Using a Chemical Genetic Screen to Enhance Our Understanding of the Antimicrobial Properties of Gallium against Escherichia coli
Abstract
:1. Introduction
2. Materials and Methods
2.1. Escherichia coli Strains
2.2. Determination of the Minimal Inhibitory Concentration and Controls
2.3. Screening
2.4. Normalization
2.5. Data Mining and Analyses
3. Results and Discussion
3.1. Genome-Wide Screen of Ga Resistant and Sensitive Hits
3.2. Ga Sensitive Systems
3.2.1. Iron Homeostasis and Transport, and Fe–Sulfur Cluster Proteins
3.2.2. Oxidative Stress
3.2.3. Deoxynucleotide and Cofactor Biosynthesis, and DNA Replication and Repair
3.3. Systems Involved in Ga Resistance
3.3.1. Fe Transport Systems
3.3.2. Amino Acid Biosynthesis
3.3.3. Lipopolysaccharides and Peptidoglycan
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Systems | Subsystems 1 |
|---|---|
| Regulation | Signaling, Sigma factor regulon, Transcription factor, and Transcription factor regulons |
| Response to stimulus | Starvation, Heat, Cold, DNA damage, pH, Detoxification, Osmotic stress, and Other |
| Cellular processes | Cell cycle and division, Cell death, Genetic transfer, Biofilm formation, Quorum sensing, Adhesion, Locomotion, Viral response, Response to bacterium, Host interactions, Symbiosis, and Other proteins |
| Energy | Glycolysis, Pentose phosphate pathway, TCA cycle, Fermentation, Aerobic and anaerobic respiration, and Other proteins |
| Other pathways | Detoxification, Inorganic nutrient metabolism, Macromolecule modification, Activation/inactivation/interconversion, and Other enzymes |
| Degradation | Amino acids, Fatty acid/lipid, Nucleotide/nucleoside, Amine, Carbohydrate/carboxylate, Secondary metabolite, Alcohol, Polymer, Cell exterior and Other proteins |
| Biosynthesis | Amino acids, Nucleotide/nucleoside, Fatty acid/lipid, Amines, Carbohydrate/carboxylates, Cofactors, Secondary metabolites, Polymer, and Other proteins |
| Cell exterior | Transport, Cell wall biogenesis and organization, Lipopolysaccharide metabolism, Pilus, Flagellar, Outer membrane, Inner membrane, Periplasm, and Cell wall components |
| Central dogma | Transcription, Translation, DNA metabolism, RNA metabolism, Protein metabolism, and Protein folding, and secretion |
References
- Chitambar, C.R. The therapeutic potential of iron-targeting gallium compounds in human disease: From basic research to clinical application. Pharmacol. Res. 2017, 115, 56–64. [Google Scholar] [CrossRef]
- Bonchi, C.; Imperi, F.; Minandri, F.; Visca, P.; Frangipani, E. Repurposing of gallium-based drugs for antibacterial therapy. BioFactors 2014, 40, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, L.R. Mechanisms of therapeutic activity for gallium. Pharmacol. Rev. 1998, 50, 665–682. [Google Scholar] [PubMed]
- Pearson, R.G. Hard and soft acids and bases. J. Am. Chem. Soc. 1963, 85, 3533–3539. [Google Scholar] [CrossRef]
- Harris, W.R.; Pecoraro, V.L. Thermodynamic binding constants for gallium transferrin. Biochemistry 1983, 22, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Weiner, R.E.; Neumann, R.D.; Mulshine, J. Transferrin dependence of Ga (NO3)3 inhibition of growth in human-derived small cell lung cancer cells. J. Cell. Biochem. 1996, 24, 276–287. [Google Scholar] [CrossRef]
- Beriault, R.; Hamel, R.; Chenier, D.; Mailloux, R.J.; Joly, H.; Appanna, V.D. The overexpression of NADPH-producing enzymes counters the oxidative stress evoked by gallium, an iron mimetic. BioMetals 2007, 20, 165–176. [Google Scholar] [CrossRef]
- Al-Aoukaty, A.; Appanna, V.D.; Falter, H. Gallium toxicity and adaptation in Pseudomonas fluorescens. FEMS Microbiol. Lett. 1992, 92, 265–272. [Google Scholar] [CrossRef]
- Kaneko, Y.; Thoendel, M.; Olakanmi, O.; Britigan, B.E.; Singh, P.K. The transition metal gallium disrupts Pseudomonas aeruginosa iron metabolism and has antimicrobial and antibiofilm activity. J. Clin. Invest. 2007, 117, 877–888. [Google Scholar] [CrossRef]
