At the Beginning of the End and in the Middle of the Beginning: Structure and Maintenance of Telomeric DNA Repeats and Interstitial Telomeric Sequences
Abstract
:1. Telomeres
1.1. Composition and Diversity of Telomeric Repeats
1.2. DNA Structures Formed by Telomeric Repeats In Vitro and In Vivo
1.3. Factors Binding to Telomeric Repeats and Structure of Telomeric Chromatin
1.3.1. Major Protein Factors Binding to Telomeric Repeats
1.3.2. Distinct RAP1 Function in Lower and Higher Eukaryotes and a Hypothesis about Evolutionary Origin of Telomeres in Budding Yeast
1.3.3. Chromatin Organization of Telomeres; Replication and Transcription of Telomeres
1.4. Regulation of Telomere Length: Alternative Mechanisms
2. Interstitial Telomeric Sequences (ITSs)
2.1. ITSs and their Proposed Origin
2.2. Genetic Instabilities Associated with ITSs
2.3. Factors Binding to ITSs and Proposed Functions of ITSs in the Genome
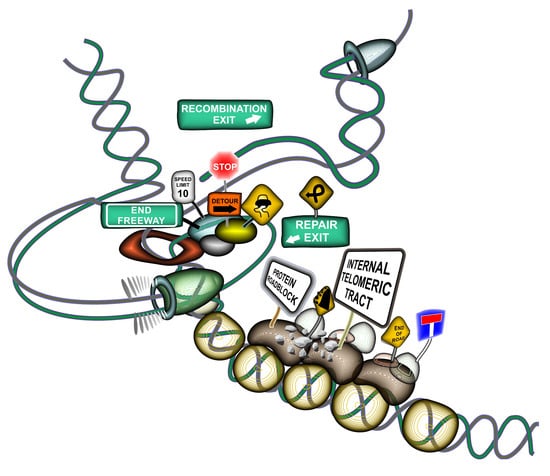
2.4. Mechanisms of ITS-Mediated Genome Instability
3. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Muller, H.J. The remaking of chromosomes. Collect. Net 1938, 13, 181–198. [Google Scholar]
- McClintock, B. The fusion of broken ends of sister half chromatids following chromatid breakage at meiotic anaphase. Miss. Agric. Exp. Stn. Res. Bull 1938, 190, 1–48. [Google Scholar]
- Muller, H.J. Induced mutations in Drosophila. Cold Spring Harb. Symp. Quant. Biol. 1941, 9, 151–167. [Google Scholar] [CrossRef]
- McClintock, B. The stability of broken ends of chromosomes in Zea Mays. Genetics 1941, 26, 234. [Google Scholar] [PubMed]
- Watson, J.D. Origin of Concatemeric T7 DNA. Nat. New Biol. 1972, 239, 197–201. [Google Scholar] [CrossRef]
- Olovnikov, A.M. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 1973, 41, 181–190. [Google Scholar] [CrossRef]
- Greider, C.W.; Blackburn, E.H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 1985, 43, 405–413. [Google Scholar] [CrossRef]
- Lundblad, V.; Blackburn, E.H. An alternative pathway for yeast telomere maintenance rescues est1-senescence. Cell 1993, 73, 347–360. [Google Scholar] [CrossRef]
- Zakian, V.A. Telomeres: Beginning to understand the end. Science 1995, 270, 1601–1607. [Google Scholar] [CrossRef]
- Wellinger, R.J.; Sen, D. The DNA structures at the ends of eukaryotic chromosomes. Eur. J. Cancer 1997, 33, 735–749. [Google Scholar] [CrossRef]
- Gomes, N.M.V.; Shay, J.W.; Wright, W.E. Telomere biology in Metazoa. FEBS Lett. 2010, 584, 3741–3751. [Google Scholar] [CrossRef]
- Vítková, M.; Král, J.; Traut, W.; Zrzavý, J.; Marec, F. The evolutionary origin of insect telomeric repeats, (TTAGG) N. Chromosom. Res. 2005, 13, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Frydrychová, R.; Grossmann, P.; Trubac, P.; Vítková, M.; Marec, F. Phylogenetic distribution of TTAGG telomeric repeats in insects. Genome 2004, 47, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, J.; Brandes, A.; Schubert, I. Telomere sequence localization and karyotype evolution in higher plants. Plant Syst. Evol. 1995, 196, 227–241. [Google Scholar] [CrossRef]
- Procházková Schrumpfová, P.; Schořová, Š.; Fajkus, J. Telomere- and Telomerase-Associated Proteins and Their Functions in the Plant Cell. Front. Plant Sci. 2016, 7, 851. [Google Scholar] [CrossRef]
- Delany, M.E.; Daniels, L.M.; Swanberg, S.E.; Taylor, H.A. Telomeres in the chicken: Genome stability and chromosome ends. Poult. Sci. 2003, 82, 917–926. [Google Scholar] [CrossRef]
- Wellinger, R.J.; Zakian, V.A. Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: Beginning to end. Genetics 2012, 191, 1073–1105. [Google Scholar] [CrossRef]
- Makarov, V.L.; Hirose, Y.; Langmore, J.P. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 1997, 88, 657–666. [Google Scholar] [CrossRef]
- Wright, W.E.; Tesmer, V.M.; Huffman, K.E.; Levene, S.D.; Shay, J.W. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997, 11, 2801–2809. [Google Scholar] [CrossRef]
- Zhao, Y.; Hoshiyama, H.; Shay, J.W.; Wright, W.E. Quantitative telomeric overhang determination using a double-strand specific nuclease. Nucleic Acids Res. 2008, 36, e14. [Google Scholar] [CrossRef]
- Yang, T.-L.B.; Song, S.; Johnson, F.B. Contributions of telomere biology to human age-related disease. In Handbook of the Biology of Aging; Elsevier: Amsterdam, The Netherlands, 2016; pp. 205–239. ISBN 9780124115965. [Google Scholar]
- Traverse, K.L.; Pardue, M.L. A spontaneously opened ring chromosome of Drosophila melanogaster has acquired He-T DNA sequences at both new telomeres. Proc. Natl. Acad. Sci. USA 1988, 85, 8116–8120. [Google Scholar] [CrossRef] [PubMed]
- Levis, R.W.; Ganesan, R.; Houtchens, K.; Tolar, L.A.; Sheen, F. Transposons in place of telomeric repeats at a Drosophila telomere. Cell 1993, 75, 1083–1093. [Google Scholar] [CrossRef]
- López, C.C.; Nielsen, L.; Edström, J.E. Terminal long tandem repeats in chromosomes form Chironomus pallidivittatus. Mol. Cell. Biol. 1996, 16, 3285–3290. [Google Scholar] [CrossRef] [PubMed]
- Villasante, A.; Abad, J.P.; Planelló, R.; Méndez-Lago, M.; Celniker, S.E.; de Pablos, B. Drosophila telomeric retrotransposons derived from an ancestral element that was recruited to replace telomerase. Genome Res. 2007, 17, 1909–1918. [Google Scholar] [CrossRef] [PubMed]
- Anzai, T.; Takahashi, H.; Fujiwara, H. Sequence-specific recognition and cleavage of telomeric repeat (TTAGG)n by endonuclease of non-long terminal repeat retrotransposon TRAS1. Mol. Cell. Biol. 2001, 21, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Osanai-Futahashi, M.; Fujiwara, H. Coevolution of telomeric repeats and telomeric repeat-specific non-LTR retrotransposons in insects. Mol. Biol. Evol. 2011, 28, 2983–2986. [Google Scholar] [CrossRef] [PubMed]
- Henderson, E.; Hardin, C.C.; Walk, S.K.; Tinoco, I.; Blackburn, E.H. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine·guanine base pairs. Cell 1987, 51, 899–908. [Google Scholar] [CrossRef]
- Williamson, J.R.; Raghuraman, M.K.K.; Cech, T.R. Monovalent cation-induced structure of telomeric DNA: The G-quartet model. Cell 1989, 59, 871–880. [Google Scholar] [CrossRef]
- Sundquist, W.I.; Klug, A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature 1989, 342, 825–829. [Google Scholar] [CrossRef]
- Wang, Y.; Patell, D.I. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure 1993, 1, 263–282. [Google Scholar] [CrossRef]
- Parkinson, G.N.; Lee, M.P.H.; Neidle, S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 2002, 417, 876–880. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, D. Sequence, stability, and structure of G-quadruplexes and their interactions with drugs. In Current Protocols in Nucleic Acid Chemistry; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2012; pp. 17.5.1–17.5.17. [Google Scholar]
- Zahler, A.M.; Williamson, J.R.; Cech, T.R.; Prescott, D.M. Inhibition of telomerase by G-quartet DMA structures. Nature 1991, 350, 718–720. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.S.; Chen, Q.; Yatsunyk, L.A.; Nicoludis, J.M.; Garcia, M.S.; Kranaster, R.; Balasubramanian, S.; Monchaud, D.; Teulade-Fichou, M.-P.; Abramowitz, L.; et al. Rudimentary G-quadruplex–based telomere capping in Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 2011, 18, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Sen, D.; Gilbert, W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 1988, 334, 364–366. [Google Scholar] [CrossRef]
- Schaffitzel, C.; Berger, I.; Postberg, J.; Hanes, J.; Lipps, H.J.; Plückthun, A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl. Acad. Sci. USA 2001, 98, 8572–8577. [Google Scholar] [CrossRef] [PubMed]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Lam, E.Y.N.; Beraldi, D.; Tannahill, D.; Balasubramanian, S. G-quadruplex structures are stable and detectable in human genomic DNA. Nat. Commun. 2013, 4, 1796. [Google Scholar] [CrossRef] [PubMed]
- Paeschke, K.; Capra, J.; Zakian, V. DNA Replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 2011, 145, 678–691. [Google Scholar] [CrossRef] [PubMed]
- Marsh, R.E.; Bierstedt, R.; Eichhorn, E.L. IUCr The crystal structure of cytosine-5-acetic acid. Acta Crystallogr. 1962, 15, 310–316. [Google Scholar] [CrossRef]
- Gehring, K.; Leroy, J.-L.; Guéron, M. A tetrameric DNA structure with protonated cytosine-cytosine base pairs. Nature 1993, 363, 561–565. [Google Scholar] [CrossRef]
- Day, H.A.; Pavlou, P.; Waller, Z.A.E. i-Motif DNA: Structure, stability and targeting with ligands. Bioorg. Med. Chem. 2014, 22, 4407–4418. [Google Scholar] [CrossRef] [PubMed]
- Griffith, J.D.; Comeau, L.; Rosenfield, S.; Stansel, R.M.; Bianchi, A.; Moss, H.; de Lange, T. Mammalian telomeres end in a large duplex loop. Cell 1999, 97, 503–514. [Google Scholar] [CrossRef]
- Strahl-Bolsinger, S.; Hecht, A.; Luo, K.; Grunstein, M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997, 11, 83–93. [Google Scholar] [CrossRef] [PubMed]
- De Bruin, D.; Zaman, Z.; Liberatore, R.A.; Ptashne, M. Telomere looping permits gene activation by a downstream UAS in yeast. Nature 2001, 409, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Luke-Glaser, S.; Poschke, H.; Luke, B. Getting in (and out of) the loop: Regulating higher order telomere structures. Front. Oncol. 2012, 2, 180. [Google Scholar] [CrossRef] [PubMed]
- Kupiec, M. Biology of telomeres: Lessons from budding yeast. FEMS Microbiol. Rev. 2014, 38, 144–171. [Google Scholar] [CrossRef] [PubMed]
- Ceccaldi, R.; Rondinelli, B.; D’Andrea, A.D. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016, 26, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Kramara, J.; Osia, B.; Malkova, A. Break-induced replication: The where, the why, and the how. Trends Genet. 2018, 34, 518–531. [Google Scholar] [CrossRef]
- Heyer, W.-D. Regulation of recombination and genomic maintenance. Cold Spring Harb. Perspect. Biol. 2015, 7, a016501. [Google Scholar] [CrossRef]
- Seol, J.-H.; Shim, E.Y.; Lee, S.E. Microhomology-mediated end joining: Good, bad and ugly. Mutat. Res. Mol. Mech. Mutagen. 2018, 809, 81–87. [Google Scholar] [CrossRef]
