Small Cell Lung Cancer through miR-101-3p/SOX9/Wnt/β-Catenin Signal Pathway. Cell Physiol. Biochem. 2019, 52, 368–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, Z.; Yin, S.; Su, Z.; Bai, M.; Zhang, H.; Hei, Z.; Cai, S. Downregulation of miR-101 contributes to epithelial-mesenchymal transition in cisplatin resistance of NSCLC cells by targeting ROCK2. Oncotarget 2016, 7, 37524–37535. [Google Scholar] [CrossRef]
- Wei, X.; Tang, C.; Lu, X.; Liu, R.; Zhou, M.; He, D.; Zheng, D.; Sun, C.; Wu, Z. MiR-101 targets DUSP1 to regulate the TGF-β secretion in sorafenib inhibits macrophage-induced growth of hepatocarcinoma. Oncotarget 2015, 6, 18389–18405. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Shao, M.-Y.; Zou, S.-C.; Xiao, Z.-F.; Chen, Z.-C. MiR-101-3p inhibits EMT to attenuate proliferation and metastasis in glioblastoma by targeting TRIM44. J. Neurooncol. 2019, 141, 19–30. [Google Scholar] [CrossRef]
- Che, Y.; Shi, X.; Shi, Y.; Jiang, X.; Ai, Q.; Shi, Y.; Gong, F.; Jiang, W. Exosomes Derived from miR-143-Overexpressing MSCs Inhibit Cell Migration and Invasion in Human Prostate Cancer by Downregulating TFF3. Mol. Ther. Nucleic Acids 2019, 18, 232–244. [Google Scholar] [CrossRef] [Green Version]
- Yusup, A.; Huji, B.; Fang, C.; Wang, F.; Dadihan, T.; Wang, H.-J.; Upur, H. Expression of trefoil factors and TWIST1 in colorectal cancer and their correlation with metastatic potential and prognosis. World J. Gastroenterol. 2017, 23, 110–120. [Google Scholar] [CrossRef]
- Lei, C.; Du, F.; Sun, L.; Li, T.; Li, T.; Min, Y.; Nie, A.; Wang, X.; Geng, L.; Lu, Y.; et al. miR-143 and miR-145 inhibit gastric cancer cell migration and metastasis by suppressing MYO6. Cell Death Dis. 2017, 8, e3101. [Google Scholar] [CrossRef] [Green Version]
- Luo, J.; Chen, J.; Li, H.; Yang, Y.; Yun, H.; Yang, S.; Mao, X. LncRNA UCA1 promotes the invasion and EMT of bladder cancer cells by regulating the miR-143/HMGB1 pathway. Oncol. Lett 2017, 14, 5556–5562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhai, L.; Ma, C.; Li, W.; Yang, S.; Liu, Z. miR-143 suppresses epithelial-mesenchymal transition and inhibits tumor growth of breast cancer through down-regulation of ERK5. Mol. Carcinog. 2016, 55, 1990–2000. [Google Scholar] [CrossRef] [PubMed]
- Bufalino, A.; Cervigne, N.K.; de Oliveira, C.E.; Fonseca, F.P.; Rodrigues, P.C.; Macedo, C.C.S.; Sobral, L.M.; Miguel, M.C.; Lopes, M.A.; Paes Leme, A.F.; et al. Low miR-143/miR-145 Cluster Levels Induce Activin A Overexpression in Oral Squamous Cell Carcinomas, Which Contributes to Poor Prognosis. PLoS ONE 2015, 10, e0136599. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Shen, S.; Zheng, X.; Ye, K.; Sun, Y.; Lu, Y.; Ge, H. Long noncoding RNA HAGLR acts as a microRNA-143-5p sponge to regulate epithelial-mesenchymal transition and metastatic potential in esophageal cancer by regulating LAMP3. FASEB J. 2019, 33, 10490–10504. [Google Scholar] [CrossRef]
- Luo, H.; Liang, C. MicroRNA-148b inhibits proliferation and the epithelial-mesenchymal transition and increases radiosensitivity in non-small cell lung carcinomas by regulating ROCK1. Exp. Ther. Med. 2018, 15, 3609–3616. [Google Scholar] [CrossRef]
- Zhou, Z.; Su, Y.; Fa, X. Restoration of BRG1 inhibits proliferation and metastasis of lung cancer by regulating tumor suppressor miR-148b. Onco Targets Ther. 2015, 8, 3603–3612. [Google Scholar] [CrossRef] [Green Version]