- Banin, E.; Lozinski, A.; Brady, K.M.; Berenshtein, E.; Butterfield, P.W.; Moshe, M.; Chevion, M.; Greenberg, E.P.; Banin, E. The potential of desferrioxamine-gallium as an Anti-Pseudomonas therapeutic agent. Proc. Natl. Acad. Sci. USA 2008, 105, 16761–16766. [Google Scholar] [CrossRef]
- Chitambar, C.R.; Narasimhan, J. Targeting iron-dependant DNA synthesis with gallium and transferrin-gallium. Pathobiology 1991, 59, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Olakanmi, O.; Gunn, J.S.; Su, S.; Soni, S.; Hassett, D.J.; Britigan, B.E. Gallium disrupts iron uptake by intracellular and extracellular Francisella strains and exhibits therapeutic efficacy in a murine pulmonary infection model. Antimicrob. Agents Chemother. 2010, 54, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Nikolova, V.; Angelova, S.; Markova, N.; Dudev, T. Gallium as a therapeutic agent: A thermodynamic evaluation of the competition between Ga3+ and Fe3+ ions in metalloproteins. J. Phys. Chem. B 2016, 120, 2241–2248. [Google Scholar] [CrossRef] [PubMed]
- Kelson, A.B.; Carnevali, M.; Truong-Le, V. Gallium-based anti-infectives: Targeting microbial iron-uptake mechanisms. Curr. Opin. Pharmacol. 2013, 13, 707–716. [Google Scholar] [CrossRef] [PubMed]
- Olakanmi, O.; Britigan, B.E.; Larry, S. Gallium disrupts iron metabolism of Mycobacteria residing within human macrophages. Infect. Immun. 2000, 68, 5619–5627. [Google Scholar] [CrossRef]
- Olakanmi, O.; Kesavalu, B.; Pasula, R.; Abdalla, M.Y.; Schlesinger, L.S.; Britigan, B.E. Gallium nitrate is efficacious in murine models of tuberculosis and inhibits key bacterial Fe-dependent enzymes. Antimicrob. Agents Chemother. 2013, 57, 6074–6080. [Google Scholar] [CrossRef]
- DeLeon, K.; Balldin, F.; Watters, C.; Hamood, A.; Griswold, J.; Sreedharan, S.; Rumbaugh, K.P. Gallium maltolate treatment eradicates Pseudomonas aeruginosa infection in thermally injured mice. Antimicrob. Agents Chemother. 2009, 53, 1331–1337. [Google Scholar] [CrossRef]
- Arnold, C.E.; Bordin, A.; Lawhon, S.D.; Libal, M.C.; Bernstein, L.R.; Cohen, N.D. Antimicrobial activity of gallium maltolate against Staphylococcus aureus and methicillin-resistant S. aureus and Staphylococcus pseudintermedius: An in vitro study. Vet. Microbiol. 2012, 155, 389–394. [Google Scholar] [CrossRef]
- Martens, R.J.; Miller, N.A.; Cohen, N.D.; Harrington, J.R.; Bernstein, L.R. Chemoprophylactic antimicrobial activity of gallium maltolate against intracellular Rhodococcus equi. J. Equine Vet. Sci. 2007, 27, 341–345. [Google Scholar] [CrossRef]
- Antunes, L.C.S.; Imperi, F.; Minandri, F.; Visca, P. In vitro and in vivo antimicrobial activities of gallium nitrate against multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 2012, 56, 5961–5970. [Google Scholar] [CrossRef]
- Gugala, N.; Lemire, J.A.; Turner, R.J. The efficacy of different anti-microbial metals at preventing the formation of, and eradicating bacterial biofilms of pathogenic indicator strains. J. Antibiot. (Tokyo) 2017, 70, 775–780. [Google Scholar] [CrossRef]
- Lemire, J.A.; Harrison, J.J.; Turner, R.J. Antimicrobial activity of metals: Mechanisms, molecular targets and applications. Nat. Rev. Microbiol. 2013, 11, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Minandri, F.; Bonchi, C.; Frangipani, E.; Imperi, F.; Visca, P. Promises and failures of gallium as an antibacterial agent. Future Microbiol. 2014, 9, 379–397. [Google Scholar] [CrossRef] [PubMed]
- Harrison, J.J.; Ceri, H.; Turner, R.J. Multimetal resistance and tolerance in microbial biofilms. Nat. Rev. Microbiol. 2007, 5, 928–938. [Google Scholar] [CrossRef] [PubMed]
- García-Contreras, R.; Lira-Silva, E.; Jasso-Chávez, R.; Hernández-González, I.L.; Maeda, T.; Hashimoto, T.; Boogerd, F.C.; Sheng, L.; Wood, T.K.; Moreno-Sánchez, R. Isolation and characterization of gallium resistant Pseudomonas aeruginosa mutants. Int. J. Med. Microbiol. 2013, 303, 574–582. [Google Scholar] [CrossRef] [PubMed]
