Digital Spindle: A New Way to Explore Mitotic Functions by Whole Cell Data Collection and a Computational Approach
Abstract
:1. Introduction
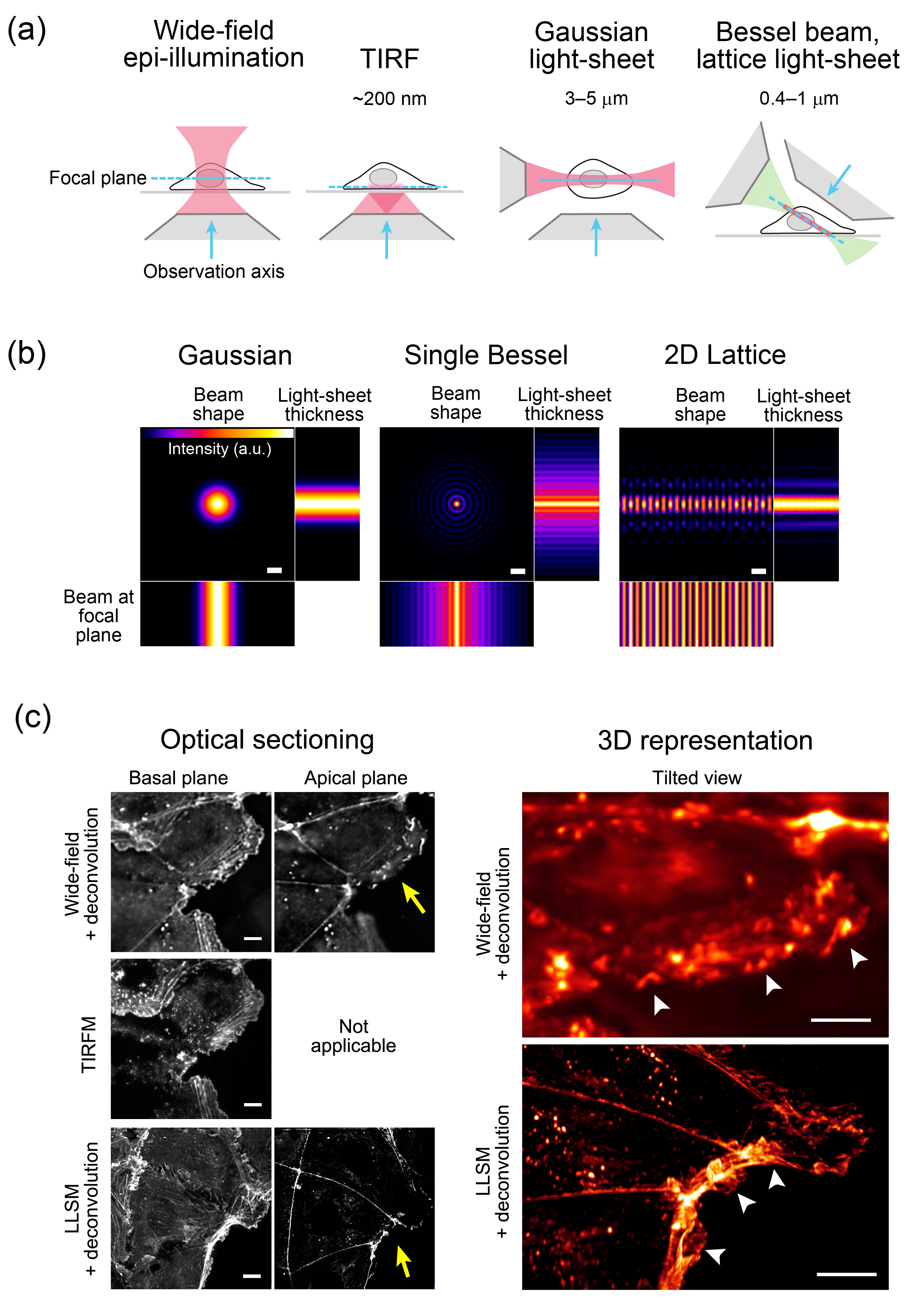
2. High Spatiotemporal Resolution, Whole-Cell, 3D Live Imaging by LLSM
2.1. Principles of LLSM
2.2. Image Collection by LLSM
2.3. LLSM with Adaptive Optics (AO-LLSM)
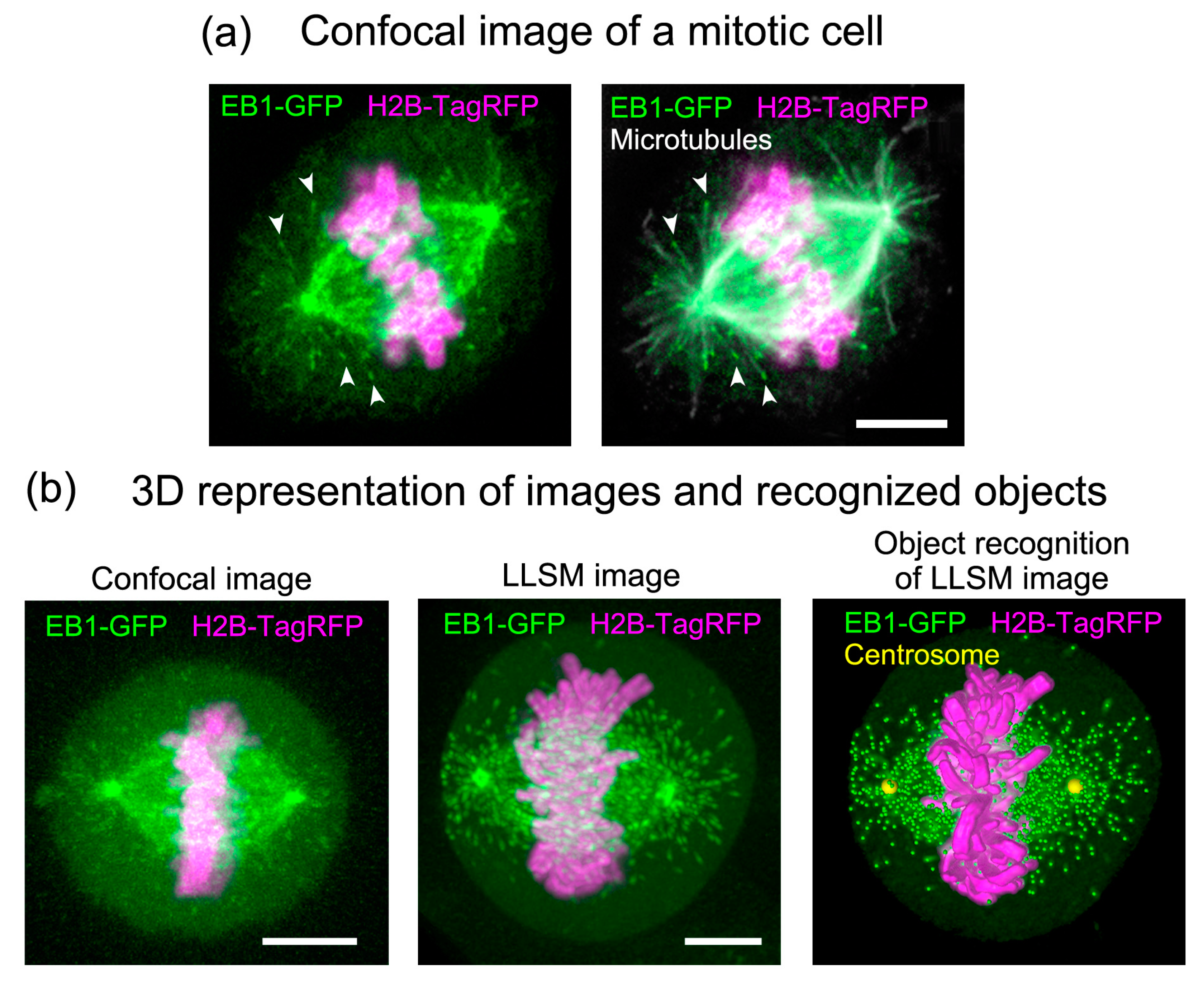
3. Image Data Processing and Analysis
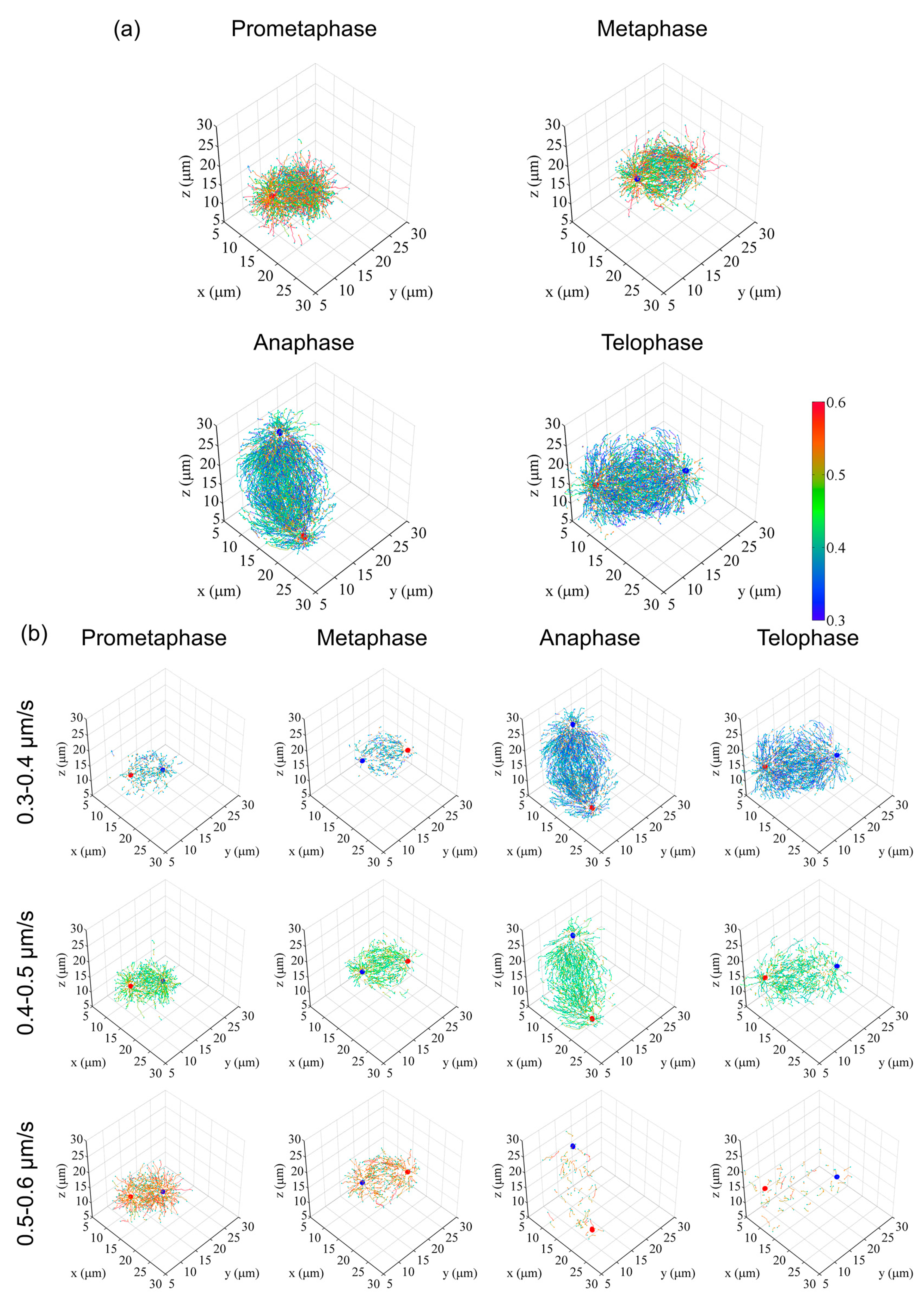
3.1. Whole-Cell 3D Movie of Mitosis and Generation of Digital Spindles
3.2. Cell Division in Developing Embryos
3.3. Digital Data Archiving and Its Utilization
3.4. Big Image Data Problem
4. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pampaloni, F.; Reynaud, E.G.; Stelzer, E.H.K. The third dimension bridges the gap between cell culture and live tissue. Nat. Rev. Mol. Cell Biol. 2007, 8, 839–845. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Lavis, L.D.; Betzig, E. Imaging live-cell dynamics and structure at the single-molecule level. Mol. Cell 2015, 58, 644–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoué, S. Video image processing greatly enhances contrast, quality, and speed in polarization-based microscopy. J. Cell Biol. 1981, 89, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Inoué, S.; Sato, H. Cell motility by labile association of molecules. The nature of mitotic spindle fibers and their role in chromosome movement. J. Gen. Physiol. 1967, 50, 259–292. [Google Scholar] [CrossRef] [Green Version]
- Salmon, E.D. Spindle Microtubules Thermodynamics of Invivo Assembly and Role in Chromosome Movement. Ann. N. Y. Acad. Sci. 1975, 253, 383–406. [Google Scholar] [CrossRef] [PubMed]
- Waterman-Storer, C.M.; Desai, A.; Bulinski, J.C.; Salmon, E.D. Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Curr. Biol. 1998, 8, 1227–1230. [Google Scholar] [CrossRef] [Green Version]
- Mitchison, T.; Evans, L.; Schulze, E.; Kirschner, M. Sites of microtubule assembly and disassembly in the mitotic spindle. Cell 1986, 45, 515–527. [Google Scholar] [CrossRef]
- Prosser, S.L.; Pelletier, L. Mitotic spindle assembly in animal cells: A fine balancing act. Nat. Rev. Mol. Cell Biol. 2017, 18, 187–201. [Google Scholar] [CrossRef]