- Yuan, L.; Liu, Y.; Qu, Y.; Liu, L.; Li, H. Exosomes Derived From MicroRNA-148b-3p-Overexpressing Human Umbilical Cord Mesenchymal Stem Cells Restrain Breast Cancer Progression. Front. Oncol. 2019, 9, 1076. [Google Scholar] [CrossRef] [Green Version]
- Li, B.-L.; Lu, W.; Qu, J.-J.; Ye, L.; Du, G.-Q.; Wan, X.-P. Loss of exosomal miR-148b from cancer-associated fibroblasts promotes endometrial cancer cell invasion and cancer metastasis. J. Cell Physiol. 2019, 234, 2943–2953. [Google Scholar] [CrossRef]
- Senfter, D.; Holzner, S.; Kalipciyan, M.; Staribacher, A.; Walzl, A.; Huttary, N.; Krieger, S.; Brenner, S.; Jäger, W.; Krupitza, G.; et al. Loss of miR-200 family in 5-fluorouracil resistant colon cancer drives lymphendothelial invasiveness in vitro. Hum. Mol. Genet. 2015, 24, 3689–3698. [Google Scholar] [CrossRef] [Green Version]
- Title, A.C.; Hong, S.-J.; Pires, N.D.; Hasenöhrl, L.; Godbersen, S.; Stokar-Regenscheit, N.; Bartel, D.P.; Stoffel, M. Genetic dissection of the miR-200-Zeb1 axis reveals its importance in tumor differentiation and invasion. Nat. Commun. 2018, 9, 4671. [Google Scholar] [CrossRef]
- Pan, Q.; Meng, L.; Ye, J.; Wei, X.; Shang, Y.; Tian, Y.; He, Y.; Peng, Z.; Chen, L.; Chen, W.; et al. Transcriptional repression of miR-200 family members by Nanog in colon cancer cells induces epithelial-mesenchymal transition (EMT). Cancer Lett. 2017, 392, 26–38. [Google Scholar] [CrossRef]
- Kundu, S.T.; Byers, L.A.; Peng, D.H.; Roybal, J.D.; Diao, L.; Wang, J.; Tong, P.; Creighton, C.J.; Gibbons, D.L. The miR-200 family and the miR-183~96~182 cluster target Foxf2 to inhibit invasion and metastasis in lung cancers. Oncogene 2016, 35, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Le, M.T.N.; Hamar, P.; Guo, C.; Basar, E.; Perdigão-Henriques, R.; Balaj, L.; Lieberman, J. miR-200-containing extracellular vesicles promote breast cancer cell metastasis. J. Clin. Investig. 2014, 124, 5109–5128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De, A.; Powers, B.; De, A.; Zhou, J.; Sharma, S.; Van Veldhuizen, P.; Bansal, A.; Sharma, R.; Sharma, M. Emblica officinalis extract downregulates pro-angiogenic molecules via upregulation of cellular and exosomal miR-375 in human ovarian cancer cells. Oncotarget 2016, 7, 31484–31500. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.-Z.; Lai, M.-F.; Li, Y.-P.; Xu, C.-H.; Zhang, H.-R.; Kuang, J.-G. Human marrow stromal cells secrete microRNA-375-containing exosomes to regulate glioma progression. Cancer Gene Ther. 2019, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.-W.; Lin, J.-S.; He, X.-X. The emerging role of miR-375 in cancer. Int. J. Cancer 2014, 135, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Selth, L.A.; Das, R.; Townley, S.L.; Coutinho, I.; Hanson, A.R.; Centenera, M.M.; Stylianou, N.; Sweeney, K.; Soekmadji, C.; Jovanovic, L.; et al. A ZEB1-miR-375-YAP1 pathway regulates epithelial plasticity in prostate cancer. Oncogene 2017, 36, 24–34. [Google Scholar] [CrossRef]
- Guo, F.; Gao, Y.; Sui, G.; Jiao, D.; Sun, L.; Fu, Q.; Jin, C. miR-375-3p/YWHAZ/β-catenin axis regulates migration, invasion, EMT in gastric cancer cells. Clin. Exp. Pharmacol. Physiol. 2019, 46, 144–152. [Google Scholar] [CrossRef]
- ClinicalTrials.gov. Circulating Exosome RNA in Lung Metastases of Primary High-Grade Osteosarcoma. 2017. Available online: https://clinicaltrials.gov/ct2/show/NCT03108677 (accessed on 11 April 2017).
- ClinicalTrials.gov. Identification and Characterization of Predictive Factors of Onset of Bone Metastases in Cancer Patients. Available online: https://clinicaltrials.gov/ct2/show/NCT03895216 (accessed on 29 March 2019).