- Rodgers, K.; McVey, M. Error-prone repair of DNA double-strand breaks. J. Cell. Physiol. 2016, 231, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Broccoli, D.; Smogorzewska, A.; Chong, L.; de Lange, T. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 1997, 17, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Fouché, N.; Cesare, A.J.; Willcox, S.; Ozgür, S.; Compton, S.A.; Griffith, J.D. The basic domain of TRF2 directs binding to DNA junctions irrespective of the presence of TTAGGG repeats. J. Biol. Chem. 2006, 281, 37486–37495. [Google Scholar] [CrossRef] [PubMed]
- Palm, W.; de Lange, T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008, 42, 301–334. [Google Scholar] [CrossRef] [PubMed]
- Feuerhahn, S.; Chen, L.; Luke, B.; Porro, A. No DDRama at chromosome ends: TRF2 takes centre stage. Trends Biochem. Sci. 2015, 40, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Cervantes, R.B.; Mandell, E.K.; Otero, J.H.; Lundblad, V. RPA-like proteins mediate yeast telomere function. Nat. Struct. Mol. Biol. 2007, 14, 208–214. [Google Scholar] [CrossRef]
- Miyake, Y.; Nakamura, M.; Nabetani, A.; Shimamura, S.; Tamura, M.; Yonehara, S.; Saito, M.; Ishikawa, F. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol. Cell 2009, 36, 193–206. [Google Scholar] [CrossRef]
- Surovtseva, Y.V.; Churikov, D.; Boltz, K.A.; Song, X.; Lamb, J.C.; Warrington, R.; Leehy, K.; Heacock, M.; Price, C.M.; Shippen, D.E. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol. Cell 2009, 36, 207–218. [Google Scholar] [CrossRef]
- Wellinger, R.J. The CST complex and telomere maintenance: The exception becomes the rule. Mol. Cell 2009, 36, 168–169. [Google Scholar] [CrossRef]
- Sun, J.; Yu, E.Y.; Yang, Y.; Confer, L.A.; Sun, S.H.; Wan, K.; Lue, N.F.; Lei, M. Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres. Genes Dev. 2009, 23, 2900–2914. [Google Scholar] [CrossRef]
- Bryan, C.; Rice, C.; Harkisheimer, M.; Schultz, D.C.; Skordalakes, E. Structure of the human telomeric Stn1-Ten1 capping complex. PLoS ONE 2013, 8, e66756. [Google Scholar] [CrossRef] [PubMed]
- Wan, B.; Tang, T.; Upton, H.; Shuai, J.; Zhou, Y.; Li, S.; Chen, J.; Brunzelle, J.S.; Zeng, Z.; Collins, K.; et al. The Tetrahymena telomerase p75–p45–p19 subcomplex is a unique CST complex. Nat. Struct. Mol. Biol. 2015, 22, 1023–1026. [Google Scholar] [CrossRef] [PubMed]
- Rice, C.; Skordalakes, E. Structure and function of the telomeric CST complex. Comput. Struct. Biotechnol. J. 2016, 14, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Murzin, A.G. OB(oligonucleotide/oligosaccharide binding)-fold: Common structural and functional solution for non-homologous sequences. EMBO J. 1993, 12, 861–867. [Google Scholar] [CrossRef] [PubMed]
- Arcus, V. OB-fold domains: A snapshot of the evolution of sequence, structure and function. Curr. Opin. Struct. Biol. 2002, 12, 794–801. [Google Scholar] [CrossRef]
- Shore, D. RAP1: A protean regulator in yeast. Trends Genet. 1994, 10, 408–412. [Google Scholar] [CrossRef]
- Gilson, E.; Gasser, S.M. Repressor activator protein 1 and its ligands: organising chromatin domains. In Nucleic Acids and Molecular Biology; Springer: Berlin/Heidelberg, Germany, 1995; pp. 308–327. [Google Scholar]
- Konig, P.; Giraldo, R.; Chapman, L.; Rhodes, D. The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell 1996, 85, 125–136. [Google Scholar] [CrossRef]
- Taylor, H.O.B.; O’Reilly, M.; Leslie, A.G.W.; Rhodes, D. How the multifunctional yeast Rap1p discriminates between DNA target sites: A crystallographic analysis. J. Mol. Biol. 2000, 303, 693–707. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Oestreich, S.; de Lange, T. Identification of human Rap1: Implications for telomere evolution. Cell 2000, 101, 471–483. [Google Scholar] [CrossRef]
- O’Connor, M.S.; Safari, A.; Liu, D.; Qin, J.; Songyang, Z. The human Rap1 protein complex and modulation of telomere length. J. Biol. Chem. 2004, 279, 28585–28591. [Google Scholar] [CrossRef]
- Kabir, S.; Sfeir, A.; de Lange, T. Taking apart Rap1: An adaptor protein with telomeric and non-telomeric functions. Cell Cycle 2010, 9, 4061–4067. [Google Scholar] [CrossRef] [PubMed]
- Sarthy, J.; Bae, N.S.; Scrafford, J.; Baumann, P. Human RAP1 inhibits non-homologous end joining at telomeres. EMBO J. 2009, 28, 3390–3399. [Google Scholar] [CrossRef] [PubMed]
- Sfeir, A.; Kabir, S.; van Overbeek, M.; Celli, G.B.; de Lange, T. Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science 2010, 327, 1657–1661. [Google Scholar] [CrossRef] [PubMed]
- Martinez, P.; Thanasoula, M.; Carlos, A.R.; Gómez-López, G.; Tejera, A.M.; Schoeftner, S.; Dominguez, O.; Pisano, D.G.; Tarsounas, M.; Blasco, M.A. Mammalian Rap1 controls telomere function and gene expression through binding to telomeric and extratelomeric sites. Nat. Cell Biol. 2010, 12, 768–780. [Google Scholar] [CrossRef]
- Yeung, F.; Ramírez, C.M.; Mateos-Gomez, P.A.; Pinzaru, A.; Ceccarini, G.; Kabir, S.; Fernández-Hernando, C.; Sfeir, A. Nontelomeric role for Rap1 in regulating metabolism and protecting against obesity. Cell Rep. 2013, 3, 1847–1856. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, M.T.; Gilson, E. Telomere maintenance, function and evolution: The yeast paradigm. Chromosom. Res. 2005, 13, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Brigati, C.; Kurtz, S.; Balderes, D.; Vidali, G.; Shore, D. An essential yeast gene encoding a TTAGGG repeat-binding protein. Mol. Cell. Biol. 1993, 13, 1306–1314. [Google Scholar] [CrossRef]
- Koering, C.E.; Fourel, G.; Binet-Brasselet, E.; Laroche, T.; Klein, F.; Gilson, E. Identification of high affinity Tbf1p-binding sites within the budding yeast genome. Nucleic Acids Res. 2000, 28, 2519–2526. [Google Scholar] [CrossRef]
- Liu, Z.P.; Tye, B.K. A yeast protein that binds to vertebrate telomeres and conserved yeast telomeric junctions. Genes Dev. 1991, 5, 49–59. [Google Scholar] [CrossRef]
- Fourel, G.; Revardel, E.; Koering, C.E.; Gilson, E. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J. 1999, 18, 2522–2537. [Google Scholar] [CrossRef]
- Gottschling, D.E.; Aparicio, O.M.; Billington, B.L.; Zakian, V.A. Position effect at S. cerevisiae telomeres: Reversible repression of Pol II transcription. Cell 1990, 63, 751–762. [Google Scholar] [CrossRef]
- Bilaud, T.; Koering, C.E.; Binet-Brasselet, E.; Ancelin, K.; Pollice, A.; Gasser, S.M.; Gilson, E. The telobox, a Myb-related telomeric DNA binding motif found in proteins from yeast, plants and human. Nucleic Acids Res. 1996, 24, 1294–1303. [Google Scholar] [CrossRef] [PubMed]
- Brevet, V.; Berthiau, A.-S.; Civitelli, L.; Donini, P.; Schramke, V.; Géli, V.; Ascenzioni, F.; Gilson, E. The number of vertebrate repeats can be regulated at yeast telomeres by Rap1-independent mechanisms. EMBO J. 2003, 22, 1697–1706. [Google Scholar] [CrossRef] [PubMed]
- Alexander, M.K.; Zakian, V.A. Rap1p telomere association is not required for mitotic stability of a C(3)TA(2) telomere in yeast. EMBO J. 2003, 22, 1688–1696. [Google Scholar] [CrossRef] [PubMed]
- Ribaud, V.; Ribeyre, C.; Damay, P.; Shore, D. DNA-end capping by the budding yeast transcription factor and subtelomeric binding protein Tbf1. EMBO J. 2012, 31, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Arnerić, M.; Lingner, J. Tel1 kinase and subtelomere-bound Tbf1 mediate preferential elongation of short telomeres by telomerase in yeast. EMBO Rep. 2007, 8, 1080–1085. [Google Scholar] [CrossRef] [PubMed]
- Preti, M.; Ribeyre, C.; Pascali, C.; Bosio, M.C.; Cortelazzi, B.; Rougemont, J.; Guarnera, E.; Naef, F.; Shore, D.; Dieci, G. The telomere-binding protein Tbf1 demarcates snoRNA gene promoters in Saccharomyces cerevisiae. Mol. Cell 2010, 38, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, Y.; Nishimura, Y.; Kurumizaka, H.; Shimizu, M. Nucleosome organization and chromatin dynamics in telomeres. Biomol. Concepts 2015, 6, 67–75. [Google Scholar] [CrossRef]
- Ichikawa, Y.; Morohashi, N.; Nishimura, Y.; Kurumizaka, H.; Shimizu, M. Telomeric repeats act as nucleosome-disfavouring sequences in vivo. Nucleic Acids Res. 2014, 42, 1541–1552. [Google Scholar] [CrossRef]
- Yarragudi, A.; Miyake, T.; Li, R.; Morse, R.H. Comparison of ABF1 and RAP1 in chromatin opening and transactivator potentiation in the budding yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 2004, 24, 9152–9164. [Google Scholar] [CrossRef]
- Badis, G.; Chan, E.T.; van Bakel, H.; Pena-Castillo, L.; Tillo, D.; Tsui, K.; Carlson, C.D.; Gossett, A.J.; Hasinoff, M.J.; Warren, C.L.; et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell 2008, 32, 878–887. [Google Scholar] [CrossRef] [PubMed]
- Kubik, S.; O’Duibhir, E.; de Jonge, W.J.; Mattarocci, S.; Albert, B.; Falcone, J.-L.; Bruzzone, M.J.; Holstege, F.C.P.; Shore, D. Sequence-directed action of RSC remodeler and general regulatory factors modulates +1 nucleosome position to facilitate transcription. Mol. Cell 2018, 71, 89–102.e5. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, N.; Moore, I.K.; Fondufe-Mittendorf, Y.; Gossett, A.J.; Tillo, D.; Field, Y.; LeProust, E.M.; Hughes, T.R.; Lieb, J.D.; Widom, J.; et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 2009, 458, 362–366. [Google Scholar] [CrossRef] [PubMed]
- Ganapathi, M.; Palumbo, M.J.; Ansari, S.A.; He, Q.; Tsui, K.; Nislow, C.; Morse, R.H. Extensive role of the general regulatory factors, Abf1 and Rap1, in determining genome-wide chromatin structure in budding yeast. Nucleic Acids Res. 2011, 39, 2032–2044. [Google Scholar] [CrossRef] [PubMed]
- Tsankov, A.; Yanagisawa, Y.; Rhind, N.; Regev, A.; Rando, O.J. Evolutionary divergence of intrinsic and trans-regulated nucleosome positioning sequences reveals plastic rules for chromatin organization. Genome Res. 2011, 21, 1851–1862. [Google Scholar] [CrossRef] [PubMed]
- Struhl, K.; Segal, E. Determinants of nucleosome positioning. Nat. Struct. Mol. Biol. 2013, 20, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Van Bakel, H.; Tsui, K.; Gebbia, M.; Mnaimneh, S.; Hughes, T.R.; Nislow, C. A compendium of nucleosome and transcript profiles reveals determinants of chromatin architecture and transcription. PLoS Genet. 2013, 9, e1003479. [Google Scholar] [CrossRef] [PubMed]
- Kubik, S.; Bruzzone, M.J.; Jacquet, P.; Falcone, J.-L.; Rougemont, J.; Shore, D. Nucleosome stability distinguishes two different promoter types at all protein-coding genes in yeast. Mol. Cell 2015, 60, 422–434. [Google Scholar] [CrossRef]