- Peeters, E.; Nelis, H.J.; Coenye, T. Resistance of planktonic and biofilm-grown Burkholderia cepacia complex isolates to the transition metal gallium. J. Antimicrob. Chemother. 2008, 61, 1062–1065. [Google Scholar] [CrossRef] [PubMed]
- Mcquillan, J.S.; Shaw, A.M. Differential gene regulation in the Ag nanoparticle and Ag-Induced silver stress response in Escherichia coli: A full transcriptomic profile. Nanotoxicology 2014, 8, 177–184. [Google Scholar] [CrossRef]
- Saulou-Bérion, C.; Gonzalez, I.; Enjalbert, B.; Audinot, J.N.; Fourquaux, I.; Jamme, F.; Cocaign-Bousquet, M.; Mercier-Bonin, M.; Girbal, L. Escherichia coli under ionic silver stress: An integrative approach to explore transcriptional, physiological and biochemical responses. PLoS ONE 2015, 10, 1–25. [Google Scholar] [CrossRef]
- Ivask, A.; Elbadawy, A.; Kaweeteerawat, C.; Boren, D.; Fischer, H.; Ji, Z.; Chang, C.H.; Liu, R.; Tolaymat, T.; Telesca, D.; et al. Toxicity mechanisms in Escherichia coli vary for silver nanoparticles and differ from ionic silver. ACS Nano 2014, 8, 374–386. [Google Scholar] [CrossRef]
- Gugala, N.; Lemire, J.; Chatfield-Reed, K.; Yan, Y.; Chua, G.; Turner, R. Using a chemical genetic screen to enhance our understanding of the antibacterial properties of silver. Genes 2018, 9, 344. [Google Scholar] [CrossRef]
- Kershaw, C.J.; Brown, N.L.; Constantinidou, C.; Patel, M.D.; Hobman, J.L. The expression profile of Escherichia coli K-12 in response to minimal, optimal and excess copper concentrations. Microbiology 2005, 151, 1187–1198. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Ishihama, A. Transcriptional response of Escherichia coli to external copper. Mol. Microbiol. 2005, 56, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Brocklehurst, K.R.; Morby, A.P. Metal-ion tolerance in Escherichia coli: Analysis of transcriptional profiles by gene-array technology. Microbiology 2000, 146, 2277–2282. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K. Transcriptional Response of Escherichia coli to External Zinc. J. Bacteriol. 2005, 187, 6333–6340. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio Collection. Mol. Syst. Biol. 2006, 2. [Google Scholar] [CrossRef] [PubMed]
- Kuzminov, A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage λ. Microbiol. Mol. Biol. Rev. 1999, 63, 751–813. [Google Scholar] [PubMed]
- Hin, A.; Tong, Y.; Evangelista, M.; Parsons, A.B.; Xu, H.; Bader, G.D.; Page, N.; Robinson, M.; Raghibizadeh, S.; Hogue, C.W.V.; et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science (80-.) 2001, 294, 2364–2369. [Google Scholar] [CrossRef]
- Wagih, O.; Usaj, M.; Baryshnikova, A.; VanderSluis, B.; Kuzmin, E.; Costanzo, M.; Myers, C.L.; Andrews, B.J.; Boone, C.M.; Parts, L. SGAtools: One-stop analysis and visualization of array-based genetic interaction screens. Nucleic Acids Res. 2013, 41, 591–596. [Google Scholar] [CrossRef] [PubMed]
- Keseler, I.M.; Mackie, A.; Santos-Zavaleta, A.; Billington, R.; Bonavides-Martínez, C.; Caspi, R.; Fulcher, C.; Gama-Castro, S.; Kothari, A.; Krummenacker, M.; et al. The EcoCyc database: Reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res. 2017, 45, D543–D550. [Google Scholar] [CrossRef]
- Bateman, A.; Martin, M.J.; O’Donovan, C.; Magrane, M.; Alpi, E.; Antunes, R.; Bely, B.; Bingley, M.; Bonilla, C.; Britto, R.; et al. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Patrick, M.W.; Quandt, M.E.; Swartzlander, B.D.; Matsumura, I. Multicopy suppression underpins metabolic evolvability. Mol. Microbiol. Evol. 2007, 24, 2716–2722. [Google Scholar] [CrossRef] [PubMed]
- Joyce, A.R.; Reed, J.L.; White, A.; Edwards, R.; Osterman, A.; Baba, T.; Mori, H.; Lesely, S.A.; Palsson, B.; Agarwalla, S. Experimental and computational assessment of conditionally essential genes in Escherichia coli. J. Bacteriol. 2006, 188, 8259–8271. [Google Scholar] [CrossRef]