- Kline-Smith, S.L.; Walczak, C.E. Mitotic spindle assembly and chromosome segregation: Refocusing on microtubule dynamics. Mol. Cell 2004, 15, 317–327. [Google Scholar] [CrossRef]
- Pines, J. Mitosis: A matter of getting rid of the right protein at the right time. Trends Cell Biol. 2006, 16, 55–63. [Google Scholar] [CrossRef]
- Chen, B.C.; Legant, W.R.; Wang, K.; Shao, L.; Milkie, D.E.; Davidson, M.W.; Janetopoulos, C.; Wu, X.S.; Hammer, J.A., 3rd; Liu, Z.; et al. Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution. Science 2014, 346, 1257998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamashita, N.; Morita, M.; Legant, W.R.; Chen, B.C.; Betzig, E.; Yokota, H.; Mimori-Kiyosue, Y. Three-dimensional tracking of plus-tips by lattice light-sheet microscopy permits the quantification of microtubule growth trajectories within the mitotic apparatus. J. Biomed. Opt. 2015, 20, 101206. [Google Scholar] [CrossRef] [PubMed]
- Mimori-Kiyosue, Y.; Shiina, N.; Tsukita, S. The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr. Biol. 2000, 10, 865–868. [Google Scholar] [CrossRef] [Green Version]
- Matov, A.; Applegate, K.; Kumar, P.; Thoma, C.; Krek, W.; Danuser, G.; Wittmann, T. Analysis of microtubule dynamic instability using a plus-end growth marker. Nat. Methods 2010, 7, 761–768. [Google Scholar] [CrossRef] [Green Version]
- Muroyama, A.; Lechler, T. A transgenic toolkit for visualizing and perturbing microtubules reveals unexpected functions in the epidermis. Elife 2017, 6. [Google Scholar] [CrossRef]
- Abe, T.; Kiyonari, H.; Shioi, G.; Inoue, K.; Nakao, K.; Aizawa, S.; Fujimori, T. Establishment of conditional reporter mouse lines at ROSA26 locus for live cell imaging. Genesis 2011, 49, 579–590. [Google Scholar] [CrossRef]
- Srayko, M.; Kaya, A.; Stamford, J.; Hyman, A.A. Identification and characterization of factors required for microtubule growth and nucleation in the early C. elegans embryo. Dev. Cell 2005, 9, 223–236. [Google Scholar] [CrossRef] [Green Version]