- Krietenstein, N.; Wal, M.; Watanabe, S.; Park, B.; Peterson, C.L.; Pugh, B.F.; Korber, P. Genomic nucleosome organization reconstituted with pure proteins. Cell 2016, 167, 709–721.e12. [Google Scholar] [CrossRef]
- Yan, C.; Chen, H.; Bai, L. Systematic study of nucleosome-displacing factors in budding yeast. Mol. Cell 2018, 71, 294–305.e4. [Google Scholar] [CrossRef]
- Bonetti, D.; Anbalagan, S.; Lucchini, G.; Clerici, M.; Longhese, M.P. Tbf1 and Vid22 promote resection and non-homologous end joining of DNA double-strand break ends. EMBO J. 2013, 32, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.; Broach, J.R. UASrpg can function as a heterochromatin boundary element in yeast. Genes Dev. 1999, 13, 1089–1101. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.; Yu, Q.; Sandmeier, J.J.; Zou, Y. Formation of boundaries of transcriptionally silent chromatin by nucleosome-excluding structures. Mol. Cell. Biol. 2004, 24, 2118–2131. [Google Scholar] [CrossRef] [PubMed]
- Donze, D.; Kamakaka, R.T. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 2001, 20, 520–531. [Google Scholar] [CrossRef] [PubMed]
- Gartenberg, M.R.; Smith, J.S. The nuts and bolts of transcriptionally silent chromatin in Saccharomyces cerevisiae. Genetics 2016, 203, 1563–1599. [Google Scholar] [CrossRef] [PubMed]
- Fourel, G.; Miyake, T.; Defossez, P.-A.; Li, R.; Gilson, E. General regulatory factors (GRFs) as genome partitioners. J. Biol. Chem. 2002, 277, 41736–41743. [Google Scholar] [CrossRef] [PubMed]
- Tommerup, H.; Dousmanis, A.; de Lange, T. Unusual chromatin in human telomeres. Mol. Cell. Biol. 1994, 14, 5777–5785. [Google Scholar] [CrossRef]
- Tardat, M.; Déjardin, J. Telomere chromatin establishment and its maintenance during mammalian development. Chromosoma 2018, 127, 3–18. [Google Scholar] [CrossRef]
- Cubiles, M.D.; Barroso, S.; Vaquero-Sedas, M.I.; Enguix, A.; Aguilera, A.; Vega-Palas, M.A. Epigenetic features of human telomeres. Nucleic Acids Res. 2018, 46, 2347–2355. [Google Scholar] [CrossRef]
- Ernst, J.; Kheradpour, P.; Mikkelsen, T.S.; Shoresh, N.; Ward, L.D.; Epstein, C.B.; Zhang, X.; Wang, L.; Issner, R.; Coyne, M.; et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 2011, 473, 43–49. [Google Scholar] [CrossRef]
- Rosenfeld, J.A.; Wang, Z.; Schones, D.E.; Zhao, K.; DeSalle, R.; Zhang, M.Q. Determination of enriched histone modifications in non-genic portions of the human genome. BMC Genom. 2009, 10, 143. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, R.J.; Kubicek, S.; Schreiber, S.L.; Karlseder, J. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat. Struct. Mol. Biol. 2010, 17, 1218–1225. [Google Scholar] [CrossRef] [PubMed]
- Vaquero-Sedas, M.I.; Gámez-Arjona, F.M.; Vega-Palas, M.A. Arabidopsis thaliana telomeres exhibit euchromatic features. Nucleic Acids Res. 2011, 39, 2007–2017. [Google Scholar] [CrossRef]
- Ivessa, A.S.; Zhou, J.-Q.; Schulz, V.P.; Monson, E.K.; Zakian, V.A. Saccharomyces Rrm3p, a 5’ to 3’ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 2002, 16, 1383–1396. [Google Scholar] [CrossRef]
- Makovets, S.; Herskowitz, I.; Blackburn, E.H. Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions. Mol. Cell. Biol. 2004, 24, 4019–4031. [Google Scholar] [CrossRef]
- Miller, K.M.; Rog, O.; Cooper, J.P. Semi-conservative DNA replication through telomeres requires Taz1. Nature 2006, 440, 824–828. [Google Scholar] [CrossRef] [PubMed]
- Sfeir, A.; Kosiyatrakul, S.T.; Hockemeyer, D.; MacRae, S.L.; Karlseder, J.; Schildkraut, C.L.; de Lange, T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 2009, 138, 90–103. [Google Scholar] [CrossRef]
- Lopes, J.; Piazza, A.; Bermejo, R.; Kriegsman, B.; Colosio, A.; Teulade-Fichou, M.-P.; Foiani, M.; Nicolas, A. G-quadruplex-induced instability during leading-strand replication. EMBO J. 2011, 30, 4033–4046. [Google Scholar] [CrossRef] [PubMed]
- Bah, A.; Gilson, E.; Wellinger, R.J. Telomerase is required to protect chromosomes with vertebrate-type T2AG3 3’ ends in Saccharomyces cerevisiae. J. Biol. Chem. 2011, 286, 27132–27138. [Google Scholar] [CrossRef] [PubMed]
- Anand, R.P.; Shah, K.A.; Niu, H.; Sung, P.; Mirkin, S.M.; Freudenreich, C.H. Overcoming natural replication barriers: Differential helicase requirements. Nucleic Acids Res. 2012, 40, 1091–1105. [Google Scholar] [CrossRef] [PubMed]
- Lormand, J.D.; Buncher, N.; Murphy, C.T.; Kaur, P.; Lee, M.Y.; Burgers, P.; Wang, H.; Kunkel, T.A.; Opresko, P.L. DNA polymerase δ stalls on telomeric lagging strand templates independently from G-quadruplex formation. Nucleic Acids Res. 2013, 41, 10323–10333. [Google Scholar] [CrossRef] [PubMed]
- Geronimo, C.L.; Zakian, V.A. Getting it done at the ends: Pif1 family DNA helicases and telomeres. DNA Repair 2016, 44, 151–158. [Google Scholar] [CrossRef]
- Croteau, D.L.; Popuri, V.; Opresko, P.L.; Bohr, V.A. Human RecQ helicases in DNA repair, recombination, and replication. Annu. Rev. Biochem. 2014, 83, 519–552. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, O.; Bourdoncle, A.; Boulé, J.-B.; Brosh, R.M.; Mergny, J.-L. G-quadruplexes and helicases. Nucleic Acids Res. 2016, 44, 1989–2006. [Google Scholar] [CrossRef] [PubMed]
- Whitby, M.C. The FANCM family of DNA helicases/translocases. DNA Repair 2010, 9, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Vannier, J.-B.; Sarek, G.; Boulton, S.J. RTEL1: Functions of a disease-associated helicase. Trends Cell Biol. 2014, 24, 416–425. [Google Scholar] [CrossRef] [PubMed]
- Poole, L.A.; Cortez, D. SMARCAL1 and telomeres: Replicating the troublesome ends. Nucleus 2016, 7, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Niu, H.; Klein, H.L. Multifunctional roles of Saccharomyces cerevisiae Srs2 protein in replication, recombination and repair. FEMS Yeast Res. 2016, 17, fow111. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, A.; Shore, D. Early replication of short telomeres in budding yeast. Cell 2007, 128, 1051–1062. [Google Scholar] [CrossRef] [PubMed]
- Davé, A.; Cooley, C.; Garg, M.; Bianchi, A. Protein phosphatase 1 recruitment by Rif1 regulates DNA replication origin firing by counteracting DDK activity. Cell Rep. 2014, 7, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Hiraga, S.-I.; Alvino, G.M.; Chang, F.; Lian, H.-Y.; Sridhar, A.; Kubota, T.; Brewer, B.J.; Weinreich, M.; Raghuraman, M.K.; Donaldson, A.D. Rif1 controls DNA replication by directing Protein Phosphatase 1 to reverse Cdc7-mediated phosphorylation of the MCM complex. Genes Dev. 2014, 28, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Mattarocci, S.; Shyian, M.; Lemmens, L.; Damay, P.; Altintas, D.M.; Shi, T.; Bartholomew, C.R.; Thomä, N.H.; Hardy, C.F.J.; Shore, D. Rif1 controls DNA replication timing in yeast through the PP1 phosphatase Glc7. Cell Rep. 2014, 7, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Hafner, L.; Lezaja, A.; Zhang, X.; Lemmens, L.; Shyian, M.; Albert, B.; Follonier, C.; Nunes, J.M.; Lopes, M.; Shore, D.; et al. Rif1 binding and control of chromosome-internal DNA replication origins is limited by telomere sequestration. Cell Rep. 2018, 23, 983–992. [Google Scholar] [CrossRef] [PubMed]
- Hafner, L.; Shore, D.; Mattarocci, S. ChECing out Rif1 action in freely cycling cells. Curr. Genet. 2018, 1–6. [Google Scholar] [CrossRef]
- Hiraga, S.; Monerawela, C.; Katou, Y.; Shaw, S.; Clark, K.R.; Shirahige, K.; Donaldson, A.D. Budding yeast Rif1 binds to replication origins and protects DNA at blocked replication forks. EMBO Rep. 2018, 19, e46222. [Google Scholar] [CrossRef]
- Arnoult, N.; Schluth-Bolard, C.; Letessier, A.; Drascovic, I.; Bouarich-Bourimi, R.; Campisi, J.; Kim, S.; Boussouar, A.; Ottaviani, A.; Magdinier, F.; et al. Replication timing of human telomeres is chromosome arm-specific, influenced by subtelomeric structures and connected to nuclear localization. PLoS Genet. 2010, 6, e1000920. [Google Scholar] [CrossRef]
- Azzalin, C.M.; Reichenbach, P.; Khoriauli, L.; Giulotto, E.; Lingner, J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 2007, 318, 798–801. [Google Scholar] [CrossRef]
- Schoeftner, S.; Blasco, M.A. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 2008, 10, 228–236. [Google Scholar] [CrossRef]
- Xu, Y.; Suzuki, Y.; Ito, K.; Komiyama, M. Telomeric repeat-containing RNA structure in living cells. Proc. Natl. Acad. Sci. USA 2010, 107, 14579–14584. [Google Scholar] [CrossRef]
- Xu, Y.; Kaminaga, K.; Komiyama, M. G-quadruplex formation by human telomeric repeats-containing RNA in Na + solution. J. Am. Chem. Soc. 2008, 130, 11179–11184. [Google Scholar] [CrossRef]
- Martadinata, H.; Phan, A.T. Structure of propeller-type parallel-stranded RNA G-quadruplexes, formed by human telomeric RNA sequences in K + solution. J. Am. Chem. Soc. 2009, 131, 2570–2578. [Google Scholar] [CrossRef] [PubMed]
- Collie, G.W.; Haider, S.M.; Neidle, S.; Parkinson, G.N. A crystallographic and modelling study of a human telomeric RNA (TERRA) quadruplex. Nucleic Acids Res. 2010, 38, 5569–5580. [Google Scholar] [CrossRef]
- Balk, B.; Maicher, A.; Dees, M.; Klermund, J.; Luke-Glaser, S.; Bender, K.; Luke, B. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat. Struct. Mol. Biol. 2013, 20, 1199–1205. [Google Scholar] [CrossRef]
- Santos-Pereira, J.M.; Aguilera, A. R loops: New modulators of genome dynamics and function. Nat. Rev. Genet. 2015, 16, 583–597. [Google Scholar] [CrossRef]
- Gan, W.; Guan, Z.; Liu, J.; Gui, T.; Shen, K.; Manley, J.L.; Li, X. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 2011, 25, 2041–2056. [Google Scholar] [CrossRef]
- Maicher, A.; Lockhart, A.; Luke, B. Breaking new ground: Digging into TERRA function. Biochim. Biophys. Acta 2014, 1839, 387–394. [Google Scholar] [CrossRef]
- Arora, R.; Azzalin, C.M. Telomere elongation chooses TERRA ALTernatives. RNA Biol. 2015, 12, 938–941. [Google Scholar] [CrossRef] [PubMed]
- Arora, R.; Lee, Y.; Wischnewski, H.; Brun, C.M.; Schwarz, T.; Azzalin, C.M. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 2014, 5, 5220. [Google Scholar] [CrossRef]
- Dilley, R.L.; Greenberg, R.A. ALTernative telomere maintenance and cancer. Trends Cancer 2015, 1, 145–156. [Google Scholar] [CrossRef]