- Mchugh, J.P.; Rodríguez-Quiñones, F.; Abdul-Tehrani, H.; Svistunenko, D.A.; Poole, R.K.; Cooper, C.E.; Andrews, S.C. Global iron-dependent gene regulation in Escherichia coli. J. Biol. Chem. 2003, 278, 29478–29486. [Google Scholar] [CrossRef] [PubMed]
- Kakuta, Y.; Horio, T.; Takahashi, Y.; Fukuyama, K. Crystal structure of Escherichia coli Fdx, an adrenodoxin-type ferredoxin involved in the assembly of iron-sulfur clusters. Biochemistry 2001, 40, 11007–11012. [Google Scholar] [CrossRef]
- Cicchillo, R.M.; Lee, K.H.; Baleanu-Gogonea, C.; Nesbitt, N.M.; Krebs, C.; Booker, S.J. Escherichia coli lipoyl synthase binds two distinct [4Fe-4S] clusters per polypeptide. Biochemistry 2004, 43, 11770–11781. [Google Scholar] [CrossRef]
- Waller, J.C.; Alvarez, S.; Naponelli, V.; Lara-Nunez, A.; Blaby, I.K.; Da Silva, V.; Ziemak, M.J.; Vickers, T.J.; Beverley, S.M.; Edison, A.S.; et al. A role for tetrahydrofolates in the metabolism of iron-sulfur clusters in all domains of life. Proc. Natl. Acad. Sci. USA 2010, 107, 10412–10417. [Google Scholar] [CrossRef] [Green Version]
- Jang, S.; Imlay, J.A. Hydrogen peroxide inactivates the Escherichia coli Isc iron-sulfur assembly system, and OxyR induces the suf system to compensate. Mol. Microbiol. 2010, 78, 1448–1467. [Google Scholar] [CrossRef]
- Laxman, S.; Sutter, B.M.; Wu, X.; Kumar, S.; Guo, X.; David, C.; Mirzaei, H.; Tu, B.P. Sulfur amino acids regulate translational capacity and metabolic homeostasis through modulation of tRNA thiolation. Cell 2014, 154, 416–429. [Google Scholar] [CrossRef]
- Miethke, M.; Marahiel, M.A. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 2007, 71, 413–451. [Google Scholar] [CrossRef] [PubMed]
- Raymond, K.N.; Dertz, E.A.; Kim, S.S. Enterobactin: An archetype for microbial iron transport. Proc. Natl. Acad. Sci. USA 2003, 100, 3584–3588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chitambar, C.R. Gallium and its competing roles with iron in biological systems. Biochim. Biophys. Acta 2016, 1863, 2044–2053. [Google Scholar] [CrossRef] [PubMed]
- Koronakis, V. TolC-the bacterial exit duct for proteins and drugs. FEBS Lett. 2003, 555, 66–71. [Google Scholar] [CrossRef]
- Komendarczyk, R.; Pullen, J. Transcriptional regulation of drug efflux genes by EvgAS, a two-component system in Escherichia coli. Microbiology 2003, 149, 2819–2828. [Google Scholar] [CrossRef]
- Bou-Abdallah, F.; Lewin, A.C.; Le Brun, N.E.; Moore, G.R.; Dennis Chasteen, N. Iron detoxification properties of Escherichia coli Bacterioferritin. Attenuation of Oxyradical Chemistry. J. Biol. Chem. 2002, 277, 37064–37069. [Google Scholar] [CrossRef] [PubMed]
- Imlay, J.A. Pathways of oxidative damage. Annu. Rev. Microbiol. 2003, 57, 395–418. [Google Scholar] [CrossRef]
- Imlay, J.A. Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 2008, 77, 755–776. [Google Scholar] [CrossRef]
- Latinwo, L.M.; Donald, C.; Ikediobi, C.; Silver, S. Effects of intracellular glutathione on sensitivity of Escherichia coli to mercury and arsenite. Biochem. Biophys. Res. Commun. 1998, 242, 67–70. [Google Scholar] [CrossRef]
- Ozyamak, E.; Black, S.S.; Walker, C.A.; MacLean, M.J.; Bartlett, W.; Miller, S.; Booth, I.R. The critical role of S-Lactoylglutathione formation during methylglyoxal detoxification in Escherichia coli. Mol. Microbiol. 2010, 78, 1577–1590. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, A.P.; Fladvad, M.; Berndt, C.; Andrésen, C.; Lillig, C.H.; Neubauer, P.; Sunnerhagen, M.; Holmgren, A.; Vlamis-Gardikas, A. A novel monothiol glutaredoxin (Grx4) from Escherichia coli can serve as a substrate for thioredoxin reductase. J. Biol. Chem. 2005, 280, 24544–24552. [Google Scholar] [CrossRef] [PubMed]
- Yeung, N.; Gold, B.; Liu, N.L.; Prathapam, R.; Sterling, H.J.; Willams, E.R.; Butland, G. The E. coli monothiol glutaredoxin GrxD forms homodimeric and heterodimeric FeS cluster containing complexes. Biochemistry 2011, 50, 8957–8969. [Google Scholar] [CrossRef] [PubMed]
