Environmental Change-Dependent Inherited Epigenetic Response
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Housing and Experimental Setup
2.2. Assessment of Differentially Methylated Regions and Comparison of Gene Methylation
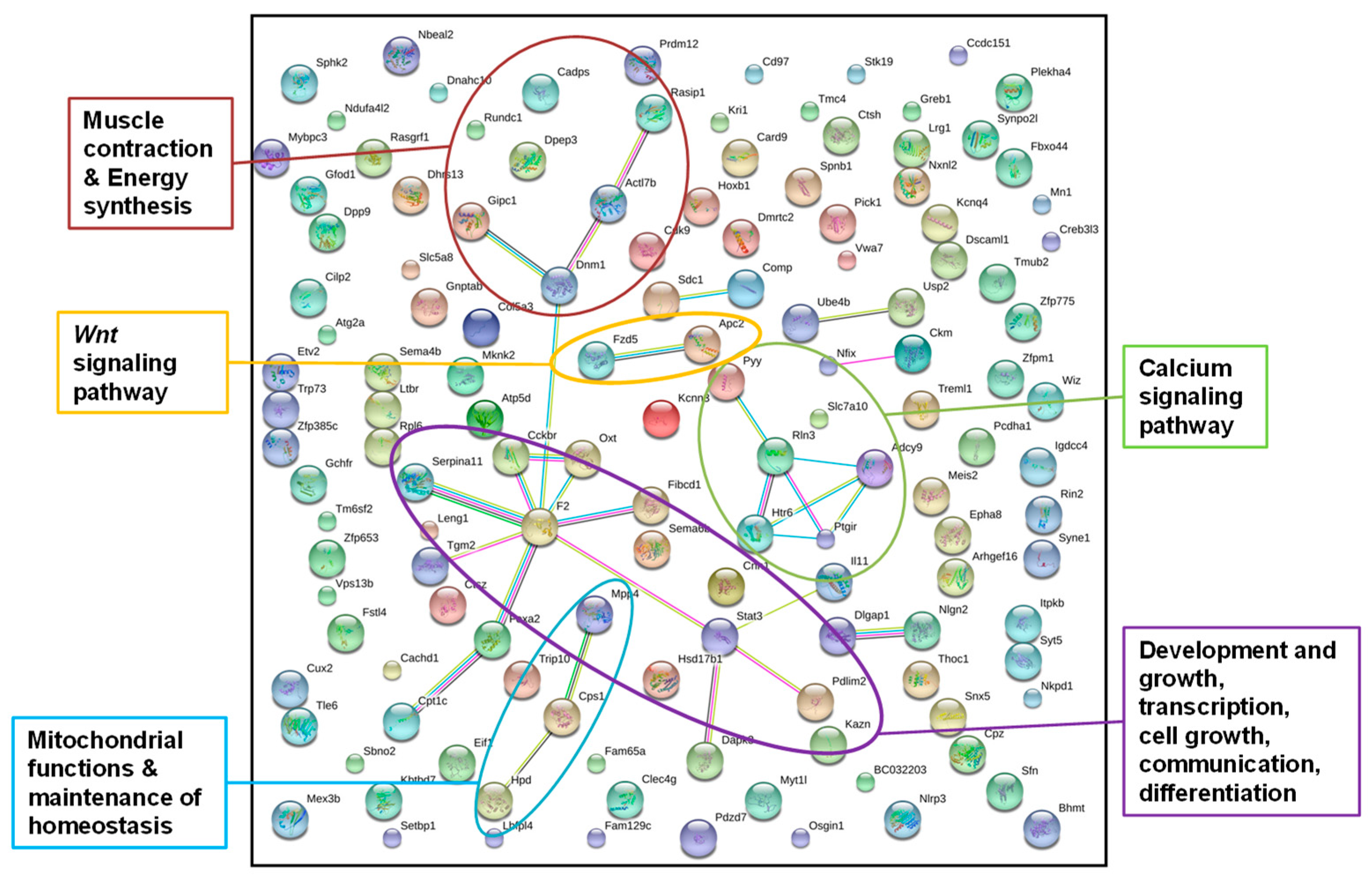
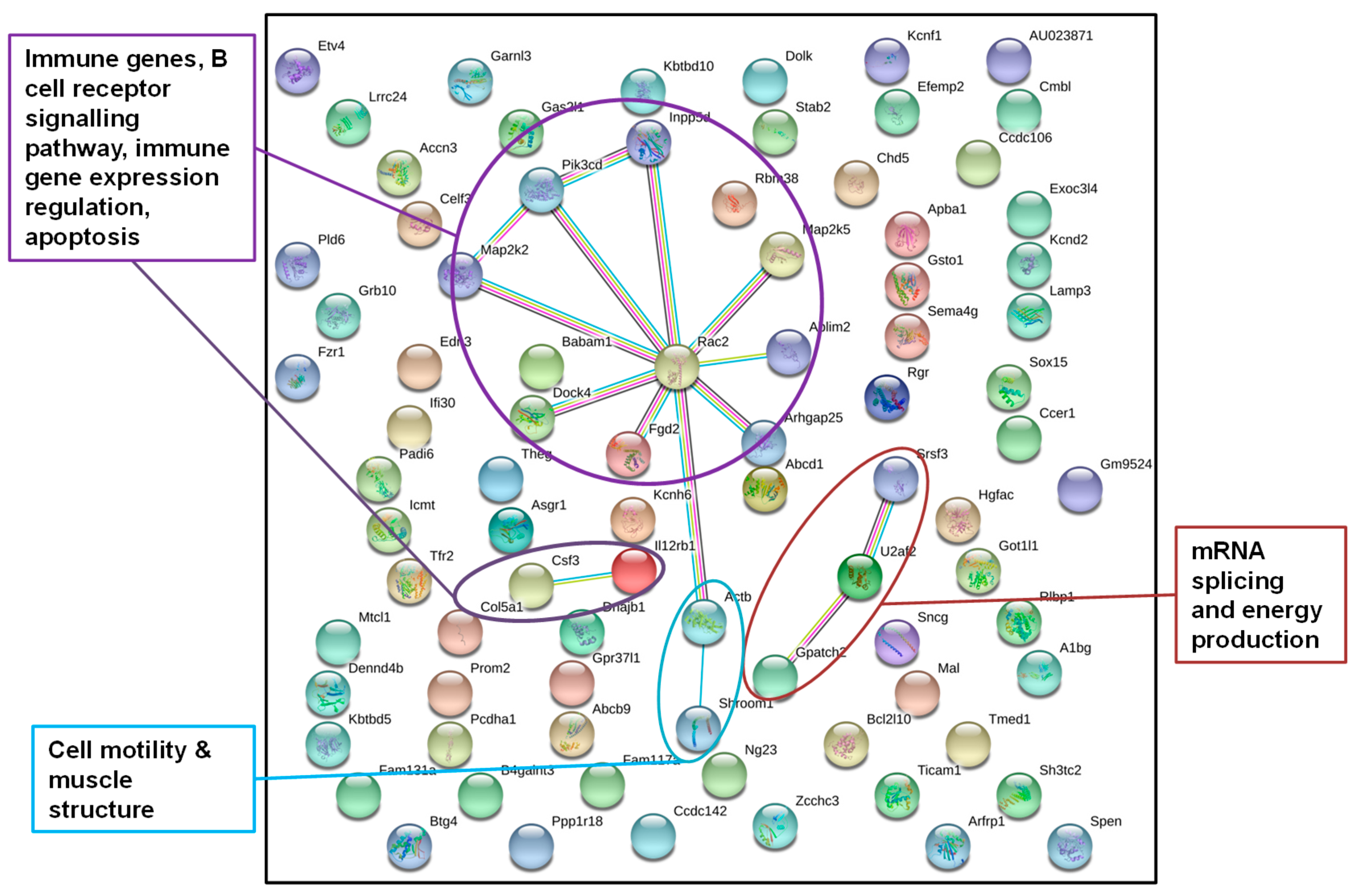
2.3 Display of Interactions of Differentially Methylated Genes
3. Results
3.1. Total Changes in DNA Methylation in Sons
3.2. Environmental Factor-Specific Epigenetic Response
3.2.1. Identification of Epigenetically Affected Pathways after Paternal Low-Protein Diet (“Experiment D”)
3.2.2. Identification of Epigenetically Affected Pathways after Paternal Exposure to a Temperature Increase (“Experiment H”)
3.2.3. Annotated Differentially Methylated Regions Shared in Response to Changes of Either One or Both Environmental Factors
3.3. Testing Hypotheses H1–H3
4. Discussion
4.1. Specific and General Response
4.2. Alternative Explanation
4.3. The Father’s Role: Roaming Males Increase Genetic and Epigenetic Diversity
4.4. Evolutionary Consequences of Epigenetic Inheritance
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- West-Eberhard, M.J. Phenotypic plasticity and the origins of diversity. Annu. Rev. Ecol. Syst. 1989, 20, 249–278. [Google Scholar] [CrossRef]
- Stearns, S.C. The Evolution of Life Histories; Oxford University Press: Oxford, UK, 1992; 249p. [Google Scholar]
- Jablonka, E.; Raz, G. Transgenerational epigenetic inheritance: Prevalence, mechanisms, and implications for the study of heredity and evolution. Q. Rev. Biol. 2009, 84, 131–176. [Google Scholar] [CrossRef] [PubMed]
- Szyf, M. Epigenetics, DNA methylation and chromatin modifying drugs. Annu. Rev. Pharmacol. Toxicol. 2009, 49, 243–263. [Google Scholar] [CrossRef] [PubMed]
- Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef] [PubMed]
- Anway, M.D.; Rekow, S.S.; Skinner, M.K. Transgenerational epigenetic programming of the embryonic testis transcriptome. Genomics 2008, 91, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Carone, B.R.; Fauquier, L.; Habib, N.; Shea, J.M.; Hart, C.E.; Li, R.; Bock, C.; Li, C.; Gu, H.; Zamore, P.D.; et al. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell 2010, 143, 1084–1096. [Google Scholar] [CrossRef] [PubMed]
- Gapp, K.; Corcoba, A.; Van Steenwyk, G.; Mansuy, I.M.; Duarte, J.M. Brain metabolic alterations in mice subjected to postnatal traumatic stress and in their offspring. J. Cereb. Blood Flow Metab. 2017, 37, 2423–2432. [Google Scholar] [CrossRef] [PubMed]
- Dolinoy, D.C.; Weidman, J.R.; Waterland, R.A.; Jirtle, R.L. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ. Health Perspect. 2006, 114, 567–572. [Google Scholar] [CrossRef] [PubMed]
- He, X.J.; Chen, T.; Zhu, J.K. Regulation and function of DNA methylation in plants and animals. Cell Res. 2011, 21, 442–465. [Google Scholar] [CrossRef] [PubMed]
- Deaton, A.M.; Bird, A. CpG islands and the regulation of transcription. Genes Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef] [PubMed]
- Hahn, M.A.; Wu, X.; Li, A.X.; Hahn, T.; Pfeifer, G.P. Relationship between gene body DNA methylation and intragenic H3K9me3 and H3K36me3 chromatin marks. PLoS ONE 2011, 6, e18844. [Google Scholar] [CrossRef] [PubMed]
- Jjingo, D.; Conley, A.B.; Soojin, V.Y.; Lunyak, V.V.; Jordan, I.K. On the presence and role of human gene-body DNA methylation. Oncotarget 2012, 3, 462–474. [Google Scholar] [CrossRef]
