Biogenesis of Developmental Master Regulatory 27nt-RNAs in Stylonychia—Can Coding RNA Turn into Non-Coding?
Abstract
:1. Introduction
2. Small ncRNA Biogenesis in Tetrahymena and Paramecium Involves Non-Coding RNA Precursors Transcribed in the Germline Micronucleus
3. Developmental Small Non-Coding RNAs Do Not Target Micronucleus-Specific Sequences in Spirotrichous Ciliates, but Safeguard the Retention of Macronucleus-Destined Sequences
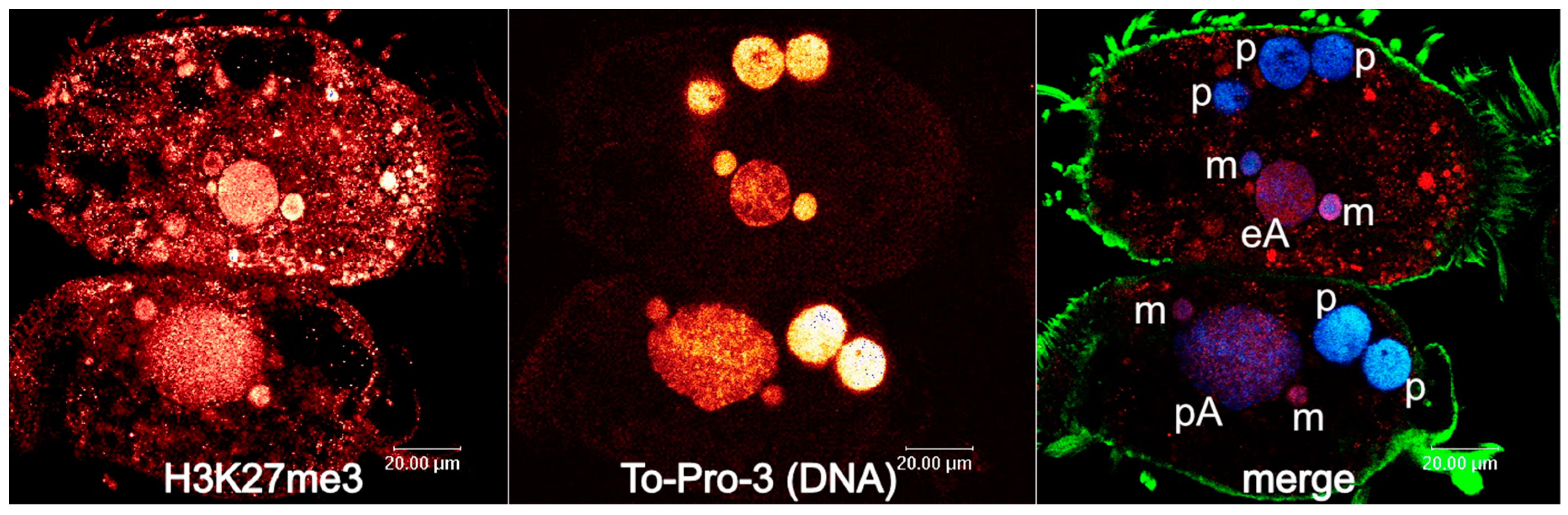
4. Developmental 27nt-RNAs in Stylonychia and Their Potential Role as Heterochromatization Preventers at Macronucleus-Destined Sequences