- Applegate, K.T.; Besson, S.; Matov, A.; Bagonis, M.H.; Jaqaman, K.; Danuser, G. plusTipTracker: Quantitative image analysis software for the measurement of microtubule dynamics. J. Struct. Biol. 2011, 176, 168–184. [Google Scholar] [CrossRef] [Green Version]
- Wan, Y.; McDole, K.; Keller, P.J. Light-Sheet Microscopy and Its Potential for Understanding Developmental Processes. Annu. Rev. Cell Dev. Biol. 2019, 35, 655–681. [Google Scholar] [CrossRef] [Green Version]
- Keller, P.J.; Schmidt, A.D.; Wittbrodt, J.; Stelzer, E.H. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 2008, 322, 1065–1069. [Google Scholar] [CrossRef] [Green Version]
- Ozawa, M.; Hiver, S.; Yamamoto, T.; Shibata, T.; Upadhyayula, S.; Mimori-Kiyosue, Y.; Takeichi, M. Adherens junction serves to generate cryptic lamellipodia required for collective migration of epithelial cells. bioRxiv 2020. [Google Scholar] [CrossRef]
- Durnin, J.; Miceli, J.J., Jr.; Eberly, J.H. Diffraction-free beams. Phys. Rev. Lett. 1987, 58, 1499–1501. [Google Scholar] [CrossRef] [PubMed]
- Durnin, J. Exact solutions for nondiffracting beams. I The scalar theory. J. Opt. Soc. Am. A-Opt. Image Sci. Vis. 1987, 4, 651–654. [Google Scholar] [CrossRef]
- Planchon, T.A.; Gao, L.; Milkie, D.E.; Davidson, M.W.; Galbraith, J.A.; Galbraith, C.G.; Betzig, E. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat. Methods 2011, 8, 417–423. [Google Scholar] [CrossRef] [Green Version]
- Fahrbach, F.O.; Gurchenkov, V.; Alessandri, K.; Nassoy, P.; Rohrbach, A. Self-reconstructing sectioned Bessel beams offer submicron optical sectioning for large fields of view in light-sheet microscopy. Opt. Express 2013, 21, 11425–11440. [Google Scholar] [CrossRef]
- Huisken, J.; Stainier, D.Y. Selective plane illumination microscopy techniques in developmental biology. Development 2009, 136, 1963–1975. [Google Scholar] [CrossRef] [Green Version]
- Mertz, J. Optical sectioning microscopy with planar or structured illumination. Nat. Methods 2011, 8, 811–819. [Google Scholar] [CrossRef]
- Krzic, U.; Gunther, S.; Saunders, T.E.; Streichan, S.J.; Hufnagel, L. Multiview light-sheet microscope for rapid in toto imaging. Nat. Methods 2012. [Google Scholar] [CrossRef]