- Weaver, I.C.G.; Cervoni, N.; Champagne, F.A.; D’Alessio, A.C.; Sharma, S.; Seckl, J.R.; Dymov, S.; Szyf, M.; Meaney, M.J. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004, 7, 847–854. [Google Scholar] [CrossRef]
- Kilvitis, H.J.; Alvarez, M.; Foust, C.M.; Schrey, A.W.; Robertson, M.; Richards, C.L. Ecological epigenetics. Adv. Exp. Med. Biol. 2014, 781, 191–210. [Google Scholar]
- Penuelas, J.; Sardans, J.; Estiarte, M.; Ogaya, R.; Carnicer, J.; Coll, M.; Barbeta, A.; Rivas-Ubach, A.; Llusià, J.; Garbulsky, M.; et al. Evidence of current impact of climate change on life: A walk from genes to the biosphere. Glob. Chang. Biol. 2013, 19, 2303–2338. [Google Scholar] [CrossRef] [PubMed]
- Herrera, C.M.; Bazaga, P. Untangling individual variation in natural populations: Ecological, genetic and epigenetic correlates of long-term inequality in herbivory. Mol. Ecol. 2011, 20, 1675–1688. [Google Scholar] [CrossRef] [PubMed]
- Kucharski, R.; Maleszka, J.; Foret, S.; Maleszka, R. Nutritional control of reproductive status in honeybees via DNA methylation. Science 2008, 319, 1827–1830. [Google Scholar] [CrossRef]
- Varriale, A.; Bernardi, G. DNA methylation and body temperature in fishes. Gene 2006, 385, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Weyrich, A.; Lenz, D.; Jeschek, M.; Chung, T.H.; Rübensam, K.; Göritz, F.; Jewgenow, K.; Fickel, J. Paternal intergenerational epigenetic response to heat exposure in male Wild guinea pigs. Mol. Ecol. 2016, 25, 1729–1740. [Google Scholar] [CrossRef] [PubMed]
- Lea, A.J.; Altmann, J.; Alberts, S.C.; Tung, J. Resource base influences genome-wide DNA methylation levels in wild baboons (Papio cynocephalus). Mol. Ecol. 2016, 25, 1681–1696. [Google Scholar] [CrossRef]