- Meganathan, R. Ubiquinone biosynthesis in microorganisms. FEMS Microbiol. Lett. 2001, 203, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Søballe, B.; Poole, R.K. Ubiquinone limits oxidative stress in Escherichia coli. Microbiology 2000, 146, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Nocentini, G. Ribonucleotide reductase inhibitors: New strategies for cancer chemotherapy. Crit. Rev. Oncol. Hematol. 1996, 22, 89–126, 1040. [Google Scholar] [CrossRef]
- Chitambar, C.R.; Seligman, P.A. Effects of different transferrin forms on transferrin receptor expression, iron uptake, and cellular proliferation of human leukemic HL60 cells. Mechanisms Responsible for the Specific Cytotoxicity of Transferrin-Gallium. J. Clin. Invest. 1986, 78, 1538–1546. [Google Scholar] [CrossRef]
- Chitambar, C.R.; Narasimhan, J.; Guy, J.; Sem, D.S.; Brien, W.J.O. Inhibition of ribonucleotide reductase by gallium in murine. Cancer Res. 1991, 51, 6199–6202. [Google Scholar]
- Smith, J.M.; Daum, H.A. Identification and nucleotide sequence of a gene encoding 5’-phosphoribosylglycinamide transformylase in Escherichia coli K12. J. Biol. Chem. 1987, 262, 10565–10569. [Google Scholar]
- Zhu, M.; Dai, X.; Guo, Z.; Yang, M.; Wang, H.; Wang, Y.-P. Manipulating the bacterial cell cycle and cell size by titrating the expression of ribonucleotide reductase. MBio 2017, 8, 6–11. [Google Scholar] [CrossRef]
- Salguero, I.; Guarino, E.; Guzmàn, E.C. RecA-dependent replication in the NrdA101(Ts) mutant of Escherichia coli under restrictive conditions. J. Bacteriol. 2011, 193, 2851–2860. [Google Scholar] [CrossRef] [PubMed]
- Guarino, E.; Jiménez-Sánchez, A.; Guzmán, E.C. Defective ribonucleoside diphosphate reductase impairs replication fork progression in Escherichia coli. J. Bacteriol. 2007, 189, 3496–3501. [Google Scholar] [CrossRef] [PubMed]
- Roux, B.; Walsh, C.T. P-Aminobenzoate Synthesis in Escherichia coli: Kinetic and mechanistic characterization of the amidotransferase PabA. Biochemistry 1992, 31, 6904–6910. [Google Scholar] [CrossRef] [PubMed]
- Bermingham, A.; Derrick, J.P. The folic acid biosynthesis pathway in bacteria: Evaluation of potential for antibacterial drug discovery. BioEssays 2002, 24, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Henderson, G.B.; Huennekens, F.M. Membrane-associated folate transport proteins. Methods Enzymol. 1986, 122, 260–269. [Google Scholar] [PubMed]
- Krakoff, I.H.; Brown, N.C.; Reichard, P. Inhibition reductase of ribonucleoside by hydroxyurea diphosphate. Cancer Res. 1968, 28, 1559–1565. [Google Scholar] [PubMed]
- Sinha, N.K.; Snustad, D.P. Mechanism of inhibition of deoxyribonucleic acid synthesis in Escherichia coli by hydroxyurea. J. Bacteriol. 1972, 112, 1321–1324. [Google Scholar]
- Rice, D.W.; Rafferty, J.B.; Artymiuk, P.J.; Lloyd, R.G. Insights into the mechanisms of homologous recombination from the structure of RuvA. Curr. Opin. Struct. Biol. 1997, 7, 798–803. [Google Scholar] [CrossRef]
- Chenault, S.S.; Earhart, C.F. Organization of genes encoding membrane proteins of the Escherichia coli ferrienterobactin permease. Mol. Microbiol. 1991, 5, 1405–1413. [Google Scholar] [CrossRef]
- Cartron, M.L.; Maddocks, S.; Gillingham, P.; Craven, C.J.; Andrews, S.C. Feo-transport of ferrous iron into bacteria. BioMetals 2006, 19, 143–157. [Google Scholar] [CrossRef]
- Templin, M.F.; Ursinus, A.; Höltje, J.V. A defect in cell wall recycling triggers autolysis during the stationary growth phase of Escherichia coli. EMBO J. 1999, 18, 4108–4117. [Google Scholar] [CrossRef] [PubMed]
- Egler, M.; Grosse, C.; Grass, G.; Nies, D.H. Role of the extracytoplasmic function protein family σ factor RpoE in metal resistance of Eschenchia coli. J. Bacteriol. 2005, 187, 2297–2307. [Google Scholar] [CrossRef] [PubMed]