- ClinicalTrials.gov. Trial of a Vaccination with Tumor Antigen-Loaded Dendritic Cell-Derived Exosomes. Available online: https://clinicaltrials.gov/ct2/show/NCT01159288 (accessed on 9 July 2010).
- ClinicalTrials.gov. Study Investigating the Ability of Plant Exosomes to Deliver Curcumin to Normal and Colon Cancer Tissue. 2011. Available online: https://clinicaltrials.gov/ct2/show/NCT01294072 (accessed on 11 February 2011).
- ClinicalTrials.gov. iExosomes in Treating Participants with Metastatic Pancreas Cancer with KrasG12D Mutation. Available online: https://clinicaltrials.gov/ct2/show/NCT03608631 (accessed on 1 August 2018).
- Kamerkar, S.; LeBleu, V.S.; Sugimoto, H.; Yang, S.; Ruivo, C.F.; Melo, S.A.; Lee, J.J.; Kalluri, R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 2017, 546, 498–503. [Google Scholar] [CrossRef]
- Theodoraki, M.-N.; Yerneni, S.S.; Brunner, C.; Theodorakis, J.; Hoffmann, T.K.; Whiteside, T.L. Plasma-derived Exosomes Reverse Epithelial-to-Mesenchymal Transition after Photodynamic Therapy of Patients with Head and Neck Cancer. Oncoscience 2018, 5, 75–87. [Google Scholar] [CrossRef] [Green Version]
- Wels, C.; Joshi, S.; Koefinger, P.; Bergler, H.; Schaider, H. Transcriptional activation of ZEB1 by Slug leads to cooperative regulation of the epithelial-mesenchymal transition-like phenotype in melanoma. J. Investig. Dermatol. 2011, 131, 1877–1885. [Google Scholar] [CrossRef] [Green Version]
- Monfared, H.; Jahangard, Y.; Nikkhah, M.; Mirnajafi-Zadeh, J.; Mowla, S.J. Potential Therapeutic Effects of Exosomes Packed With a miR-21-Sponge Construct in a Rat Model of Glioblastoma. Front. Oncol. 2019, 9, 782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krichevsky, A.M.; Gabriely, G. miR-21: A small multi-faceted RNA. J. Cell Mol. Med. 2009, 13, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Costa, P.M.; Cardoso, A.L.; Nóbrega, C.; Pereira de Almeida, L.F.; Bruce, J.N.; Canoll, P.; Pedroso de Lima, M.C. MicroRNA-21 silencing enhances the cytotoxic effect of the antiangiogenic drug sunitinib in glioblastoma. Hum. Mol. Genet. 2013, 22, 904–918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kyuno, D.; Zhao, K.; Bauer, N.; Ryschich, E.; Zöller, M. Therapeutic Targeting Cancer-Initiating Cell Markers by Exosome miRNA: Efficacy and Functional Consequences Exemplified for claudin7 and EpCAM. Transl. Oncol. 2019, 12, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Philip, R.; Heiler, S.; Mu, W.; Büchler, M.W.; Zöller, M.; Thuma, F. Claudin-7 promotes the epithelial-mesenchymal transition in human colorectal cancer. Oncotarget 2015, 6, 2046–2063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Zhou, C.-Z.; Zhu, R.; Fan, H.; Liu, X.-X.; Duan, X.-Y.; Tang, Q.; Shou, Z.-X.; Zuo, D.-M. miR-200b-containing microvesicles attenuate experimental colitis associated intestinal fibrosis by inhibiting epithelial-mesenchymal transition. J. Gastroenterol. Hepatol. 2017, 32, 1966–1974. [Google Scholar] [CrossRef]
- Almanza, G.; Rodvold, J.J.; Tsui, B.; Jepsen, K.; Carter, H.; Zanetti, M. Extracellular vesicles produced in B cells deliver tumor suppressor miR-335 to breast cancer cells disrupting oncogenic programming in vitro and in vivo. Sci. Rep. 2018, 8, 17581. [Google Scholar] [CrossRef]