- Navarro-Martin, L.; Viñas, J.; Ribas, L.; Díaz, N.; Gutiérrez, A.; Di Croce, L.; Piferrer, F. DNA methylation of the gonadal aromatase (cyp19a) promoter is involved in temperature-dependent sex ratio shifts in the European sea bass. PloS Genet. 2011, 7, e1002447. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, N.; Lance, V. Temperature Dependent Sex Determination in Vertebrates; Smithsonian Books: Washington, DC, USA, 2004. [Google Scholar]
- Schrey, A.W.; Richards, C.L.; Meller, V.; Sollars, V.; Ruden, D.M. The role of epigenetics in evolution: The extended synthesis. Genet. Res. Int. 2012, 2012, 286164. [Google Scholar] [CrossRef] [PubMed]
- Dickins, T.E.; Rahman, Q. The extended evolutionary synthesis and the role of soft inheritance in evolution. Proc. Biol. Sci. 2012, 279, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Weyrich, A.; Jeschek, M.; Schrapers, K.T.; Lenz, D.; Chung, T.H.; Rübensam, K.; Yasar, S.; Schneemann, M.; Ortmann, S.; Jewgenow, K.; et al. Diet changes alter paternally inherited epigenetic pattern in male Wild guinea pigs. Environ. Epigenet. 2018, 4, dvy011. [Google Scholar] [CrossRef] [PubMed]
- Asher, M.; Lippmann, T.; Epplen, J.T.; Kraus, C.; Trillmich, F.; Sachser, N. Large males dominate: Ecology, social organization, and mating system of wild cavies, the ancestors of the guinea pig. Behav. Ecol. Sociobiol. 2008, 62, 1509–1521. [Google Scholar] [CrossRef]
- Bernal, N. 2016. Cavia aperea. The IUCN Red List of Threatened Species 2016: e.T86257782A22189256. Cambridge, UK, 2015; p. 2. Available online: http://dx.doi.org/10.2305/IUCN.UK.2016-2.RLTS.T86257782A22189256.en (accessed on 20 December 2018).