- Truong, T.V.; Supatto, W.; Koos, D.S.; Choi, J.M.; Fraser, S.E. Deep and fast live imaging with two-photon scanned light-sheet microscopy. Nat. Methods 2011, 8, 757–760. [Google Scholar] [CrossRef] [Green Version]
- Ahrens, M.B.; Orger, M.B.; Robson, D.N.; Li, J.M.; Keller, P.J. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat Methods 2013, 10, 413–420. [Google Scholar] [CrossRef]
- Booth, M.J. Adaptive optics in microscopy. Philos. Trans. A Math Phys. Eng. Sci. 2007, 365, 2829–2843. [Google Scholar] [CrossRef]
- Royer, L.A.; Lemon, W.C.; Chhetri, R.K.; Wan, Y.; Coleman, M.; Myers, E.W.; Keller, P.J. Adaptive light-sheet microscopy for long-term, high-resolution imaging in living organisms. Nat. Biotechnol. 2016, 34, 1267–1278. [Google Scholar] [CrossRef]
- Wang, K.; Milkie, D.E.; Saxena, A.; Engerer, P.; Misgeld, T.; Bronner, M.E.; Mumm, J.; Betzig, E. Rapid adaptive optical recovery of optimal resolution over large volumes. Nat. Methods 2014, 11, 625–628. [Google Scholar] [CrossRef] [Green Version]
- Ji, N.; Milkie, D.E.; Betzig, E. Adaptive optics via pupil segmentation for high-resolution imaging in biological tissues. Nat Methods 2010, 7, 141–147. [Google Scholar] [CrossRef]
- Liu, T.L.; Upadhyayula, S.; Milkie, D.E.; Singh, V.; Wang, K.; Swinburne, I.A.; Mosaliganti, K.R.; Collins, Z.M.; Hiscock, T.W.; Shea, J.; et al. Observing the cell in its native state: Imaging subcellular dynamics in multicellular organisms. Science 2018, 360. [Google Scholar] [CrossRef] [Green Version]
- Tillberg, P.W.; Chen, F.; Piatkevich, K.D.; Zhao, Y.; Yu, C.C.; English, B.P.; Gao, L.; Martorell, A.; Suk, H.J.; Yoshida, F.; et al. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat. Biotechnol. 2016, 34, 987–992. [Google Scholar] [CrossRef] [Green Version]
- Gao, R.; Asano, S.M.; Upadhyayula, S.; Pisarev, I.; Milkie, D.E.; Liu, T.L.; Singh, V.; Graves, A.; Huynh, G.H.; Zhao, Y.; et al. Cortical column and whole-brain imaging with molecular contrast and nanoscale resolution. Science 2019, 363. [Google Scholar] [CrossRef] [PubMed]
- Laster, S.M.; Mackenzie, J.M., Jr. Bleb formation and F-actin distribution during mitosis and tumor necrosis factor-induced apoptosis. Microsc. Res. Tech. 1996, 34, 272–280. [Google Scholar] [CrossRef]