- Fujiwara, T.; Eguchi, T.; Sogawa, C.; Ono, K.; Murakami, J.; Ibaragi, S.; Asaumi, J.-I.; Calderwood, S.K.; Okamoto, K.; Kozaki, K.-I. Carcinogenic epithelial-mesenchymal transition initiated by oral cancer exosomes is inhibited by anti-EGFR antibody cetuximab. Oral Oncol. 2018, 86, 251–257. [Google Scholar] [CrossRef]
- Yang, T.; Martin, P.; Fogarty, B.; Brown, A.; Schurman, K.; Phipps, R.; Yin, V.P.; Lockman, P.; Bai, S. Exosome delivered anticancer drugs across the blood-brain barrier for brain cancer therapy in Danio rerio. Pharm. Res. 2015, 32, 2003–2014. [Google Scholar] [CrossRef]
- Vázquez-Ríos, A.J.; Molina-Crespo, Á.; Bouzo, B.L.; López-López, R.; Moreno-Bueno, G.; de la Fuente, M. Exosome-mimetic nanoplatforms for targeted cancer drug delivery. J. Nanobiotechnol. 2019, 17, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Name | Cancer Type | Target | Signaling Pathway | Reference | |
|---|---|---|---|---|---|
| miRNA | miR-665 | Hepatocellular carcinoma | PTPRB | Hippo pathway | [23,24] |
| miR-34c | Nasopharyngeal carcinoma | β-catenin, SOX9, SATB2, MAP3K2, DANCR, RAB27B | Wnt/β-catenin pathway | [92,93,94,95,96,97] | |
| miR-301a | GBM | TCEAL7, p63, ZEB1, ZEB2, E-cadherin | [31,32] | ||
| miR-92a | Colorectal cancer | FBXW7, MOAP1 | [36] | ||
| miR-155 | Breast cancer | FOXO3a, SGK3, p85a | [38,39,40,41,42,43] | ||
| miR-30c | Esophageal cancer | circPRMT5, BCL9 | [44,45,46] | ||
| miR-1260b | Lung adenocarcinoma | SOCS6, YY1 | [33] | ||
| miR-375 | Gastric cancer | 14-3-3ζ, YAP, SNAI1, SLC31A1 | [120,121,122,123,124] | ||
| miR-34a | OSCC | STAT3, LEF1, ZEB1, SNAI1 | NOTCH signaling pathway | [87,88,89,90,91] | |
| miR-1260b | NSCLC | KIT, MEK, ERK | ERK pathway | [34,35] | |
| miR-21 | OSCC | HIF-1a, HIF-2a, PTEN | [64] | ||
| miR-92a | NSCLC | PTEN, PI3K, AKT | PI3K/AKT pathway | [37] | |
| Protein | AREG | Chronic myelogenous leukemia | EGFR, SNAI1, YAP/TAZ | Hippo pathway | [25,26] |
| linc-ROR | Breast cancer | miRNP, miR-205, ZEB2 | Wnt/β-catenin pathway | [69] | |
| Cir-ABCC1 | Colorectal cancer | Unclear | [47] | ||
| UBR2 | Gastric cancer | p53 | [60] | ||
| TNC | PDAC | E-cadherin, CD44, MMP-7, MET | [61] | ||
| TNC | Breast cancer | p-Y418, p-FAK, SRC | [62] | ||
| Wnt1 | Colorectal cancer | TCF/LEF | [48] | ||
| Wnt3a | Colorectal cancer | TCF/LEF | [49] | ||
| Wnt4 | Colorectal cancer | HIF1α | [54] | ||
| Wnt5a | Breast cancer | p38 | [50,57,58] | ||
| Wnt10b | Breast cancer | p85a | [51] | ||
| Wnt11a | Breast cancer | PCP | [52,53] | ||
| HOTAIR | Bladder cancer | PTEN, miR-29b, DNMT3b | MAPK/ERK pathway | [81,82,83,84,85] | |
| Cbl | Liver cancerGastric cancer | Rab27a, AKT | [65,66,67] | ||
| lncRNA-Sox2ot | PDAC | Sox2ot, miR-200 | Sox2ot pathway | [68] | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.; Lee, S.; Shin, E.; Seong, K.M.; Jin, Y.W.; Youn, H.; Youn, B. The Emerging Roles of Exosomes as EMT Regulators in Cancer. Cells 2020, 9, 861. https://doi.org/10.3390/cells9040861
Kim H, Lee S, Shin E, Seong KM, Jin YW, Youn H, Youn B. The Emerging Roles of Exosomes as EMT Regulators in Cancer. Cells. 2020; 9(4):861. https://doi.org/10.3390/cells9040861
Chicago/Turabian StyleKim, Hyunwoo, Sungmin Lee, Eunguk Shin, Ki Moon Seong, Young Woo Jin, HyeSook Youn, and BuHyun Youn. 2020. "The Emerging Roles of Exosomes as EMT Regulators in Cancer" Cells 9, no. 4: 861. https://doi.org/10.3390/cells9040861