- Nies, D.H. Bacterial tranistion metal homeostasis. In Molecular Microbiology of Heavy Metals; Springer: Berlin/Heidelberg, Germany, 2007; pp. 118–142. [Google Scholar]
- Jones, C.M.; Niederweis, M. Role of porins in iron uptake by Mycobacterium smegmatis. J. Bacteriol. 2010, 192, 6411–6417. [Google Scholar] [CrossRef] [PubMed]
- Rigal, A.; Bouveret, E.; Lloubes, R.; Lazdunski, C.; Benedetti, H. The TolB protein interacts with the porins of Escherichia coli. J. Bacteriol. 1997, 179, 7274–7279. [Google Scholar] [CrossRef] [PubMed]
- Sirko, A.; Hryniewicz, M.; Hulanicka, D.; Böck, A. Sulfate and thiosulfate transport in Escherichia coli K-12: Nucleotide sequence and expression of the CysTWAM gene cluster. J. Bacteriol. 1990, 172, 3351–3357. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.S.; Adhikari, P.; Nowalk, A.J.; Chen, C.Y.; Mietzner, T.A. The HFbpABC transporter from Haemophilus influenzae Functions as a Binding-Protein-Dependent ABC transporter with high specificity and affinity for ferric iron. J. Bacteriol. 2004, 186, 6220–6229. [Google Scholar] [CrossRef] [PubMed]
- Stojiljkovic, I.; Kumar, V.; Srinivasan, N. Non-iron metalloporphyrins: Potent antibacterial compounds that exploit Haem/Hb uptake systems of pathogenic bacteria. Mol. Microbiol. 1999, 31, 429–442. [Google Scholar] [CrossRef]
- Xu, J.Z.; Yang, H.K.; Zhang, W.G. NADPH Metabolism: A survey of its theoretical characteristics and manipulation strategies in amino acid biosynthesis. Crit. Rev. Biotechnol. 2018, 38, 1061–1076. [Google Scholar] [CrossRef]
- LaRossa, R.A.; Schloss, J.V. The sulfonylurea herbicide sulfometuron methyl is an extremely potent and selective inhibitor of acetolactate synthase in Salmonella typhimurium. J. Biol. Chem. 1984, 259, 8753–8757. [Google Scholar] [CrossRef]
- Watson, M.D.; Wild, J.; Umbarger, H.E. Positive control of IlvC expression in Escherichia coli K-12; Identification and Mapping of Regulatory Gene IlvY. J. Bacteriol. 1979, 139, 1014–1020. [Google Scholar]
- Stohs, S.J.; Bagchi, D. Oxidative mechanisms in the toxicity of metal ions. Free Radic. Biol. Med. 1995, 18, 321–336. [Google Scholar] [CrossRef] [Green Version]
- Ercal, N.; Gurer-Orhan, H.; Aykin-Burns, N. Toxic metals and oxidative stress part I: Mechanisms involved in metal induced oxidative damage. Curr. Top. Med. Chem. 2001, 1, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Cabeen, M.T.; Jacobs-Wagner, C. Bacterial cell shape. Nat. Rev. Microbiol. 2005, 3, 601–610. [Google Scholar] [CrossRef]
- De La Fuente-Núñez, C.; Korolik, V.; Bains, M.; Nguyen, U.; Breidenstein, E.B.M.; Horsman, S.; Lewenza, S.; Burrows, L.; Hancock, R.E.W. Inhibition of bacterial biofilm formation and swarming motility by a small synthetic cationic peptide. Antimicrob. Agents Chemother. 2012, 56, 2696–2704. [Google Scholar] [CrossRef] [PubMed]
- Caparrós, M.; Quintela, J.C.; de Pedro, M.A. Variability of peptidoglycan surface density in Escherichia coli. FEMS Microbiol. Lett. 1994, 121, 71–76. [Google Scholar] [CrossRef]
- Gronow, S.; Brabetz, W.; Brade, H. Comparative functional characterization in vitro of heptosyltransferase I (WaaC) and II (WaaF) from Escherichia coli. Eur. J. Biochem. 2000, 267, 6602–6611. [Google Scholar] [CrossRef] [PubMed]
- Beher, M.; Schnaitman, C. Regulation of the OmpA outer membrane protein of Escherichia coli. J. Bacteriol. 1981, 147, 972–985. [Google Scholar]
- Lazzaroni, J.C.; Portalier, R.C. Genetic and biochemical characterization of periplasmic leaky mutants of Escherichia coli K-12. J. Bacteriol. 1981, 145, 1351–1358. [Google Scholar] [CrossRef]
- Beveridge, T.J.; Koval, S.F. Binding of Metals to Cell Envelopes of Escherichia coli binding of metals to cell envelopes of Escherichia coli K-12. Appl. Environ. Microbiol. 1981, 42, 325–335. [Google Scholar]
- Hoyle, B.D.; Beveridge, T.J. Metal binding by the peptidoglycan sacculus of Escherichia coli K-12. Can. J. Microbiol. 1984, 30, 204–211. [Google Scholar] [CrossRef]