- Boss, J. Mitosis in cultures of newt tissues. IV. The cell surface in late anaphase and the movements of ribonucleoprotein. Exp. Cell Res. 1955, 8, 181–187. [Google Scholar] [CrossRef]
- Keller, P.J.; Schmidt, A.D.; Santella, A.; Khairy, K.; Bao, Z.; Wittbrodt, J.; Stelzer, E.H. Fast, high-contrast imaging of animal development with scanned light sheet-based structured-illumination microscopy. Nat. Methods 2010, 7, 637–642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomer, R.; Khairy, K.; Amat, F.; Keller, P.J. Quantitative high-speed imaging of entire developing embryos with simultaneous multiview light-sheet microscopy. Nat. Methods 2012. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Hossain, M.J.; Heriche, J.K.; Politi, A.Z.; Walther, N.; Koch, B.; Wachsmuth, M.; Nijmeijer, B.; Kueblbeck, M.; Martinic-Kavur, M.; et al. Experimental and computational framework for a dynamic protein atlas of human cell division. Nature 2018, 561, 411–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, Q.; Busetto, A.G.; Fededa, J.P.; Buhmann, J.M.; Gerlich, D.W. Unsupervised modeling of cell morphology dynamics for time-lapse microscopy. Nat. Methods 2012, 9, 711–713. [Google Scholar] [CrossRef] [PubMed]
- Driscoll, M.K.; Welf, E.S.; Jamieson, A.R.; Dean, K.M.; Isogai, T.; Fiolka, R.; Danuser, G. Robust and automated detection of subcellular morphological motifs in 3D microscopy images. Nat. Methods 2019, 16, 1037–1044. [Google Scholar] [CrossRef]
- Duchi, S.; Piccinini, F.; Pierini, M.; Bevilacqua, A.; Torre, M.L.; Lucarelli, E.; Santi, S. A new holistic 3D non-invasive analysis of cellular distribution and motility on fibroin-alginate microcarriers using light sheet fluorescent microscopy. PLoS ONE 2017, 12, e0183336. [Google Scholar] [CrossRef]
- Piccinini, F.; Tesei, A.; Zanoni, M.; Bevilacqua, A. ReViMS: Software tool for estimating the volumes of 3-D multicellular spheroids imaged using a light sheet fluorescence microscope. Biotechniques 2017, 63, 227–229. [Google Scholar] [CrossRef] [Green Version]