- Weyrich, A.; Benz, S.; Karl, S.; Jeschek, M.; Jewgenow, K.; Fickel, J. Paternal heat exposure causes DNA methylation and gene expression changes of Stat3 in Wild guinea pig sons. Ecol. Evol. 2016, 6, 2657–2666. [Google Scholar] [CrossRef] [PubMed]
- Hingst, O.; Blottner, S. Quantification of apoptosis (programmed cell-death) in mammalian testis by DNA-fragmentation ELISA. Theriogenology 1995, 44, 313–319. [Google Scholar] [CrossRef]
- Holt, W.V. Postnatal development of the testes in the cuis, Galea Musteloides. Lab. Anim. 1977, 11, 87–91. [Google Scholar] [CrossRef]
- Meissner, A.; Gnirke, A.; Bell, G.W.; Ramsahoye, B.; Lander, E.S.; Jaenisch, R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005, 33, 5868–5877. [Google Scholar] [CrossRef]
- Snel, B.; Lehmann, G.; Bork, P.; Huynen, M.A. STRING: A web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000, 28, 3442–3444. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Bollati, V.; Baccarelli, A. Environmental epigenetics. Heredity 2010, 105, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Baccarelli, A.; Bollati, V. Epigenetics and environmental chemicals. Curr. Opin. Pediatr. 2009, 21, 243–251. [Google Scholar] [CrossRef]
- Skinner, M.K. Environmental stress and epigenetic transgenerational inheritance. BMC Med. 2014, 12, 153. [Google Scholar] [CrossRef] [PubMed]
- Hauser, R.; Singh, N.P.; Chen, Z.; Pothier, L.; Altshul, L. Lack of an association between environmental exposure to polychlorinated biphenyls and p,p’-DDE and DNA damage in human sperm measured using the neutral comet assay. Hum. Reprod. 2003, 18, 2525–2533. [Google Scholar] [CrossRef] [PubMed]
- Logan, C.Y.; Nusse, R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004, 20, 781–810. [Google Scholar] [CrossRef]
- Tortora, G.; Derrickson, B. Principles of Anatomy and Physiology; Wiley: Hoboken, NJ, USA, 2012. [Google Scholar]
- Falk, M.H.; Issels, R.D. Hyperthermia in oncology. Int. J. Hyperth. 2001, 17, 1–18. [Google Scholar] [CrossRef]
- Pagani, E.; Falcinelli, S.; Pepponi, R.; Turriziani, M.; Caporaso, P.; Caporali, S.; Bonmassar, E.; D’Atri, S. Combined effect of temozolomide and hyperthermia on human melanoma cell growth and O-6-methylguanine-DNA methyltransferase activity. Int. J. Oncol. 2007, 30, 443–451. [Google Scholar] [PubMed]
- Sharpe, R.M. Environmental/lifestyle effects on spermatogenesis. Philos. Trans. R. Soc. B-Biol. Sci. 2010, 365, 1697–1712. [Google Scholar] [CrossRef]
- Slotkin, R.K.; Martienssen, R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007, 8, 272–285. [Google Scholar] [CrossRef]
- Shen, J.S.J.; Dushoff, J.; Bewick, A.J.; Chain, F.J.; Evans, B.J. Genomic dynamics of transposable elements in the Western clawed frog (Silurana tropicalis). Genome Biol. Evol. 2013, 5, 998–1009. [Google Scholar] [CrossRef] [PubMed]
- Hamamoto, R.; Toyokawa, G.; Nakakido, M.; Ueda, K.; Nakamura, Y. SMYD2-dependent HSP90 methylation promotes cancer cell proliferation by regulating the chaperone complex formation. Cancer Lett. 2014, 351, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Fickel, J.; Bubliy, O.A.; Stache, A.; Noventa, T.; Jirsa, A.; Heurich, M. Crossing the border? Structure of the red deer (Cervus elaphus) population from the Bavarian-Bohemian forest ecosystem. Mamm. Biol. 2012, 77, 211–220. [Google Scholar] [CrossRef]
- Ng, S.F.; Lin, R.C.; Laybutt, D.R.; Barres, R.; Owens, J.A.; Morris, M.J. Chronic high-fat diet in fathers programs β-cell dysfunction in female rat offspring. Nature 2010, 467, 963–966. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Yang, C.R.; Wei, Y.P.; Zhao, Z.A.; Hou, Y.; Schatten, H.; Sun, Q.Y. Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proc. Natl. Acad. Sci. USA 2014, 111, 1873–1878. [Google Scholar] [CrossRef] [PubMed]