- Cusanelli, E.; Romero, C.A.P.; Chartrand, P. Telomeric noncoding RNA TERRA is induced by telomere shortening to nucleate telomerase molecules at short telomeres. Mol. Cell 2013, 51, 780–791. [Google Scholar] [CrossRef] [PubMed]
- Moravec, M.; Wischnewski, H.; Bah, A.; Hu, Y.; Liu, N.; Lafranchi, L.; King, M.C.; Azzalin, C.M. TERRA promotes telomerase-mediated telomere elongation in Schizosaccharomyces pombe. EMBO Rep. 2016, 17, 999–1012. [Google Scholar] [CrossRef]
- Graf, M.; Bonetti, D.; Lockhart, A.; Serhal, K.; Kellner, V.; Maicher, A.; Jolivet, P.; Teixeira, M.T.; Luke, B. Telomere length determines TERRA and R-Loop regulation through the cell cycle. Cell 2017, 170, 72–85.e14. [Google Scholar] [CrossRef]
- Deng, Z.; Norseen, J.; Wiedmer, A.; Riethman, H.; Lieberman, P.M. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell 2009, 35, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Bah, A.; Azzalin, C.M. The telomeric transcriptome: From fission yeast to mammals. Int. J. Biochem. Cell Biol. 2012, 44, 1055–1059. [Google Scholar] [CrossRef] [PubMed]
- Rippe, K.; Luke, B. TERRA and the state of the telomere. Nat. Struct. Mol. Biol. 2015, 22, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Luke, B.; Panza, A.; Redon, S.; Iglesias, N.; Li, Z.; Lingner, J. The Rat1p 5’ to 3’ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol. Cell 2008, 32, 465–477. [Google Scholar] [CrossRef]
- Redon, S.; Reichenbach, P.; Lingner, J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010, 38, 5797–5806. [Google Scholar] [CrossRef] [PubMed]
- Farnung, B.O.; Brun, C.M.; Arora, R.; Lorenzi, L.E.; Azzalin, C.M. Telomerase efficiently elongates highly transcribing telomeres in human cancer cells. PLoS ONE 2012, 7, e35714. [Google Scholar] [CrossRef]
- Cusanelli, E.; Chartrand, P. Telomeric repeat-containing RNA TERRA: A noncoding RNA connecting telomere biology to genome integrity. Front. Genet. 2015, 6, 143. [Google Scholar] [CrossRef]
- Vrbsky, J.; Akimcheva, S.; Watson, J.M.; Turner, T.L.; Daxinger, L.; Vyskot, B.; Aufsatz, W.; Riha, K. siRNA–mediated methylation of Arabidopsis telomeres. PLoS Genet. 2010, 6, e1000986. [Google Scholar] [CrossRef] [PubMed]
- Bah, A.; Wischnewski, H.; Shchepachev, V.; Azzalin, C.M. The telomeric transcriptome of Schizosaccharomyces pombe. Nucleic Acids Res. 2012, 40, 2995–3005. [Google Scholar] [CrossRef] [PubMed]
- Majerová, E.; Mandáková, T.; Vu, G.T.H.; Fajkus, J.; Lysak, M.A.; Fojtová, M. Chromatin features of plant telomeric sequences at terminal vs. internal positions. Front. Plant Sci. 2014, 5, 593. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Liang, P.; Liu, D.; Huang, J.; Songyang, Z. Telomere regulation in pluripotent stem cells. Protein Cell 2014, 5, 194–202. [Google Scholar] [CrossRef]
- Liu, L. Linking telomere regulation to stem cell pluripotency. Trends Genet. 2017, 33, 16–33. [Google Scholar] [CrossRef]
- Bodnar, A.G.; Ouellette, M.; Frolkis, M.; Holt, S.E.; Chiu, C.P.; Morin, G.B.; Harley, C.B.; Shay, J.W.; Lichtsteiner, S.; Wright, W.E. Extension of life-span by introduction of telomerase into normal human cells. Science 1998, 279, 349–352. [Google Scholar] [CrossRef]
- Grossi, S.; Puglisi, A.; Dmitriev, P.V.; Lopes, M.; Shore, D. Pol12, the B subunit of DNA polymerase alpha, functions in both telomere capping and length regulation. Genes Dev. 2004, 18, 992–1006. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Zakian, V.A. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 2000, 14, 1777–1788. [Google Scholar]
- Diede, S.J.; Gottschling, D.E. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell 1999, 99, 723–733. [Google Scholar] [CrossRef]
- Carson, M.J.; Hartwell, L. CDC17: An essential gene that prevents telomere elongation in yeast. Cell 1985, 42, 249–257. [Google Scholar] [CrossRef]
- Adams, A.K.; Holm, C. Specific DNA replication mutations affect telomere length in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996, 16, 4614–4620. [Google Scholar] [CrossRef]
- Adams Martin, A.; Dionne, I.; Wellinger, R.J.; Holm, C. The function of DNA polymerase alpha at telomeric G tails is important for telomere homeostasis. Mol. Cell. Biol. 2000, 20, 786–796. [Google Scholar] [CrossRef] [PubMed]
- Parenteau, J.; Wellinger, R.J. Differential processing of leading- and lagging-strand ends at Saccharomyces cerevisiae telomeres revealed by the absence of Rad27p nuclease. Genetics 2002, 162, 1583–1594. [Google Scholar] [PubMed]
- Casteel, D.E.; Zhuang, S.; Zeng, Y.; Perrino, F.W.; Boss, G.R.; Goulian, M.; Pilz, R.B. A DNA polymerase alpha- primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J. Biol. Chem. 2009, 284, 5807–5818. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Stewart, J.A.; Kasbek, C.; Zhao, Y.; Wright, W.E.; Price, C.M. Human CST has independent functions during telomere duplex replication and C-strand fill-in. Cell Rep. 2012, 2, 1096–1103. [Google Scholar] [CrossRef]
- Chen, L.-Y.; Redon, S.; Lingner, J. The human CST complex is a terminator of telomerase activity. Nature 2012, 488, 540–544. [Google Scholar] [CrossRef] [PubMed]
- Price, C.M.; Boltz, K.A.; Chaiken, M.F.; Stewart, J.A.; Beilstein, M.A.; Shippen, D.E. Evolution of CST function in telomere maintenance. Cell Cycle 2010, 9, 3157–3165. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.A.; Wang, F.; Chaiken, M.F.; Kasbek, C.; Chastain, P.D.; Wright, W.E.; Price, C.M. Human CST promotes telomere duplex replication and general replication restart after fork stalling. EMBO J. 2012, 31, 3537–3549. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Takai, H.; de Lange, T. Telomeric 3′ overhangs derive from resection by Exo1 and Apollo and fill-in by POT1b-associated CST. Cell 2012, 150, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-Y.; Lingner, J. CST for the grand finale of telomere replication. Nucleus 2013, 4, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Goulian, M.; Heard, C.J. The mechanism of action of an accessory protein for DNA polymerase alpha/primase. J. Biol. Chem. 1990, 265, 13231–13239. [Google Scholar]
- Goulian, M.; Heard, C.J.; Grimm, S.L. Purification and properties of an accessory protein for DNA polymerase alpha/primase. J. Biol. Chem. 1990, 265, 13221–13230. [Google Scholar] [PubMed]
- Chandra, A.; Hughes, T.R.; Nugent, C.I.; Lundblad, V. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 2001, 15, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Pennock, E.; Buckley, K.; Lundblad, V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 2001, 104, 387–396. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Tan, C.R.; Li, S. Sequential phosphorylation of CST subunits by different cyclin-Cdk1 complexes orchestrate telomere replication. Cell Cycle 2017, 16, 1271–1287. [Google Scholar] [CrossRef] [PubMed]
- Tseng, S.-F.; Lin, J.-J.; Teng, S.-C. The telomerase-recruitment domain of the telomere binding protein Cdc13 is regulated by Mec1p/Tel1p-dependent phosphorylation. Nucleic Acids Res. 2006, 34, 6327–6336. [Google Scholar] [CrossRef]
- Shen, Z.-J.; Hsu, P.-H.; Su, Y.-T.; Yang, C.-W.; Kao, L.; Tseng, S.-F.; Tsai, M.-D.; Teng, S.-C. PP2A and Aurora differentially modify Cdc13 to promote telomerase release from telomeres at G2/M phase. Nat. Commun. 2014, 5, 5312. [Google Scholar] [CrossRef]
- Hang, L.E.; Liu, X.; Cheung, I.; Yang, Y.; Zhao, X. SUMOylation regulates telomere length homeostasis by targeting Cdc13. Nat. Struct. Mol. Biol. 2011, 18, 920–926. [Google Scholar] [CrossRef]
- Greider, C.W. Regulating telomere length from the inside out: The replication fork model. Genes Dev. 2016, 30, 1483–1491. [Google Scholar] [CrossRef]
- Marcand, S.; Gilson, E.; Shore, D. A protein-counting mechanism for telomere length regulation in yeast. Science 1997, 275, 986–990. [Google Scholar] [CrossRef]
- Lundblad, V. Telomere maintenance without telomerase. Oncogene 2002, 21, 522–531. [Google Scholar] [CrossRef]
- Cesare, A.J.; Reddel, R.R. Alternative lengthening of telomeres: Models, mechanisms and implications. Nat. Rev. Genet. 2010, 11, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Bryan, T.M.; Englezou, A.; Gupta, J.; Bacchetti, S.; Reddel, R.R. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995, 14, 4240–4248. [Google Scholar] [CrossRef] [PubMed]
- Bryan, T.M.; Reddel, R.R. Telomere dynamics and telomerase activity in in vitro immortalised human cells. Eur. J. Cancer 1997, 33, 767–773. [Google Scholar] [CrossRef]
- Cerone, M.A.; Londono-Vallejo, J.A.; Bacchetti, S. Telomere maintenance by telomerase and by recombination can coexist in human cells. Hum. Mol. Genet. 2001, 10, 1945–1952. [Google Scholar] [CrossRef]
- Samassekou, O.; Malina, A.; Hébert, J.; Yan, J. Presence of alternative lengthening of telomeres associated circular extrachromosome telomere repeats in primary leukemia cells of chronic myeloid leukemia. J. Hematol. Oncol. 2013, 6, 26. [Google Scholar] [CrossRef]
- Slatter, T.L.; Tan, X.; Yuen, Y.C.; Gunningham, S.; Ma, S.S.; Daly, E.; Packer, S.; Devenish, C.; Royds, J.A.; Hung, N.A. The alternative lengthening of telomeres pathway may operate in non-neoplastic human cells. J. Pathol. 2012, 226, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Neumann, A.A.; Watson, C.M.; Noble, J.R.; Pickett, H.A.; Tam, P.P.L.; Reddel, R.R. Alternative lengthening of telomeres in normal mammalian somatic cells. Genes Dev. 2013, 27, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Bailey, S.M.; Okuka, M.; Muñoz, P.; Li, C.; Zhou, L.; Wu, C.; Czerwiec, E.; Sandler, L.; Seyfang, A.; et al. Telomere lengthening early in development. Nat. Cell Biol. 2007, 9, 1436–1441. [Google Scholar] [CrossRef] [PubMed]
- Růčková, E.; Friml, J.; Procházková Schrumpfová, P.; Fajkus, J. Role of alternative telomere lengthening unmasked in telomerase knock-out mutant plants. Plant Mol. Biol. 2008, 66, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.-L.; Bradley, J.D.; Attardi, L.D.; Blackburn, E.H. In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature 1990, 344, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Singer, M.S.; Gottschling, D.E. TLC1: Template RNA component of Saccharomyces cerevisiae telomerase. Science 1994, 266, 404–409. [Google Scholar] [CrossRef] [PubMed]