- Zanoni, M.; Piccinini, F.; Arienti, C.; Zamagni, A.; Santi, S.; Polico, R.; Bevilacqua, A.; Tesei, A. 3D tumor spheroid models for in vitro therapeutic screening: A systematic approach to enhance the biological relevance of data obtained. Sci. Rep. 2016, 6, 19103. [Google Scholar] [CrossRef]
- Vogel, S.; Schafer, C.; Hess, S.; Folz-Donahue, K.; Nelles, M.; Minassian, A.; Schwarz, M.K.; Kukat, C.; Ehrlich, M.; Zaehres, H.; et al. The in vivo timeline of differentiation of engrafted human neural progenitor cells. Stem Cell Res. 2019, 37, 101429. [Google Scholar] [CrossRef]
- Doerr, J.; Schwarz, M.K.; Wiedermann, D.; Leinhaas, A.; Jakobs, A.; Schloen, F.; Schwarz, I.; Diedenhofen, M.; Braun, N.C.; Koch, P.; et al. Whole-brain 3D mapping of human neural transplant innervation. Nat. Commun. 2017, 8, 14162. [Google Scholar] [CrossRef] [Green Version]
- Eliceiri, K.W.; Berthold, M.R.; Goldberg, I.G.; Ibanez, L.; Manjunath, B.S.; Martone, M.E.; Murphy, R.F.; Peng, H.; Plant, A.L.; Roysam, B.; et al. Biological imaging software tools. Nat. Methods 2012, 9, 697–710. [Google Scholar] [CrossRef] [Green Version]
- Stephens, D.J.; Allan, V.J. Light microscopy techniques for live cell imaging. Science 2003, 300, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Fatehullah, A.; Tan, S.H.; Barker, N. Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 2016, 18, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Heo, I.; Clevers, H. Disease Modeling in Stem Cell-Derived 3D Organoid Systems. Trends Mol. Med. 2017, 23, 393–410. [Google Scholar] [CrossRef] [PubMed]
- Nicodemus, G.D.; Bryant, S.J. Cell encapsulation in biodegradable hydrogels for tissue engineering applications. Tissue Eng. Part B Rev. 2008, 14, 149–165. [Google Scholar] [CrossRef] [PubMed]
- Magno, V.; Meinhardt, A.; Werner, C. Polymer Hydrogels to Guide Organotypic and Organoid Cultures. Adv. Funct. Mater. 2020, 2000097. [Google Scholar] [CrossRef]
- Bhise, N.S.; Ribas, J.; Manoharan, V.; Zhang, Y.S.; Polini, A.; Massa, S.; Dokmeci, M.R.; Khademhosseini, A. Organ-on-a-chip platforms for studying drug delivery systems. J. Control. Release 2014, 190, 82–93. [Google Scholar] [CrossRef] [Green Version]
- Skardal, A.; Shupe, T.; Atala, A. Organoid-on-a-chip and body-on-a-chip systems for drug screening and disease modeling. Drug Discov. Today 2016, 21, 1399–1411. [Google Scholar] [CrossRef]