5. A Minimalistic Model for the Biogenesis of 27nt-RNAs—Is mRNA/pre-mRNA the Substrate?
6. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Grewal, S.I. RNAi-dependent formation of heterochromatin and its diverse functions. Curr. Opin. Genet. Dev. 2010, 20, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Wassenegger, M. The role of the RNAi machinery in heterochromatin formation. Cell 2005, 122, 13–16. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, K. RNA-directed epigenetic regulation of DNA rearrangements. Essays Biochem. 2010, 48, 89–100. [Google Scholar] [PubMed]
- Noto, T.; Mochizuki, K. Whats, hows and whys of programmed DNA elimination in Tetrahymena. Open Biol. 2017, 7, 170172. [Google Scholar] [CrossRef] [PubMed]
- Ammermann, D.; Steinbruck, G.; von Berger, L.; Hennig, W. The development of the macronucleus in the ciliated protozoan Stylonychia mytilus. Chromosoma 1974, 45, 401–429. [Google Scholar] [CrossRef]
- Prescott, D.M. The DNA of ciliated protozoa. Microbiol. Rev. 1994, 58, 233–267. [Google Scholar] [CrossRef]
- Betermier, M.; Duharcourt, S. Programmed Rearrangement in Ciliates: Paramecium. Microbiol. Spectr. 2014, 2, 1–20. [Google Scholar]
- Heyse, G.; Jonsson, F.; Chang, W.J.; Lipps, H.J. RNA-dependent control of gene amplification. Proc. Natl. Acad. Sci. USA 2010, 107, 22134–22139. [Google Scholar] [CrossRef]
- Nowacki, M.; Haye, J.E.; Fang, W.; Vijayan, V.; Landweber, L.F. RNA-mediated epigenetic regulation of DNA copy number. Proc. Natl. Acad. Sci. USA 2010, 107, 22140–22144. [Google Scholar] [CrossRef]
- Fang, W.; Wang, X.; Bracht, J.R.; Nowacki, M.; Landweber, L.F. Piwi-interacting RNAs protect DNA against loss during Oxytricha genome rearrangement. Cell 2012, 151, 1243–1255. [Google Scholar] [CrossRef]
- Fetzer, C.P.; Hogan, D.J.; Lipps, H.J. A PIWI homolog is one of the proteins expressed exclusively during macronuclear development in the ciliate Stylonychia lemnae. Nucleic Acids Res. 2002, 30, 4380–4386. [Google Scholar] [CrossRef] [PubMed]
- Juranek, S.A.; Rupprecht, S.; Postberg, J.; Lipps, H.J. snRNA and heterochromatin formation are involved in DNA excision during macronuclear development in stichotrichous ciliates. Eukaryot. Cell 2005, 4, 1934–1941. [Google Scholar] [CrossRef] [PubMed]
- Postberg, J.; Jonsson, F.; Weil, P.P.; Bulic, A.; Juranek, S.A.; Lipps, H.J. 27nt-RNAs guide histone variant deposition via ‘RNA-induced DNA replication interference’ and thus transmit parental genome partitioning in Stylonychia. Epigenet. Chromatin 2018, 11, 31. [Google Scholar] [CrossRef] [PubMed]
- Zahler, A.M.; Neeb, Z.T.; Lin, A.; Katzman, S. Mating of the stichotrichous ciliate Oxytricha trifallax induces production of a class of 27 nt small RNAs derived from the parental macronucleus. PLoS ONE 2012, 7, e42371. [Google Scholar] [CrossRef] [PubMed]
- Paeschke, K.; Juranek, S.; Simonsson, T.; Hempel, A.; Rhodes, D.; Lipps, H.J. Telomerase recruitment by the telomere end binding protein-β facilitates G-quadruplex DNA unfolding in ciliates. Nat. Struct. Mol. Biol. 2008, 15, 598–604. [Google Scholar] [CrossRef] [PubMed]
- Schaffitzel, C.; Berger, I.; Postberg, J.; Hanes, J.; Lipps, H.J.; Pluckthun, A. In Vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl. Acad. Sci. USA 2001, 98, 8572–8577. [Google Scholar] [CrossRef]
- Postberg, J.; Tsytlonok, M.; Sparvoli, D.; Rhodes, D.; Lipps, H.J. A telomerase-associated RecQ protein-like helicase resolves telomeric G-quadruplex structures during replication. Gene 2012, 497, 147–154. [Google Scholar] [CrossRef]
- Aeschlimann, S.H.; Jonsson, F.; Postberg, J.; Stover, N.A.; Petera, R.L.; Lipps, H.J.; Nowacki, M.; Swart, E.C. The draft assembly of the radically organized Stylonychia lemnae macronuclear genome. Genome Biol. Evol. 2014, 6, 1707–1723. [Google Scholar] [CrossRef]