- Arunmanee, W.; Pathania, M.; Solovyova, A.S.; Le Brun, A.P.; Ridley, H.; Baslé, A.; van den Berg, B.; Lakey, J.H. Gram-negative Trimeric Porins Have Specific LPS binding sites that are essential for porin biogenesis. Proc. Natl. Acad. Sci. USA 2016, 113, 5034–5043. [Google Scholar] [CrossRef] [PubMed]
- Bolla, J.M.; Lazdunski, C.; Pagès, J.M. The assembly of the major outer membrane protein OmpF of Escherichia coli depends on lipid synthesis. EMBO J. 1988, 7, 3595–3599. [Google Scholar] [CrossRef] [PubMed]

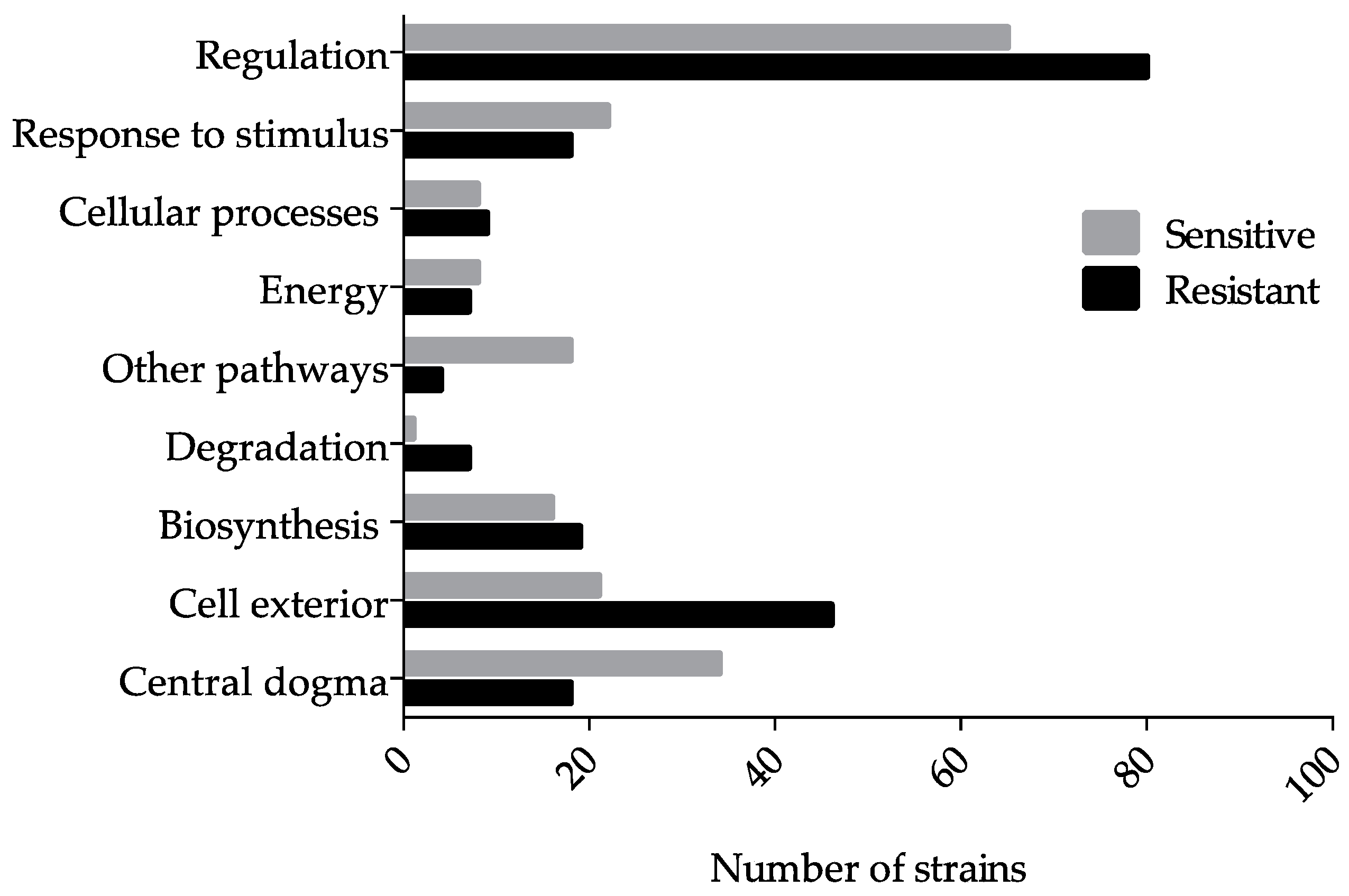



| System | Subsystem | Gene 1 | Score 2,3 |
|---|---|---|---|
| Central dogma | Transcription | evgA | −0.166 |
| hns | −0.175 | ||
| lgoR | −0.401 | ||
| nagC | −0.191 | ||
| rseA | −0.26 | ||
| ulaR | −0.556 | ||
| Translation | bipA | −0.204 | |
| DNA metabolism | holC | −0.327 | |
| holD | −0.217 | ||
| ruvC | −0.184 | ||
| intR | −0.27 | ||
| recA | −0.309 | ||
| recD | −0.199 | ||
| RNA metabolism | rbfA | −0.35 | |
| rim | −0.298 | ||
| mnmA | −0.212 | ||
| rnt | −0.322 | ||
| ygfZ | −0.373 | ||
| evgA | −0.166 | ||
| hns | −0.175 | ||
| lgoR | −0.401 | ||
| nagC | −0.191 | ||
| rseA | −0.269 | ||
| sspA | −0.214 | ||
| ulaR | −0.556 | ||
| Protein metabolism | lipA | −0.318 | |
| pphA | −0.198 | ||
| slyD | −0.273 | ||
| Protein folding and secretion | slyD | −0.273 | |
| Cell exterior | Transport | zunC | −0.361 |
| tolC | −0.539 | ||
| ugpC | −0.29 | ||
| Pilus | ybgO | −0.163 | |
| Flagellum | fliG | −0.235 | |
| Outer membrane | tolC | −0.539 | |
| Plasma membrane | clsA | −0.171 | |
| cysQ | −0.203 | ||
| fdnI | −0.251 | ||
| fliG | −0.235 | ||
| gspA | −0.199 | ||
| hokA | −0.181 | ||
| nuoK | −0.247 | ||
| rseA | −0.269 | ||
| ubiG | −0.265 | ||
| ugpC | −0.29 | ||
| znuC | −0.361 | ||
| Periplasm | tolC | −0.539 | |
| yebF | −0.268 | ||
| Biosynthesis | Amino acid | dmI | −0.418 |
| metL | −0.189 | ||
| mtn | −0.329 | ||
| Nucleoside and nucleotide | purT | −0.216 | |
| Fatty acid/lipid | clsA | −0.171 | |
| Carbohydrate | mdh | −0.287 | |
| Secondary metabolites | mtn | −0.329 | |
| fdx | −0.168 | ||
| Cofactor | fdx | −0.168 | |
| gshA | −0.165 | ||
| lipA | −0.318 | ||
| pabA | −0.224 | ||
| pabC | −0.258 | ||
| ubiG | −0.265 | ||
| Other | metL | −0.189 | |
| Degradation | Amino acid | astD | −0.301 |
| Nucleoside and nucleotide | mtn | −0.329 | |
| Amine | purT | −0.216 | |
| Carbohydrate | garK | −0.173 | |
| dmlA | −0.418 | ||
| Energy | Glycolysis | gpmA | −0.175 |
| Tricarboxylic acid cycle | mdh | −0.287 | |
| Fermentation | mdh | −0.287 | |
| Aerobic respiration | nuoK | −0.247 | |
| Anaerobic respiration | fdnI | −0.251 | |
| nuoK | −0.247 | ||
| Other | mdh | −0.287 | |
| nuoK | −0.247 | ||
| Cellular processes | Biofilm | hns | −0.175 |
| Adhesion | ybgO | −0.163 | |
| Locomotion | fliG | −0.235 | |
| recA | −0.309 | ||
| Viral response | intR | −0.27 | |
| Host interaction | intR | −0.27 | |
| slyD | −0.273 | ||
| Symbiosis | slyD | −0.273 | |
| Response to stimulus | Starvation | sspA | −0.29 |
| ugpC | −0.214 | ||
| Heat | bipA | −0.204 | |
| gloB | −0.297 | ||
| slyD | −0.273 | ||
| Cold | bipA | −0.204 | |
| rbfA | −0.35 | ||
| DNA damage | rbfA | −0.35 | |
| recA | −0.39 | ||
| recD | −0.199 | ||
| ruvC | −0.184 | ||
| Osmotic stress | gshA | −0.165 | |
| ubiG | −0.265 | ||
| Other | evgA | −0.166 | |
| fliG | −0.235 | ||
| grxD | −0.266 | ||
| holC | −0.327 | ||
| holD | −0.217 | ||
| pphA | −0.198 | ||
| rseA | −0.269 | ||
| sspA | −0.214 | ||
| tolC | −0.539 | ||
| ugpC | −0.29 | ||
| Other pathways | Inorganic nutrient metabolism | fdnI | −0.251 |
| nuoK | −0.247 | ||
| Detoxification | gloB | −0.297 | |
| grxD | −0.266 | ||
| Macromolecule modification | mnmA | −0.212 | |
| rnt | −0.322 | ||
| Other enzymes | bfr | −0.17 | |
| cysQ | −0.203 | ||
| pphA | −0.198 | ||
| recD | −0.199 | ||
| ruvC | −0.184 | ||
| slyD | −0.273 |