- McEachern, M.J.; Blackburn, E.H. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev. 1996, 10, 1822–1834. [Google Scholar] [CrossRef] [PubMed]
- Teng, S.-C.; Zakian, V.A. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999, 19, 8083–8093. [Google Scholar] [CrossRef]
- Le, S.; Moore, J.K.; Haber, J.E.; Greider, C.W. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 1999, 152, 143–152. [Google Scholar] [PubMed]
- Chen, Q.; Ijpma, A.; Greider, C.W. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 2001, 21, 1819–1827. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Pryde, F.E.; Lester, D.; Maddison, R.L.; Borts, R.H.; Hickson, I.D.; Louis, E.J. SGS1 is required for telomere elongation in the absence of telomerase. Curr. Biol. 2001, 11, 125–129. [Google Scholar] [CrossRef]
- Cohen, H.; Sinclair, D.A. Recombination-mediated lengthening of terminal telomeric repeats requires the Sgs1 DNA helicase. Proc. Natl. Acad. Sci. USA 2001, 98, 3174–3179. [Google Scholar] [CrossRef]
- Lydeard, J.R.; Jain, S.; Yamaguchi, M.; Haber, J.E. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 2007, 448, 820–823. [Google Scholar] [CrossRef]
- Teng, S.C.; Chang, J.; McCowan, B.; Zakian, V.A. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell 2000, 6, 947–952. [Google Scholar] [CrossRef]
- Johnson, F.B.; Marciniak, R.A.; McVey, M.; Stewart, S.A.; Hahn, W.C.; Guarente, L. The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J. 2001, 20, 905–913. [Google Scholar] [CrossRef]
- Tsai, Y.-L.; Tseng, S.-F.; Chang, S.-H.; Lin, C.-C.; Teng, S.-C. Involvement of replicative polymerases, Tel1p, Mec1p, Cdc13p, and the Ku complex in telomere-telomere recombination. Mol. Cell. Biol. 2002, 22, 5679–5687. [Google Scholar] [CrossRef]
- Maringele, L.; Lydall, D. Telomerase- and recombination-independent immortalization of budding yeast. Genes Dev. 2004, 18, 2663–2675. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Mogen, J.L.; Chavez, A.; Johnson, F.B. Sgs1 RecQ helicase inhibits survival of Saccharomyces cerevisiae cells lacking telomerase and homologous recombination. J. Biol. Chem. 2008, 283, 29847–29858. [Google Scholar] [CrossRef] [PubMed]
- Grandin, N.; Damon, C.; Charbonneau, M. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 2001, 20, 1173–1183. [Google Scholar] [CrossRef] [PubMed]
- Murnane, J.P.; Sabatier, L.; Marder, B.A.; Morgan, W.F. Telomere dynamics in an immortal human cell line. EMBO J. 1994, 13, 4953–4962. [Google Scholar] [CrossRef] [PubMed]
- Londoño-Vallejo, J.A.; Der-Sarkissian, H.; Cazes, L.; Bacchetti, S.; Reddel, R.R. Alternative lengthening of telomeres is characterized by high rates of telomeric exchange. Cancer Res. 2004, 64, 2324–2327. [Google Scholar] [CrossRef] [PubMed]
- Cesare, A.J.; Griffith, J.D. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol. Cell. Biol. 2004, 24, 9948–9957. [Google Scholar] [CrossRef] [PubMed]
- Nabetani, A.; Ishikawa, F. Unusual telomeric DNAs in human telomerase-negative immortalized cells. Mol. Cell. Biol. 2009, 29, 703–713. [Google Scholar] [CrossRef]
- Yeager, T.R.; Neumann, A.A.; Englezou, A.; Huschtscha, L.I.; Noble, J.R.; Reddel, R.R. Telomerase-negative immortalized human cells contain a novel type of Promyelocytic Leukemia (PML) body. Cancer Res. 1999, 59, 4175–4179. [Google Scholar]
- Dilley, R.L.; Verma, P.; Cho, N.W.; Winters, H.D.; Wondisford, A.R.; Greenberg, R.A. Break-induced telomere synthesis underlies alternative telomere maintenance. Nature 2016, 539, 54–58. [Google Scholar] [CrossRef]
- Roumelioti, F.F.-M.; Sotiriou, S.K.; Katsini, V.; Chiourea, M.; Halazonetis, T.D.; Gagos, S. Alternative lengthening of human telomeres is a conservative DNA replication process with features of break-induced replication. EMBO Rep. 2016, 17, 1731–1737. [Google Scholar] [CrossRef]
- Min, J.; Wright, W.E.; Shay, J.W. Alternative lengthening of telomeres mediated by mitotic DNA synthesis engages break-induced replication processes. Mol. Cell. Biol. 2017. [Google Scholar] [CrossRef]
- Sobinoff, A.P.; Allen, J.A.; Neumann, A.A.; Yang, S.F.; Walsh, M.E.; Henson, J.D.; Reddel, R.R.; Pickett, H.A. BLM and SLX4 play opposing roles in recombination-dependent replication at human telomeres. EMBO J. 2017, 36, 2907–2919. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Exposito, L.; Bournique, E.; Bergoglio, V.; Bose, A.; Barroso-Gonzalez, J.; Zhang, S.; Roncaioli, J.L.; Lee, M.; Wallace, C.T.; Watkins, S.C.; et al. Proteomic profiling reveals a specific role for translesion DNA polymerase η in the alternative lengthening of telomeres. Cell Rep. 2016, 17, 1858–1871. [Google Scholar] [CrossRef] [PubMed]
- Sobinoff, A.P.; Pickett, H.A. Alternative lengthening of telomeres: DNA repair pathways converge. Trends Genet. 2017, 33, 921–932. [Google Scholar] [CrossRef] [PubMed]
- Heaphy, C.M.; de Wilde, R.F.; Jiao, Y.; Klein, A.P.; Edil, B.H.; Shi, C.; Bettegowda, C.; Rodriguez, F.J.; Eberhart, C.G.; Hebbar, S.; et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science 2011, 333, 425. [Google Scholar] [CrossRef]
- Lovejoy, C.A.; Li, W.; Reisenweber, S.; Thongthip, S.; Bruno, J.; de Lange, T.; De, S.; Petrini, J.H.J.; Sung, P.A.; Jasin, M.; et al. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 2012, 8, e1002772. [Google Scholar] [CrossRef]
- Schwartzentruber, J.; Korshunov, A.; Liu, X.-Y.; Jones, D.T.W.; Pfaff, E.; Jacob, K.; Sturm, D.; Fontebasso, A.M.; Quang, D.-A.K.; Tönjes, M.; et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 2012, 482, 226–231. [Google Scholar] [CrossRef]
- Haase, S.; Garcia-Fabiani, M.B.; Carney, S.; Altshuler, D.; Núñez, F.J.; Méndez, F.M.; Núñez, F.; Lowenstein, P.R.; Castro, M.G. Mutant ATRX: Uncovering a new therapeutic target for glioma. Expert Opin. Ther. Targets 2018, 22, 599–613. [Google Scholar] [CrossRef]
- O’Sullivan, R.J.; Arnoult, N.; Lackner, D.H.; Oganesian, L.; Haggblom, C.; Corpet, A.; Almouzni, G.; Karlseder, J. Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nat. Struct. Mol. Biol. 2014, 21, 167–174. [Google Scholar] [CrossRef]
- Episkopou, H.; Draskovic, I.; Van Beneden, A.; Tilman, G.; Mattiussi, M.; Gobin, M.; Arnoult, N.; Londoño-Vallejo, A.; Decottignies, A. Alternative lengthening of telomeres is characterized by reduced compaction of telomeric chromatin. Nucleic Acids Res. 2014, 42, 4391–4405. [Google Scholar] [CrossRef] [PubMed]
- Déjardin, J.; Kingston, R.E. Purification of proteins associated with specific genomic loci. Cell 2009, 136, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Conomos, D.; Stutz, M.D.; Hills, M.; Neumann, A.A.; Bryan, T.M.; Reddel, R.R.; Pickett, H.A. Variant repeats are interspersed throughout the telomeres and recruit nuclear receptors in ALT cells. J. Cell Biol. 2012, 199, 893–906. [Google Scholar] [CrossRef] [PubMed]
- Marzec, P.; Armenise, C.; Pérot, G.; Roumelioti, F.-M.M.; Basyuk, E.; Gagos, S.; Chibon, F.; Déjardin, J. Nuclear-receptor-mediated telomere insertion leads to genome instability in ALT cancers. Cell 2015, 160, 913–927. [Google Scholar] [CrossRef] [PubMed]
- Conomos, D.; Reddel, R.R.; Pickett, H.A. NuRD-ZNF827 recruitment to telomeres creates a molecular scaffold for homologous recombination. Nat. Struct. Mol. Biol. 2014, 21, 760–770. [Google Scholar] [CrossRef]
- Iglesias, N.; Redon, S.; Pfeiffer, V.; Dees, M.; Lingner, J.; Luke, B. Subtelomeric repetitive elements determine TERRA regulation by Rap1/Rif and Rap1/Sir complexes in yeast. EMBO Rep. 2011, 12, 587–593. [Google Scholar] [CrossRef]
- Yu, T.-Y.; Kao, Y.; Lin, J.-J. Telomeric transcripts stimulate telomere recombination to suppress senescence in cells lacking telomerase. Proc. Natl. Acad. Sci. USA 2014, 111, 3377–3382. [Google Scholar] [CrossRef]
- Wells, R.A.; Germino, G.G.; Krishna, S.; Buckle, V.J.; Reeders, S.T. Telomere-related sequences at interstitial sites in the human genome. Genomics 1990, 8, 699–704. [Google Scholar] [CrossRef]
- Ruiz-Herrera, A.; García, F.; Azzalin, C.; Giulotto, E.; Egozcue, J.; Ponsà, M.; Garcia, M. Distribution of intrachromosomal telomeric sequences (ITS) on Macaca fascicularis (Primates) chromosomes and their implication for chromosome evolution. Hum. Genet. 2002, 110, 578–586. [Google Scholar] [CrossRef]
- Ruiz-Herrera, A.; García, F.; Giulotto, E.; Attolini, C.; Egozcue, J.; Ponsà, M.; Garcia, M. Evolutionary breakpoints are co-localized with fragile sites and intrachromosomal telomeric sequences in primates. Cytogenet. Genome Res. 2005, 108, 234–247. [Google Scholar] [CrossRef]
- Ruiz-Herrera, A.; Nergadze, S.G.; Santagostino, M.; Giulotto, E. Telomeric repeats far from the ends: Mechanisms of origin and role in evolution. Cytogenet. Genome Res. 2008, 122, 219–228. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Liu, J.; Torres, G.A.; Zhang, H.; Jiang, J.; Xie, C. Interstitial telomeric repeats are enriched in the centromeres of chromosomes in Solanum species. Chromosom. Res. 2013, 21, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Bolzán, A.D. Interstitial telomeric sequences in vertebrate chromosomes: Origin, function, instability and evolution. Mutat. Res. Mutat. Res. 2017, 773, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Weber, B.; Collins, C.; Robbins, C.; Magenis, R.E.; Delaney, A.D.; Gray, J.W.; Hayden, M.R. Characterization and organization of DNA sequences adjacent to the human telomere associated repeat (TTAGGG)n. Nucleic Acids Res. 1990, 18, 3353–3361. [Google Scholar] [CrossRef] [PubMed]
- Meyne, J.; Baker, R.J.; Hobart, H.H.; Hsu, T.C.; Ryder, O.A.; Ward, O.G.; Wiley, J.E.; Wurster-Hill, D.H.; Yates, T.L.; Moyzis, R.K. Distribution of non-telomeric sites of the (TTAGGG)n telomeric sequence in vertebrate chromosomes. Chromosoma 1990, 99, 3–10. [Google Scholar] [CrossRef]
- Weber, B.; Allen, L.; Magenis, R.E.; Goodfellow, P.J.; Smith, L.; Hayden, M.R. Intrachromosomal location of the telomeric repeat (TTAGGG)n. Mamm. Genome 1991, 1, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Cox, A.V.; Bennett, S.T.; Parokonny, A.S.; Kenton, A.; Callimassia, M.A.; Bennett, M.D. Comparison of plant telomere locations using a PCR-generated synthetic probe. Ann. Bot. 1993, 72, 239–247. [Google Scholar] [CrossRef]