- Jonsson, F.; Postberg, J.; Lipps, H.J. The unusual way to make a genetically active nucleus. DNA Cell Biol. 2009, 28, 71–78. [Google Scholar] [CrossRef]
- Wen, J.; Maercker, C.; Lipps, H.J. Sequential excision of internal eliminated DNA sequences in the differentiating macronucleus of the hypotrichous ciliate Stylonychia lemnae. Nucleic Acids Res. 1996, 24, 4415–4419. [Google Scholar] [CrossRef]
- Meyer, G.F.; Lipps, H.J. The formation of polytene chromosomes during macronuclear development of the hypotrichous ciliate Stylonychia mytilus. Chromosoma 1981, 82, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Mollenbeck, M.; Zhou, Y.; Cavalcanti, A.R.; Jonsson, F.; Higgins, B.P.; Chang, W.J.; Juranek, S.; Doak, T.G.; Rozenberg, G.; Lipps, H.J.; et al. The pathway to detangle a scrambled gene. PLoS ONE 2008, 3, e2330. [Google Scholar] [CrossRef] [PubMed]
- Forcob, S.; Bulic, A.; Jonsson, F.; Lipps, H.J.; Postberg, J. Differential expression of histone H3 genes and selective association of the variant H3.7 with a specific sequence class in Stylonychia macronuclear development. Epigenet. Chromatin 2014, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Postberg, J.; Heyse, K.; Cremer, M.; Cremer, T.; Lipps, H.J. Spatial and temporal plasticity of chromatin during programmed DNA-reorganization in Stylonychia macronuclear development. Epigenet. Chromatin 2008, 1, 3. [Google Scholar] [CrossRef] [PubMed]
- Meyer, G.F.; Lipps, H.J. Chromatin elimination in the hypotrichous ciliate Stylonychia mytilus. Chromosoma 1980, 77, 285–297. [Google Scholar] [CrossRef]
- Fuhrmann, G.; Jonsson, F.; Weil, P.P.; Postberg, J.; Lipps, H.J. RNA-template dependent de novo telomere addition. RNA Biol. 2016, 13, 733–739. [Google Scholar] [CrossRef]
- Jonsson, F.; Steinbruck, G.; Lipps, H.J. Both subtelomeric regions are required and sufficient for specific DNA fragmentation during macronuclear development in Stylonychia lemnae. Genome Biol. 2001, 2, research0005-1. [Google Scholar] [CrossRef]
- Rivera, C.; Saavedra, F.; Alvarez, F.; Diaz-Celis, C.; Ugalde, V.; Li, J.; Forne, I.; Gurard-Levin, Z.A.; Almouzni, G.; Imhof, A.; et al. Methylation of histone H3 lysine 9 occurs during translation. Nucleic Acids Res. 2015, 43, 9097–9106. [Google Scholar] [CrossRef]
- Postberg, J.; Alexandrova, O.; Lipps, H.J. Synthesis of pre-rRNA and mRNA is directed to a chromatin-poor compartment in the macronucleus of the spirotrichous ciliate Stylonychia lemnae. Chromosome Res. 2006, 14, 161–175. [Google Scholar] [CrossRef]
- Postberg, J.; Alexandrova, O.; Cremer, T.; Lipps, H.J. Exploiting nuclear duality of ciliates to analyse topological requirements for DNA replication and transcription. J. Cell Sci. 2005, 118 Pt 17, 3973–3983. [Google Scholar] [CrossRef]
- Sapra, G.R.; Ammermann, D. RNA synthesis and acquisition of actinomycin D insensitivity during conjugation in Stylonychia mytilus. Exp. Cell Res. 1973, 78, 168–174. [Google Scholar] [CrossRef]
- Sapra, G.R.; Ammermann, D. An analysis of the development program in relation to RNA metabolism in the ciliate Stylonychia mytilus. Dev. Biol. 1974, 36, 105–112. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Postberg, J.; Weil, P.P.; Pembaur, A. Biogenesis of Developmental Master Regulatory 27nt-RNAs in Stylonychia—Can Coding RNA Turn into Non-Coding? Genes 2019, 10, 940. https://doi.org/10.3390/genes10110940
Postberg J, Weil PP, Pembaur A. Biogenesis of Developmental Master Regulatory 27nt-RNAs in Stylonychia—Can Coding RNA Turn into Non-Coding? Genes. 2019; 10(11):940. https://doi.org/10.3390/genes10110940
Chicago/Turabian StylePostberg, Jan, Patrick Philipp Weil, and Anton Pembaur. 2019. "Biogenesis of Developmental Master Regulatory 27nt-RNAs in Stylonychia—Can Coding RNA Turn into Non-Coding?" Genes 10, no. 11: 940. https://doi.org/10.3390/genes10110940