| System | Subsystem | Gene 1 | Score 2,3 |
|---|---|---|---|
| Central dogma | Transcription | ilvY | 0.215 |
| metR | 0.372 | ||
| odhR | 0.353 | ||
| DNA metabolism | hofM | 0.62 | |
| xerD | 0.168 | ||
| cas2 | 0.177 | ||
| RNA metabolism | symE | 0.177 | |
| ilvY | 0.215 | ||
| metR | 0.372 | ||
| pdhR | 0.353 | ||
| Protein metabolism | mrcB | 0.249 | |
| Protein folding and secretion | yraI | 0.18 | |
| Cell exterior | Transport | cysU | 0.362 |
| fepG | 0.312 | ||
| tonB | 0.341 | ||
| caiT | 0.403 | ||
| yiaO | 0.6 | ||
| par | 0.266 | ||
| Cell wall biogenesis | alr | 0.353 | |
| evnC | 0.203 | ||
| mrcB | 0.249 | ||
| yraI | 0.18 | ||
| Lipopolysaccharide metabolism | cspG | 0.204 | |
| rfaC | 0.201 | ||
| Outer membrane | par | 0.266 | |
| pqiC | 0.345 | ||
| Plasma membrane | atpE | 0.172 | |
| atpH | 0.176 | ||
| caiT | 0.403 | ||
| cycU | 0.362 | ||
| envU | 0.203 | ||
| fepG | 0.312 | ||
| mrcB | 0.249 | ||
| pqiC | 0.345 | ||
| tonB | 0.341 | ||
| torC | 0.259 | ||
| rfaC | 0.201 | ||
| yaaU | 0.237 | ||
| yafU | 0.214 | ||
| yifK | 0.18 | ||
| Periplasm | ansB | 0.204 | |
| asr | 0.247 | ||
| envC | 0.203 | ||
| mrcB | 0.249 | ||
| pqiC | 0.345 | ||
| tolB | 0.2 | ||
| tonB | 0.341 | ||
| torC | 0.259 | ||
| yiaO | 0.6 | ||
| yral | 0.18 | ||
| Cell wall component | mrcB | 0.249 | |
| torC | 0.259 | ||
| Biosynthesis | Amino acid | alr | 0.353 |
| avtA | 0.384 | ||
| leuA | 0.302 | ||
| leuC | 0.205 | ||
| metA | 0.241 | ||
| proB | 0.258 | ||
| trpB | 0.611 | ||
| trpD | 0.273 | ||
| Fatty acid/lipid | rfaC | 0.201 | |
| Carbohydrate | cpsG | 0.204 | |
| rfaC | 0.201 | ||
| Cofactor, prosthetic groups, electron carrier | bioF | 0.183 | |
| bioH | 0.194 | ||
| coaA | 0.193 | ||
| thiE | 0.226 | ||
| Cell structure | mrcB | 0.249 | |
| Other | aroF | 0.236 | |
| Degradation | Amino acid | alr | 0.353 |
| ansB | 0.204 | ||
| Fatty acid/lipid | atoA | 0.246 | |
| Energy | Glycolysis | pykF | 0.169 |
| Fermentation | pykF | 0.169 | |
| Anaerobic respiration | torC | 0.259 | |
| Adenosine triphosphate biosynthesis | atpE | 0.172 | |
| atpH | 0.176 | ||
| Other | hydN | 0.249 | |
| Cellular processes | Cell cycle/division | envC | 0.203 |
| tolB | 0.2 | ||
| xerD | 0.168 | ||
| Cell death | envC | 0.203 | |
| Adhesion | tonB | 0.341 | |
| Viral response | cas2 | 0.177 | |
| tonB | 0.341 | ||
| Symbiosis | tonB | 0.341 | |
| Response to stimulus | Heat | pykF | 0.169 |
| DNA damage | par | 0.266 | |
| symE | 0.177 | ||
| yiaO | 0.6 | ||
| pH | oxc | 0.519 | |
| Other | asr | 0.247 | |
| caiT | 0.403 | ||
| cas2 | 0.177 | ||
| envC | 0.203 | ||
| mrcB | 0.249 | ||
| tolB | 0.2 | ||
| tonB | 0.341 | ||
| torC | 0.259 | ||
| xerD | 0.168 | ||
| yaaU | 0.237 | ||
| Other pathways | Other enzymes | oxc | 0.519 |
| sepG | 0.201 |
| Gene | Score without HA | Score with HA 1,2 |
|---|---|---|
| ruvA | N/A | −0.257 |
| recA | −0.309 | −0.299 |
| ruvC | −0.184 | −0.299 |
| holC | −0.327 | −0.351 |
| recD | −0.199 | −0.561 |
| Gene | Score without SMM | Score with SMM 1,2 |
|---|---|---|
| leuA | 0.302 | 0.341 |
| ilvY | 0.215 | 0.3 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gugala, N.; Chatfield-Reed, K.; Turner, R.J.; Chua, G. Using a Chemical Genetic Screen to Enhance Our Understanding of the Antimicrobial Properties of Gallium against Escherichia coli. Genes 2019, 10, 34. https://doi.org/10.3390/genes10010034
Gugala N, Chatfield-Reed K, Turner RJ, Chua G. Using a Chemical Genetic Screen to Enhance Our Understanding of the Antimicrobial Properties of Gallium against Escherichia coli. Genes. 2019; 10(1):34. https://doi.org/10.3390/genes10010034
Chicago/Turabian StyleGugala, Natalie, Kate Chatfield-Reed, Raymond J. Turner, and Gordon Chua. 2019. "Using a Chemical Genetic Screen to Enhance Our Understanding of the Antimicrobial Properties of Gallium against Escherichia coli" Genes 10, no. 1: 34. https://doi.org/10.3390/genes10010034