- Azzalin, C.M.; Mucciolo, E.; Bertoni, L.; Giulotto, E. Fluorescence in situ hybridization with a synthetic (T2AG3)n polynucleotide detects several intrachromosomal telomere-like repeats on human chromosomes. Cytogenet. Cell Genet. 1997, 78, 112–115. [Google Scholar] [CrossRef]
- Mondello, C.; Pirzio, L.; Azzalin, C.M.; Giulotto, E. Instability of interstitial telomeric sequences in the human genome. Genomics 2000, 68, 111–117. [Google Scholar] [CrossRef]
- Azzalin, C.M.; Nergadze, S.G.; Giulotto, E. Human intrachromosomal telomeric-like repeats: Sequence organization and mechanisms of origin. Chromosoma 2001, 110, 75–82. [Google Scholar] [CrossRef]
- Uchida, W.; Matsunaga, S.; Sugiyama, R.; Kawano, S. Interstitial telomere-like repeats in the Arabidopsis thaliana genome. Genes Genet. Syst. 2002, 77, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Flint, J.; Bates, G.P.; Clark, K.; Dorman, A.; Willingham, D.; Roe, B.A.; Micklem, G.; Higgs, D.R.; Louis, E.J. Sequence comparison of human and yeast telomeres identifies structurally distinct subtelomeric domains. Hum. Mol. Genet. 1997, 6, 1305–1313. [Google Scholar] [CrossRef] [PubMed]
- Ambrosini, A.; Paul, S.; Hu, S.; Riethman, H. Human subtelomeric duplicon structure and organization. Genome Biol. 2007, 8, R151. [Google Scholar] [CrossRef] [PubMed]
- IJdo, J.W.; Baldini, A.; Ward, D.C.; Reeders, S.T.; Wells, R.A. Origin of human chromosome 2: An ancestral telomere-telomere fusion. Proc. Natl. Acad. Sci. USA 1991, 88, 9051–9055. [Google Scholar] [CrossRef]
- Fan, Y.; Linardopoulou, E.; Friedman, C.; Williams, E.; Trask, B.J. Genomic structure and evolution of the ancestral chromosome fusion site in 2q13-2q14.1 and paralogous regions on other human chromosomes. Genome Res. 2002, 12, 1651–1662. [Google Scholar] [CrossRef]
- Slijepcevic, P. Telomeres and mechanisms of Robertsonian fusion. Chromosoma 1998, 107, 136–140. [Google Scholar] [CrossRef]
- Sánchez-Guillén, R.A.; Capilla, L.; Reig-Viader, R.; Martínez-Plana, M.; Pardo-Camacho, C.; Andrés-Nieto, M.; Ventura, J.; Ruiz-Herrera, A. On the origin of Robertsonian fusions in nature: Evidence of telomere shortening in wild house mice. J. Evol. Biol. 2015, 28, 241–249. [Google Scholar] [CrossRef]
- Rovatsos, M.T.; Marchal, J.A.; Romero-Fernández, I.; Fernández, F.J.; Giagia-Athanosopoulou, E.B.; Sánchez, A. Rapid, independent, and extensive amplification of telomeric repeats in pericentromeric regions in karyotypes of arvicoline rodents. Chromosom. Res. 2011, 19, 869–882. [Google Scholar] [CrossRef]
- Rovatsos, M.; Kratochvíl, L.; Altmanová, M.; Johnson Pokorná, M. Interstitial telomeric motifs in squamate reptiles: When the exceptions outnumber the rule. PLoS ONE 2015, 10, e0134985. [Google Scholar] [CrossRef]
- Nergadze, S.G.; Santagostino, M.A.; Salzano, A.; Mondello, C.; Giulotto, E. Contribution of telomerase RNA retrotranscription to DNA double-strand break repair during mammalian genome evolution. Genome Biol. 2007, 8, R260. [Google Scholar] [CrossRef]
- Lin, K.W.; Yan, J. Endings in the middle: Current knowledge of interstitial telomeric sequences. Mutat. Res. 2008, 658, 95–110. [Google Scholar] [CrossRef] [PubMed]
- Nergadze, S.G.; Rocchi, M.; Azzalin, C.M.; Mondello, C.; Giulotto, E. Insertion of telomeric repeats at intrachromosomal break sites during primate evolution. Genome Res. 2004, 14, 1704–1710. [Google Scholar] [CrossRef]
- Simonet, T.; Zaragosi, L.-E.; Philippe, C.; Lebrigand, K.; Schouteden, C.; Augereau, A.; Bauwens, S.; Ye, J.; Santagostino, M.; Giulotto, E.; et al. The human TTAGGG repeat factors 1 and 2 bind to a subset of interstitial telomeric sequences and satellite repeats. Cell Res. 2011, 21, 1028–1038. [Google Scholar] [CrossRef] [PubMed]
- Wood, A.M.; Danielsen, J.M.R.; Lucas, C.A.; Rice, E.L.; Scalzo, D.; Shimi, T.; Goldman, R.D.; Smith, E.D.; Le Beau, M.M.; Kosak, S.T. TRF2 and lamin A/C interact to facilitate the functional organization of chromosome ends. Nat. Commun. 2014, 5, 5467. [Google Scholar] [CrossRef] [PubMed]
- Jia, P.; Chastain, M.; Zou, Y.; Her, C.; Chai, W. Human MLH1 suppresses the insertion of telomeric sequences at intra-chromosomal sites in telomerase-expressing cells. Nucleic Acids Res. 2017, 45, 1219–1232. [Google Scholar] [CrossRef] [PubMed]
- Jia, P.; Chai, W. The MLH1 ATPase domain is needed for suppressing aberrant formation of interstitial telomeric sequences. DNA Repair 2018, 65, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Sýkorová, E.; Lim, K.Y.; Kunická, Z.; Chase, M.W.; Bennett, M.D.; Fajkus, J.; Leitch, A.R. Telomere variability in the monocotyledonous plant order Asparagales. Proc. Biol. Sci. 2003, 270, 1893–1904. [Google Scholar] [CrossRef] [PubMed]
- Souza, G.; Vanzela, A.L.L.; Crosa, O.; Guerra, M. Interstitial telomeric sites and Robertsonian translocations in species of Ipheion and Nothoscordum (Amaryllidaceae). Genetica 2016, 144, 157–166. [Google Scholar] [CrossRef]
- Sýkorová, E.; Lim, K.Y.; Fajkus, J.; Leitch, A.R. The signature of the Cestrum genome suggests an evolutionary response to the loss of (TTTAGGG)n telomeres. Chromosoma 2003, 112, 164–172. [Google Scholar] [CrossRef]
- Peška, V.; Fajkus, P.; Fojtová, M.; Dvořáčková, M.; Hapala, J.; Dvořáček, V.; Polanská, P.; Leitch, A.R.; Sýkorová, E.; Fajkus, J. Characterisation of an unusual telomere motif (TTTTTTAGGG)n in the plant Cestrum elegans (Solanaceae), a species with a large genome. Plant J. 2015, 82, 644–654. [Google Scholar] [CrossRef]
- Dumas, F.; Cuttaia, H.; Sineo, L. Chromosomal distribution of interstitial telomeric sequences in nine neotropical primates (Platyrrhini): Possible implications in evolution and phylogeny. J. Zool. Syst. Evol. Res. 2016, 54, 226–236. [Google Scholar] [CrossRef]
- Mazzoleni, S.; Schillaci, O.; Sineo, L.; Dumas, F. Distribution of interstitial telomeric sequences in Primates and the Pygmy tree shrew (Scandentia). Cytogenet. Genome Res. 2017, 151, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, L.; Evans, J.W.; Wilks, R.; Lucas, J.N.; Brown, J.M.; Giaccia, A.J. Chromosomal radiosensitivity at intrachromosomal telomeric sites. Genes. Chromosomes Cancer 1993, 8, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Bertoni, L.; Attolini, C.; Faravelli, M.; Simi, S.; Giulotto, E. Intrachromosomal telomere-like DNA sequences in Chinese hamster. Mamm. Genome 1996, 7, 853–855. [Google Scholar] [CrossRef] [PubMed]
- Musio, A.; Rainaldi, G.; Sbrana, I. Spontaneous and aphidicolin-sensitive fragile site 3cen co-localizes with the (TTAGGG)n telomeric sequence in Chinese hamster cells. Cytogenet. Genome Res. 1996, 75, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Slijepcevic, P.; Xiao, Y.; Dominguez, I.; Natarajan, A.T. Spontaneous and radiation-induced chromosomal breakage at interstitial telomeric sites. Chromosoma 1996, 104, 596–604. [Google Scholar] [CrossRef] [PubMed]
- Camats, N.; Ruiz-Herrera, A.; Parrilla, J.J.; Acien, M.; Payá, P.; Giulotto, E.; Egozcue, J.; García, F.; Garcia, M. Genomic instability in rat: Breakpoints induced by ionising radiation and interstitial telomeric-like sequences. Mutat. Res. 2006, 595, 156–166. [Google Scholar] [CrossRef]
- Schneider, C.H.; Gross, M.C.; Terencio, M.L.; Artoni, R.F.; Vicari, M.R.; Martins, C.; Feldberg, E. Chromosomal evolution of neotropical cichlids: The role of repetitive DNA sequences in the organization and structure of karyotype. Rev. Fish Biol. Fish. 2013, 23, 201–214. [Google Scholar] [CrossRef]
- Barros, A.V.; Wolski, M.A.V.; Nogaroto, V.; Almeida, M.C.; Moreira-Filho, O.; Vicari, M.R. Fragile sites, dysfunctional telomere and chromosome fusions: What is 5S rDNA role? Gene 2017, 608, 20–27. [Google Scholar] [CrossRef]
- Glugoski, L.; Giuliano-Caetano, L.; Moreira-Filho, O.; Vicari, M.R.; Nogaroto, V. Co-located hAT transposable element and 5S rDNA in an interstitial telomeric sequence suggest the formation of Robertsonian fusion in armored catfish. Gene 2018, 650, 49–54. [Google Scholar] [CrossRef]
- Rosato, M.; Álvarez, I.; Feliner, G.N.; Rosselló, J.A. Inter- and intraspecific hypervariability in interstitial telomeric-like repeats (TTTAGGG)n in Anacyclus (Asteraceae). Ann. Bot. 2018, 122, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Bosco, N.; de Lange, T. A TRF1-controlled common fragile site containing interstitial telomeric sequences. Chromosoma 2012, 121, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Bolzán, A.D.; Bianchi, M.S. Telomeres, interstitial telomeric repeat sequences, and chromosomal aberrations. Mutat. Res. 2006, 612, 189–214. [Google Scholar] [CrossRef]
- Sánchez, J.; Bianchi, M.S.; Bolzán, A.D. Relationship between heterochromatic interstitial telomeric sequences and chromosome damage induced by the radiomimetic compound streptonigrin in Chinese hamster ovary cells. Mutat. Res. 2010, 684, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Bolzán, A.D. Chromosomal aberrations involving telomeres and interstitial telomeric sequences. Mutagenesis 2012, 27, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Swier, V.J.; Anwarali Khan, F.A.; Baker, R.J. Do Time, heterochromatin, NORs, or chromosomal rearrangements correlate with distribution of interstitial telomeric repeats in Sigmodon (cotton rats)? J. Hered. 2012, 103, 493–502. [Google Scholar] [CrossRef] [PubMed]
- Hastie, N.D.; Allshire, R.C. Human telomeres: Fusion and interstitial sites. Trends Genet. 1989, 5, 326–331. [Google Scholar] [CrossRef]
- Yen, C.H.; Pazik, J.; Elliott, R.W. A polymorphic interstitial telomere array near the center of mouse chromosome 8. Mamm. Genome 1996, 7, 218–221. [Google Scholar] [CrossRef] [PubMed]
- Samassekou, O.; Yan, J. Polymorphism in a human chromosome-specific interstitial telomere-like sequence at 22q11.2. Cytogenet. Genome Res. 2011, 134, 174–181. [Google Scholar] [CrossRef]
- Pluta, A.F.; Zakian, V.A. Recombination occurs during telomere formation in yeast. Nature 1989, 337, 429–433. [Google Scholar] [CrossRef]
- Ashley, T.; Ward, D.C. A “hot spot” of recombination coincides with an interstitial telomeric sequence in the Armenian hamster. Cytogenet. Cell Genet. 1993, 62, 169–171. [Google Scholar] [CrossRef] [PubMed]