- Vassoler, F.M.; White, S.L.; Schmidt, H.D.; Sadri-Vakili, G.; Pierce, R.C. Epigenetic inheritance of a cocaine-resistance phenotype. Nat. Neurosci. 2013, 16, 42–47. [Google Scholar] [CrossRef] [PubMed]



| Gene Name (Ensembl ID) | Full Gene Name | Regulatory Region | Gene Ontology (GO) Term |
|---|---|---|---|
| Dock6 (ENSCPOG00000026620) | Dedicator of Cytokinesis 6 | CDS | Positive regulation of hydrolyze guanosine triphosphate (GTP) GTPase activity, guanyl-nucleotide exchange factor activity, small GTPase-mediated signal transduction, cytoplasm |
| Eps8l2 (ENSCPOG00000003802) | Epidermal growth factor receptor kinase substrate 8-like protein 2 | CDS | Cytoplasm, plasma membrane, protein complex, protein binding, positive regulation of GTPase activity, actin binding, extracellular exosome, Rho guanyl-nucleotide exchange factor activity, regulation of Rho protein signal transduction, actin filament binding, Rho protein signal transduction, ruffle, ruffle membrane, vesicle, Rac guanyl-nucleotide exchange factor activity, Rac protein signal transduction, positive regulation of ruffle assembly |
| Hmcn2 (ENSCPOG00000013220) | Hemicentin 2 | CDS | Calcium ion binding, protein binding, cell junction, cell cortex, response to stimulus |
| Icam5 (ENSCPOG00000000999) | Intercellular adhesion molecule 5 | CDS | Plasma membrane, integral component of plasma membrane, protein binding, single organismal cell-cell adhesion cell adhesion, phagocytosis, integrin binding |
| Kcns1 (ENSCPOG00000013646) | Potassium voltage-gated channel subfamily S member 1 | CDS | Transmembrane transport, protein homo-oligomerization, perinuclear region of cytoplasm, protein binding, membrane voltage-gated potassium channel complex, potassium ion transmembrane transport, potassium ion transport, ion transport, ion channel activity, delayed rectifier potassium channel activity, regulation of delayed rectifier potassium channel activity, potassium channel regulator activity |
| Klhl10 (ENSCPOG00000002818) | Kelch-Like Family Member 10, Testicular Tissue Protein Li 104 | CDS | Cytoplasm, homeostasis of number of cells within a tissue, protein binding, ubiquitin-protein transferase activity, protein ubiquitination, spermatid development, Cul3-RING ubiquitin ligase complex, cell morphogenesis, male gonad development, fertilization, male genitalia morphogenesis |
| Map3k6 (ENSCPOG00000012359) | Mitogen-Activated Protein Kinase 6 | CDS | Protein kinase activity, ATP binding, protein phosphorylation, MAP kinase, kinase activity, activation of MAPKK activity, magnesium ion binding |
| Mmp9 (ENSCPOG00000007559) | Matrix Metallopeptidase 9 | CDS | Proteolysis, metalloendopeptidase activity, ossification, collagen catabolic process, leukocyte migration |
| Myh14 (ENSCPOG00000002223) | Myosin-14 | CDS | ATP binding, metabolic process, protein binding, actin filament binding, sensory perception of sound, calmodulin binding, actomyosin structure organization, neuronal action potential, axon, motor activity, myosin complex, regulation of cell shape, stress fiber, skeletal muscle tissue development, vocalization behavior, skeletal muscle contraction, mitochondrion morphogenesis, actin-dependent ATPase activity, actin filament-based movement, microfilament motor activity, actomyosin, myosin filament |