- Gut, G.; Herrmann, M.D.; Pelkmans, L. Multiplexed protein maps link subcellular organization to cellular states. Science 2018, 361. [Google Scholar] [CrossRef] [Green Version]
- Schubert, W.; Bonnekoh, B.; Pommer, A.J.; Philipsen, L.; Bockelmann, R.; Malykh, Y.; Gollnick, H.; Friedenberger, M.; Bode, M.; Dress, A.W. Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nat. Biotechnol. 2006, 24, 1270–1278. [Google Scholar] [CrossRef]
- Gerdes, M.J.; Sevinsky, C.J.; Sood, A.; Adak, S.; Bello, M.O.; Bordwell, A.; Can, A.; Corwin, A.; Dinn, S.; Filkins, R.J.; et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc. Natl. Acad. Sci. USA 2013, 110, 11982–11987. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.R.; Fallahi-Sichani, M.; Sorger, P.K. Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat. Commun. 2015, 6, 8390. [Google Scholar] [CrossRef] [PubMed]
- Kiuchi, T.; Higuchi, M.; Takamura, A.; Maruoka, M.; Watanabe, N. Multitarget super-resolution microscopy with high-density labeling by exchangeable probes. Nat. Methods 2015, 12, 743–746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stack, E.C.; Wang, C.; Roman, K.A.; Hoyt, C.C. Multiplexed immunohistochemistry, imaging, and quantitation: A review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods 2014, 70, 46–58. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamashita, N.; Morita, M.; Yokota, H.; Mimori-Kiyosue, Y. Digital Spindle: A New Way to Explore Mitotic Functions by Whole Cell Data Collection and a Computational Approach. Cells 2020, 9, 1255. https://doi.org/10.3390/cells9051255
Yamashita N, Morita M, Yokota H, Mimori-Kiyosue Y. Digital Spindle: A New Way to Explore Mitotic Functions by Whole Cell Data Collection and a Computational Approach. Cells. 2020; 9(5):1255. https://doi.org/10.3390/cells9051255
Chicago/Turabian StyleYamashita, Norio, Masahiko Morita, Hideo Yokota, and Yuko Mimori-Kiyosue. 2020. "Digital Spindle: A New Way to Explore Mitotic Functions by Whole Cell Data Collection and a Computational Approach" Cells 9, no. 5: 1255. https://doi.org/10.3390/cells9051255