- Fernández, J.L.; Gosálvez, J.; Goyanes, V. High frequency of mutagen-induced chromatid exchanges at interstitial telomere-like DNA sequence blocks of Chinese hamster cells. Chromosome Res. 1995, 3, 281–284. [Google Scholar] [CrossRef] [PubMed]
- Day, J.P.; Limoli, C.L.; Morgan, W.F. Recombination involving interstitial telomere repeat-like sequences promotes chromosomal instability in Chinese hamster cells. Carcinogenesis 1998, 19, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Wood, A.M.; Laster, K.; Rice, E.L.; Kosak, S.T. A beginning of the end: New insights into the functional organization of telomeres. Nucleus 2015, 6, 172–178. [Google Scholar] [CrossRef]
- Kilburn, A.E.; Shea, M.J.; Sargent, R.G.; Wilson, J.H. Insertion of a telomere repeat sequence into a mammalian gene causes chromosome instability. Mol. Cell. Biol. 2001, 21, 126–135. [Google Scholar] [CrossRef]
- Aksenova, A.Y.; Greenwell, P.W.; Dominska, M.; Shishkin, A.A.; Kim, J.C.; Petes, T.D.; Mirkin, S.M. Genome rearrangements caused by interstitial telomeric sequences in yeast. Proc. Natl. Acad. Sci. USA 2013, 110, 19866–19871. [Google Scholar] [CrossRef]
- Aksenova, A.Y.; Han, G.; Shishkin, A.A.; Volkov, K.V.; Mirkin, S.M. Expansion of Interstitial Telomeric Sequences in Yeast. Cell Rep. 2015, 13, 1545–1551. [Google Scholar] [CrossRef] [PubMed]
- Berger, R.; Bernard, O.A. Jumping translocations. Genes Chromosomes Cancer 2007, 46, 717–723. [Google Scholar] [CrossRef]
- Park, V.M.; Gustashaw, K.M.; Wathen, T.M. The presence of interstitial telomeric sequences in constitutional chromosome abnormalities. Am. J. Hum. Genet. 1992, 50, 914–923. [Google Scholar] [PubMed]
- Rossi, E.; Floridia, G.; Casali, M.; Danesino, C.; Chiumello, G.; Bernardi, F.; Magnani, I.; Papi, L.; Mura, M.; Zuffardi, O. Types, stability, and phenotypic consequences of chromosome rearrangements leading to interstitial telomeric sequences. J. Med. Genet. 1993, 30, 926–931. [Google Scholar] [CrossRef]
- Devriendt, K.; Petit, P.; Matthijs, G.; Vermeesch, J.R.; Holvoet, M.; De Muelenaere, A.; Marynen, P.; Cassiman, J.J.; Fryns, J.P. Trisomy 15 rescue with jumping translocation of distal 15q in Prader-Willi syndrome. J. Med. Genet. 1997, 34, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Vermeesch, J.R.; Petit, P.; Speleman, F.; Devriendt, K.; Fryns, J.-P.P.; Marynen, P. Interstitial telomeric sequences at the junction site of a jumping translocation. Hum. Genet. 1997, 99, 735–737. [Google Scholar] [CrossRef] [PubMed]
- Mignon-Ravix, C.; Depetris, D.; Luciani, J.J.; Cuoco, C.; Krajewska-Walasek, M.; Missirian, C.; Collignon, P.; Delobel, B.; Croquette, M.-F.; Moncla, A.; et al. Recurrent rearrangements in the proximal 15q11-q14 region: A new breakpoint cluster specific to unbalanced translocations. Eur. J. Hum. Genet. 2007, 15, 432–440. [Google Scholar] [CrossRef]
- Fortin, F.; Beaulieu Bergeron, M.; Fetni, R.; Lemieux, N. Frequency of chromosome healing and interstitial telomeres in 40 cases of constitutional abnormalities. Cytogenet. Genome Res. 2009, 125, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Lefort, G.; Blanchet, P.; Chaze, A.M.; Girardet, A.; Sarda, P.; Demaille, J.; Pellestor, F. Cytogenetic and molecular study of a jumping translocation in a baby with Dandy-Walker malformation. J. Med. Genet. 2001, 38, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, S.; Fujita, K.; Mori, H.; Omine, M.; Ishikawa, F. Shortened telomeres involved in a case with a jumping translocation at 1q21. Blood 1998, 91, 1514–1519. [Google Scholar]
- Cuthbert, G.; McCullough, S.; Finney, R.; Breese, G.; Bown, N. Jumping translocation at 11q23 with MLL gene rearrangement and interstitial telomeric sequences. Genes Chromosomes Cancer 1999, 24, 295–298. [Google Scholar] [CrossRef]
- Busson Le Coniat, M.; Brizard, F.; Smadja, N.V.; Maarek, O.; Der Sarkissian, H.; Berger, R. Interstitial telomere repeats in translocations of hematopoietic disorders. Leukemia 2000, 14, 1630–1633. [Google Scholar] [CrossRef] [PubMed]
- Boutouil, M.; Fetni, R.; Qu, J.; Dallaire, L.; Richer, C.L.; Lemieux, N. Fragile site and interstitial telomere repeat sequences at the fusion point of a de novo (Y;13) translocation. Hum. Genet. 1996, 98, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Lévy, J.; Receveur, A.; Jedraszak, G.; Chantot-Bastaraud, S.; Renaldo, F.; Gondry, J.; Andrieux, J.; Copin, H.; Siffroi, J.-P.; Portnoï, M.-F. Involvement of interstitial telomeric sequences in two new cases of mosaicism for autosomal structural rearrangements. Am. J. Med. Genet. A 2015, 167A, 428–433. [Google Scholar] [CrossRef]
- Marlet, L.; Alix, E.; Till, M.; Raskin-Champion, F.; Attia, J.; Boggio, D.; Sanlaville, D.; Schluth-Bolard, C. Prenatal diagnosis of trisomy 2p due to terminal 2p duplication including interstitial telomeric sequences. Cytogenet. Genome Res. 2017, 153, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Schleiermacher, G.; Bourdeaut, F.; Combaret, V.; Picrron, G.; Raynal, V.; Aurias, A.; Ribeiro, A.; Janoueix-Lerosey, I.; Delattre, O. Stepwise occurrence of a complex unbalanced translocation in neuroblastoma leading to insertion of a telomere sequence and late chromosome 17q gain. Oncogene 2005, 24, 3377–3384. [Google Scholar] [CrossRef] [PubMed]
- Bodvarsdottir, S.K.; Steinarsdottir, M.; Bjarnason, H.; Eyfjord, J.E. Dysfunctional telomeres in human BRCA2 mutated breast tumors and cell lines. Mutat. Res. Mol. Mech. Mutagen. 2012, 729, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Goto, G.H.; Zencir, S.; Hirano, Y.; Ogi, H.; Ivessa, A.; Sugimoto, K. Binding of multiple Rap1 proteins stimulates chromosome breakage induction during DNA replication. PLoS Genet. 2015, 11, e1005283. [Google Scholar] [CrossRef]
- Larcher, M.V.; Pasquier, E.; MacDonald, R.S.; Wellinger, R.J. Ku Binding on telomeres occurs at sites distal from the physical chromosome ends. PLoS Genet. 2016, 12, e1006479. [Google Scholar] [CrossRef]
- Yang, D.; Xiong, Y.; Kim, H.; He, Q.; Li, Y.; Chen, R.; Songyang, Z. Human telomeric proteins occupy selective interstitial sites. Cell Res. 2011, 21, 1013–1027. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Lenain, C.; Bauwens, S.; Rizzo, A.; Saint-Léger, A.; Poulet, A.; Benarroch, D.; Magdinier, F.; Morere, J.; Amiard, S.; et al. TRF2 and Apollo cooperate with topoisomerase 2α to protect human telomeres from replicative damage. Cell 2010, 142, 230–242. [Google Scholar] [CrossRef]
- Mignon-Ravix, C.; Depetris, D.; Delobel, B.; Croquette, M.-F.; Mattei, M.-G. A human interstitial telomere associates in vivo with specific TRF2 and TIN2 proteins. Eur. J. Hum. Genet. 2002, 10, 107–112. [Google Scholar] [CrossRef]
- Krutilina, R.I.; Oei, S.-L.; Buchlow, G.; Yau, P.M.; Zalensky, A.O.; Zalenskaya, I.A.; Bradbury, E.M.; Tomilin, N.V. A negative regulator of telomere-length protein TRF1 is associated with interstitial (TTAGGG)n blocks in immortal Chinese hamster ovary cells. Biochem. Biophys. Res. Commun. 2001, 280, 471–475. [Google Scholar] [CrossRef]
- Krutilina, R.I.; Smirnova, A.N.; Mudrak, O.S.; Pleskach, N.M.; Svetlova, M.P.; Oei, S.-L.; Yau, P.M.; Bradbury, E.M.; Zalensky, A.O.; Tomilin, N.V. Protection of internal (TTAGGG)n repeats in Chinese hamster cells by telomeric protein TRF1. Oncogene 2003, 22, 6690–6698. [Google Scholar] [CrossRef]
- Gu, P.; Wang, Y.; Bisht, K.K.; Wu, L.; Kukova, L.; Smith, E.M.; Xiao, Y.; Bailey, S.M.; Lei, M.; Nandakumar, J.; et al. Pot1 OB-fold mutations unleash telomere instability to initiate tumorigenesis. Oncogene 2017, 36, 1939–1951. [Google Scholar] [CrossRef]
- Ilic, A.; Lu, S.; Bhatia, V.; Begum, F.; Klonisch, T.; Agarwal, P.; Xu, W.; Davie, J.R. Ubiquitin C-terminal hydrolase isozyme L1 is associated with shelterin complex at interstitial telomeric sites. Epigenetics Chromatin 2017, 10, 54. [Google Scholar] [CrossRef] [PubMed]
- Robin, J.D.; Ludlow, A.T.; Batten, K.; Magdinier, F.; Stadler, G.; Wagner, K.R.; Shay, J.W.; Wright, W.E. Telomere position effect: Regulation of gene expression with progressive telomere shortening over long distances. Genes Dev. 2014, 28, 2464–2476. [Google Scholar] [CrossRef] [PubMed]
- Robin, J.D.; Ludlow, A.T.; Batten, K.; Gaillard, M.-C.; Stadler, G.; Magdinier, F.; Wright, W.E.; Shay, J.W. SORBS2 transcription is activated by telomere position effect-over long distance upon telomere shortening in muscle cells from patients with facioscapulohumeral dystrophy. Genome Res. 2015, 25, 1781–1790. [Google Scholar] [CrossRef]
- Grunstein, M. Molecular model for telomeric heterochromatin in yeast. Curr. Opin. Cell Biol. 1997, 9, 383–387. [Google Scholar] [CrossRef]
- Kim, W.; Ludlow, A.T.; Min, J.; Robin, J.D.; Stadler, G.; Mender, I.; Lai, T.-P.; Zhang, N.; Wright, W.E.; Shay, J.W. Regulation of the human telomerase gene TERT by telomere position effect—over long distances (TPE-OLD): Implications for aging and cancer. PLoS Biol. 2016, 14, e2000016. [Google Scholar] [CrossRef]
- Shay, J.W. Telomeres and aging. Curr. Opin. Cell Biol. 2018, 52, 1–7. [Google Scholar] [CrossRef]
- Mukherjee, A.K.; Sharma, S.; Sengupta, S.; Saha, D.; Kumar, P.; Hussain, T.; Srivastava, V.; Roy, S.D.; Shay, J.W.; Chowdhury, S. Telomere length-dependent transcription and epigenetic modifications in promoters remote from telomere ends. PLoS Genet. 2018, 14, e1007782. [Google Scholar] [CrossRef]
- Berthiau, A.-S.; Yankulov, K.; Bah, A.; Revardel, E.; Luciano, P.; Wellinger, R.J.; Géli, V.; Gilson, E. Subtelomeric proteins negatively regulate telomere elongation in budding yeast. EMBO J. 2006, 25, 846–856. [Google Scholar] [CrossRef]
- Huettel, B.; Kanno, T.; Daxinger, L.; Bucher, E.; van der Winden, J.; Matzke, A.J.M.; Matzke, M. RNA-directed DNA methylation mediated by DRD1 and Pol IVb: A versatile pathway for transcriptional gene silencing in plants. Biochim. Biophys. Acta Gene Struct. Expr. 2007, 1769, 358–374. [Google Scholar] [CrossRef]
- Matzke, M.A.; Mosher, R.A. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014, 15, 394–408. [Google Scholar] [CrossRef] [PubMed]