| Notch4 (ENSCPOG00000000591) | Neurogenic locus notch homolog 4 | CDS | Integral component of membrane, calcium ion binding, protein binding, multicellular organismal development, cell differentiation, Notch signaling pathway, regulation of developmental process, mammary gland development, endothelial cell differentiation, endothelial cell morphogenesis |
| Otud6a (ENSCPOG00000012916) | OTU domain containing 6 | CDS | Skeletal system morphogenesis |
| Pclo (ENSCPOG00000009376) | Piccolo Presynaptic Cytomatrix Protein | CDS | Calcium ion binding, intracellular, metal ion binding, protein binding, cell junction, presynaptic active zone, synapse assembly, cytoskeleton organization, insulin secretion, extracellular exosome, calcium-dependent phospholipid binding, regulation of exocytosis, synapse, postsynaptic density, cAMP-mediated signaling, profilin binding, synaptic vesicle targeting |
| Plekhh3 (ENSCPOG00000010080) | Pleckstrin Homology, MyTH4 And FERM Domain Containing H3 | CDS | Signal transduction, cytoskeleton |
| Sh3gl1 (ENSCPOG00000023236) | SH3 Domain Containing GRB2 Like 1, Endophilin A2 | CDS | Cytoplasm, protein binding, cell junction, early endosome, membrane, identical protein binding, lipid binding, endocytosis, podosome, cell projection, phosphatase binding, GTPase binding |
| Sigirr (ENSCPOG00000025549) | Single Immunoglobulin and Toll-Interleukin 1 Receptor (TIR) Domain | CDS | Integral component of membrane, signal transduction, protein binding, membrane, negative regulation of cytokine-mediated signaling pathway, acute-phase response, negative regulation of sequence-specific DNA binding transcription factor activity, negative regulation of chemokine biosynthetic process |
| Slc46a2 (ENSCPOG00000001823) | Solute Carrier Family 46 Member 2 | CDS | Integral component of membrane, transmembrane transport, molecular function, plasma membrane, cell surface, transporter activity, T cell homeostasis, regulation of T cell differentiation, negative regulation of T cell apoptotic process, thymus development |
| Sox13 (ENSCPOG00000006604) | CDS | Nucleus, regulation of transcription, DNA-templated, sequence-specific DNA binding | |
| Med26 (ENSCPOG00000008968) | Mediator complex subunit 26 (adopted from mouse) | CDS | Nucleus, DNA binding, transcription, DNA-templated, regulation of transcription from RNA polymerase II promoter, transcription initiation from RNA polymerase II promoter, nucleoplasm RNA polymerase II transcription cofactor activity, mediator complex, transcription coactivator activity |
| Sncg (ENSCPOG00000023463) | Synuclein Gamma | Promoter | Cytoplasm, synaptic transmission, perinuclear region of cytoplasm, protein binding, synapse organization, extracellular exosome, microtubule organizing center, axon, neuronal cell body spindle, adult locomotory behavior, protein secretion, regulation of neurotransmitter secretion, regulation of dopamine secretion |
| Svp-1 (ENSCPOG00000025237) | Seminal vesicle polypeptide | Promoter | Copulation, DNA binding, transcription |
| Unknown (ENSCPOG00000022766) | Promoter |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weyrich, A.; Lenz, D.; Fickel, J. Environmental Change-Dependent Inherited Epigenetic Response. Genes 2019, 10, 4. https://doi.org/10.3390/genes10010004
Weyrich A, Lenz D, Fickel J. Environmental Change-Dependent Inherited Epigenetic Response. Genes. 2019; 10(1):4. https://doi.org/10.3390/genes10010004
Chicago/Turabian StyleWeyrich, Alexandra, Dorina Lenz, and Jörns Fickel. 2019. "Environmental Change-Dependent Inherited Epigenetic Response" Genes 10, no. 1: 4. https://doi.org/10.3390/genes10010004