- Marcomini, I.; Shimada, K.; Delgoshaie, N.; Yamamoto, I.; Seeber, A.; Cheblal, A.; Horigome, C.; Naumann, U.; Gasser, S.M. Asymmetric processing of DNA ends at a double-strand break leads to unconstrained dynamics and ectopic translocation. Cell Rep. 2018, 24, 2614–2628. [Google Scholar] [CrossRef] [PubMed]
- Moore, A.; Dominska, M.; Greenwell, P.; Aksenova, A.Y.; Mirkin, S.; Petes, T. Genetic control of genomic alterations induced in yeast by interstitial telomeric sequences. Genetics 2018, 209, 425–438. [Google Scholar] [CrossRef]
- Adam, R.D.; Nash, T.E.; Wellems, T.E. Telomeric location of Giardia rDNA genes. Mol. Cell. Biol. 1991, 11, 3326–3330. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.L.; Blackburn, E.H. Developmentally programmed healing of chromosomes by telomerase in Tetrahymena. Cell 1991, 67, 823–832. [Google Scholar] [CrossRef]
- Butler, D.K. Ribosomal DNA is a site of chromosome breakage in aneuploid strains of Neurospora. Genetics 1992, 131, 581–592. [Google Scholar]
- Salvadori, S.; Deiana, A.; Elisabetta, C.; Floridia, G.; Rossi, E.; Zuffardi, O. Colocalization of (TTAGGG)n telomeric sequences and ribosomal genes in Atlantic eels. Chromosom. Res. 1995, 3, 54–58. [Google Scholar] [CrossRef]
- Abuín, M.; Martínez, P.; Sánchez, L. Localization of the repetitive telomeric sequence (TTAGGG) n in four salmonid species. Genome 1996, 39, 1035–1038. [Google Scholar] [CrossRef]
- Copenhaver, G.P.; Pikaard, C.S. RFLP and physical mapping with an rDNA-specific endonuclease reveals that nucleolus organizer regions of Arabidopsis thaliana adjoin the telomeres on chromosomes 2 and 4. Plant J. 1996, 9, 259–272. [Google Scholar] [CrossRef]
- Liu, W.; Fredga, K. Telomeric (TTAGGG)n sequences are associated with nucleolus organizer regions (NORs) in the wood lemming. Chromosom. Res. 1999, 7, 235–240. [Google Scholar] [CrossRef]
- Stimpson, K.M.; Sullivan, L.L.; Kuo, M.E.; Sullivan, B.A. Nucleolar organization, ribosomal DNA array stability, and acrocentric chromosome integrity are linked to telomere function. PLoS ONE 2014, 9, e92432. [Google Scholar] [CrossRef]
- Villasante, A.; Abad, J.P.; Mendez-Lago, M. Centromeres were derived from telomeres during the evolution of the eukaryotic chromosome. Proc. Natl. Acad. Sci. USA 2007, 104, 10542–10547. [Google Scholar] [CrossRef]
- Rocchi, M.; Archidiacono, N.; Schempp, W.; Capozzi, O.; Stanyon, R. Centromere repositioning in mammals. Heredity 2012, 108, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Mirkin, E.V.; Mirkin, S.M. Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev. 2007, 71, 13–35. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Morse, R.H. Chromatin opening and transactivator potentiation by RAP1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999, 19, 5279–5288. [Google Scholar] [CrossRef] [PubMed]
- Morse, R.H. RAP, RAP, open up! New wrinkles for RAP1 in yeast. Trends Genet. 2000, 16, 51–53. [Google Scholar] [CrossRef]
- Xie, K.T.; Wang, G.; Thompson, A.C.; Wucherpfennig, J.I.; Reimchen, T.E.; MacColl, A.D.C.; Schluter, D.; Bell, M.A.; Vasquez, K.M.; Kingsley, D.M. DNA fragility in the parallel evolution of pelvic reduction in stickleback fish. Science 2019, 363, 81–84. [Google Scholar] [CrossRef]
- Labib, K.; Hodgson, B. Replication fork barriers: Pausing for a break or stalling for time? EMBO Rep. 2007, 8, 346–353. [Google Scholar] [CrossRef]
- Hodgson, B.; Calzada, A.; Labib, K. Mrc1 and Tof1 regulate DNA replication forks in different ways during normal S phase. Mol. Biol. Cell 2007, 18, 3894–3902. [Google Scholar] [CrossRef]
- Leman, A.R.; Dheekollu, J.; Deng, Z.; Lee, S.W.; Das, M.M.; Lieberman, P.M.; Noguchi, E. Timeless preserves telomere length by promoting efficient DNA replication through human telomeres. Cell Cycle 2012, 11, 2337–2347. [Google Scholar] [CrossRef]
- Gadaleta, M.C.; Das, M.M.; Tanizawa, H.; Chang, Y.-T.; Noma, K.; Nakamura, T.M.; Noguchi, E. Swi1Timeless prevents repeat instability at fission yeast telomeres. PLoS Genet. 2016, 12, e1005943. [Google Scholar] [CrossRef] [PubMed]
- Mazzoccoli, G.; Piepoli, A.; Carella, M.; Panza, A.; Pazienza, V.; Benegiamo, G.; Palumbo, O.; Ranieri, E. Altered expression of the clock gene machinery in kidney cancer patients. Biomed. Pharmacother. 2012, 66, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Fu, A.; Leaderer, D.; Zheng, T.; Chen, K.; Zhu, Y. Potential cancer-related role of circadian gene TIMELESS suggested by expression profiling and in vitro analyses. BMC Cancer 2013, 13, 498. [Google Scholar] [CrossRef] [PubMed]
- Relles, D.; Sendecki, J.; Chipitsyna, G.; Hyslop, T.; Yeo, C.J.; Arafat, H.A. Circadian gene expression and clinicopathologic correlates in pancreatic cancer. J. Gastrointest. Surg. 2013, 17, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Baldeyron, C.; Brisson, A.; Tesson, B.; Némati, F.; Koundrioukoff, S.; Saliba, E.; De Koning, L.; Martel, E.; Ye, M.; Rigaill, G.; et al. TIPIN depletion leads to apoptosis in breast cancer cells. Mol. Oncol. 2015, 9, 1580–1598. [Google Scholar] [CrossRef] [PubMed]
- Chi, L.; Zou, Y.; Qin, L.; Ma, W.; Hao, Y.; Tang, Y.; Luo, R.; Wu, Z. TIMELESS contributes to the progression of breast cancer through activation of MYC. Breast Cancer Res. 2017, 19, 53. [Google Scholar] [CrossRef]
- Zhang, W.; He, W.; Shi, Y.; Zhao, J.; Liu, S.; Zhang, F.; Yang, J.; Xie, C.; Zhang, Y. Aberrant TIMELESS expression is associated with poor clinical survival and lymph node metastasis in early-stage cervical carcinoma. Int. J. Oncol. 2017, 50, 173–184. [Google Scholar] [CrossRef]
- Parenteau, J.; Wellinger, R.J. Accumulation of single-stranded DNA and destabilization of telomeric repeats in yeast mutant strains carrying a deletion of RAD27. Mol. Cell. Biol. 1999, 19, 4143–4152. [Google Scholar] [CrossRef] [PubMed]
- Saharia, A.; Guittat, L.; Crocker, S.; Lim, A.; Steffen, M.; Kulkarni, S.; Stewart, S.A. Flap endonuclease 1 contributes to telomere stability. Curr. Biol. 2008, 18, 496–500. [Google Scholar] [CrossRef] [PubMed]
- Moser, B.A.; Subramanian, L.; Chang, Y.-T.; Noguchi, C.; Noguchi, E.; Nakamura, T.M. Differential arrival of leading and lagging strand DNA polymerases at fission yeast telomeres. EMBO J. 2009, 28, 810–820. [Google Scholar] [CrossRef] [PubMed]
- Gatbonton, T.; Imbesi, M.; Nelson, M.; Akey, J.M.; Ruderfer, D.M.; Kruglyak, L.; Simon, J.A.; Bedalov, A. Telomere length as a quantitative trait: Genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet. 2006, 2, e35. [Google Scholar]
- Fallet, E.; Jolivet, P.; Soudet, J.; Lisby, M.; Gilson, E.; Teixeira, M.T. Length-dependent processing of telomeres in the absence of telomerase. Nucleic Acids Res. 2014, 42, 3648–3665. [Google Scholar] [CrossRef]
- Johnson, R.E.; Henderson, S.T.; Petes, T.D.; Prakash, S.; Bankmann, M.; Prakash, L. Saccharomyces cerevisiae RAD5-encoded DNA repair protein contains DNA helicase and zinc-binding sequence motifs and affects the stability of simple repetitive sequences in the genome. Mol. Cell. Biol. 1992, 12, 3807–3818. [Google Scholar] [CrossRef] [PubMed]
- Luke-Glaser, S.; Luke, B. The Mph1 helicase can promote telomere uncapping and premature senescence in budding yeast. PLoS ONE 2012, 7, e42028. [Google Scholar] [CrossRef] [PubMed]
- Štafa, A.; Donnianni, R.A.; Timashev, L.A.; Lam, A.F.; Symington, L.S. Template switching during break-induced replication is promoted by the mph1 helicase in Saccharomyces cerevisiae. Genetics 2014, 196, 1017–1028. [Google Scholar] [CrossRef]
- Motegi, A.; Liaw, H.-J.; Lee, K.-Y.; Roest, H.P.; Maas, A.; Wu, X.; Moinova, H.; Markowitz, S.D.; Ding, H.; Hoeijmakers, J.H.J.; et al. Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc. Natl. Acad. Sci. USA 2008, 105, 12411–12416. [Google Scholar] [CrossRef] [PubMed]
- Unk, I.; Hajdú, I.; Blastyák, A.; Haracska, L. Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Repair 2010, 9, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Vannier, J.-B.; Sandhu, S.; Petalcorin, M.I.R.; Wu, X.; Nabi, Z.; Ding, H.; Boulton, S.J. RTEL1 is a replisome-associated helicase that promotes telomere and genome-wide replication. Science 2013, 342, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Vannier, J.-B.; Pavicic-Kaltenbrunner, V.; Petalcorin, M.I.R.; Ding, H.; Boulton, S.J. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell 2012, 149, 795–806. [Google Scholar] [CrossRef] [PubMed]
- Ballew, B.J.; Joseph, V.; De, S.; Sarek, G.; Vannier, J.-B.; Stracker, T.; Schrader, K.A.; Small, T.N.; O’Reilly, R.; Manschreck, C.; et al. A recessive founder mutation in regulator of telomere elongation helicase 1, RTEL1, underlies severe immunodeficiency and features of Hoyeraal Hreidarsson syndrome. PLoS Genet. 2013, 9, e1003695. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Glousker, G.; Molczan, A.; Fox, A.J.; Lamm, N.; Dheekollu, J.; Weizman, O.-E.; Schertzer, M.; Wang, Z.; Vladimirova, O.; et al. Inherited mutations in the helicase RTEL1 cause telomere dysfunction and Hoyeraal-Hreidarsson syndrome. Proc. Natl. Acad. Sci. USA 2013, 110, E3408–E3416. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Drosopoulos, W.C.; Sethi, L.; Madireddy, A.; Schildkraut, C.L.; Zhang, D. FANCM, BRCA1, and BLM cooperatively resolve the replication stress at the ALT telomeres. Proc. Natl. Acad. Sci. USA 2017, 114, E5940–E5949. [Google Scholar] [CrossRef] [PubMed]
- Neil, A.J.; Kim, J.C.; Mirkin, S.M. Precarious maintenance of simple DNA repeats in eukaryotes. BioEssays 2017, 39, 1700077. [Google Scholar] [CrossRef] [PubMed]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aksenova, A.Y.; Mirkin, S.M. At the Beginning of the End and in the Middle of the Beginning: Structure and Maintenance of Telomeric DNA Repeats and Interstitial Telomeric Sequences. Genes 2019, 10, 118. https://doi.org/10.3390/genes10020118
Aksenova AY, Mirkin SM. At the Beginning of the End and in the Middle of the Beginning: Structure and Maintenance of Telomeric DNA Repeats and Interstitial Telomeric Sequences. Genes. 2019; 10(2):118. https://doi.org/10.3390/genes10020118
Chicago/Turabian StyleAksenova, Anna Y., and Sergei M. Mirkin. 2019. "At the Beginning of the End and in the Middle of the Beginning: Structure and Maintenance of Telomeric DNA Repeats and Interstitial Telomeric Sequences" Genes 10, no. 2: 118. https://doi.org/10.3390/genes10020118