Molecular Cloning and Characterization of Three Glucosinolate Transporter (GTR) Genes from Chinese Kale
Abstract
:1. Introduction
2. Results
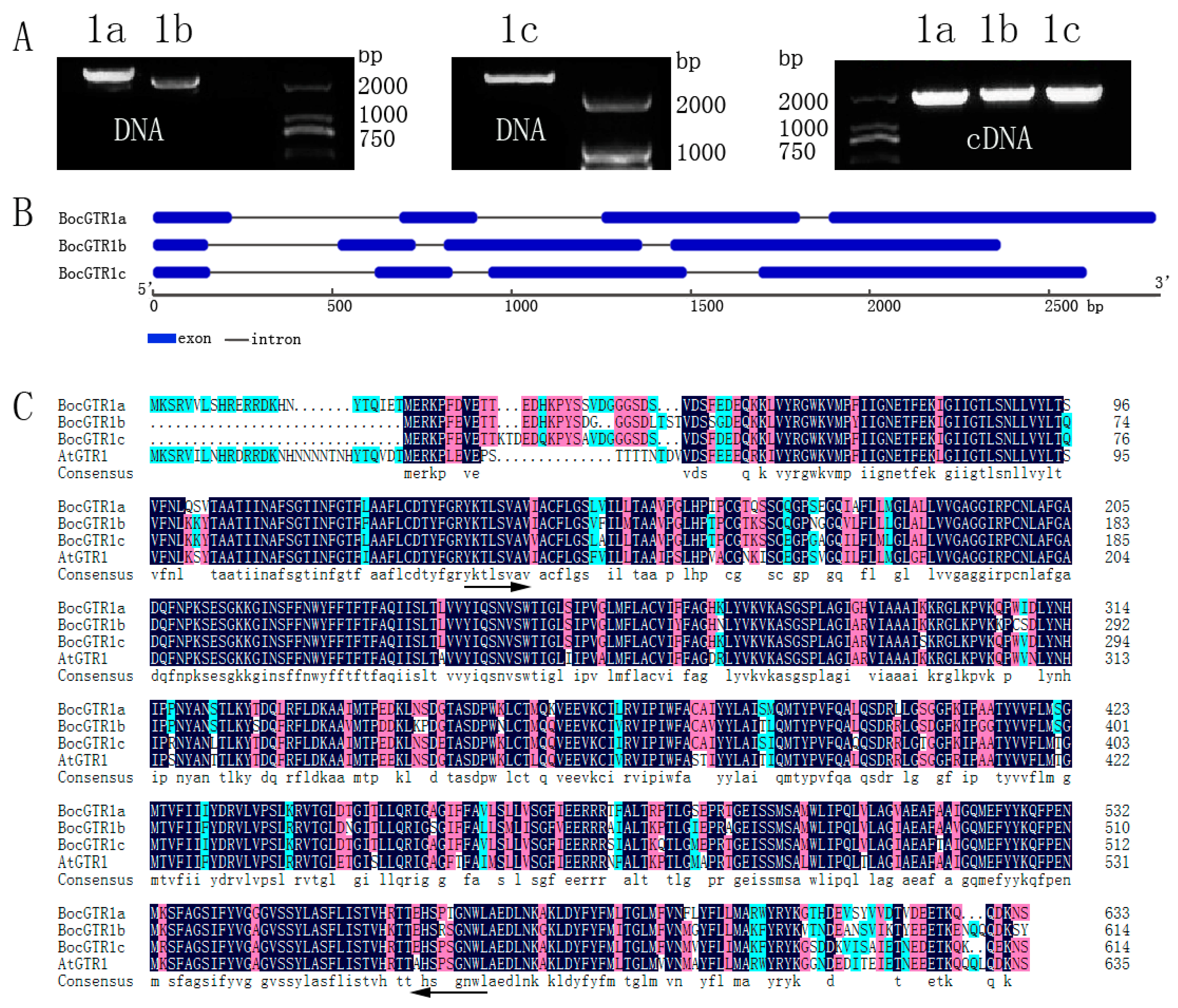
2.1. Molecular Cloning of Three BocGTR1s
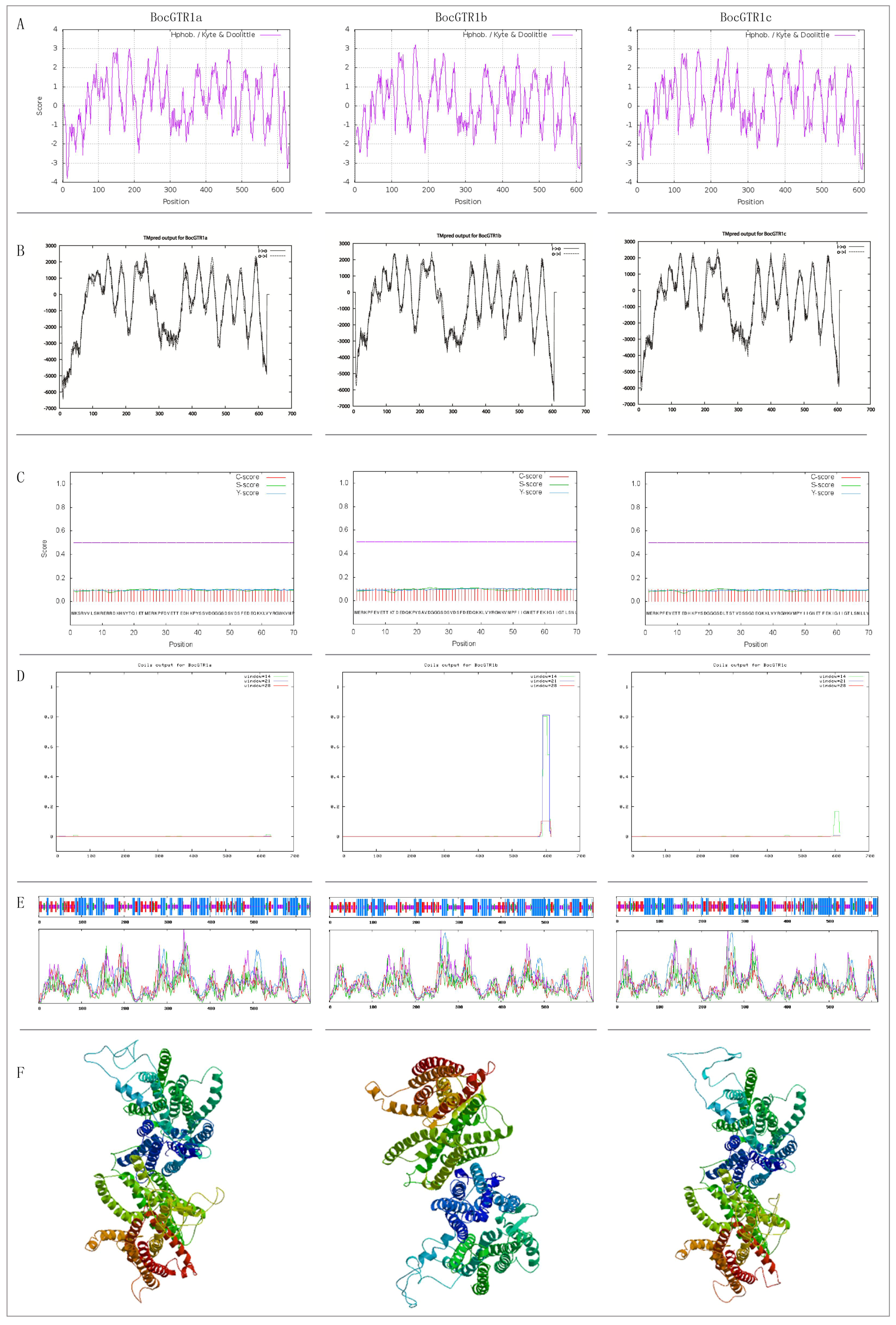
2.2. Bioinformatics Analysis of Three BocGTR1s
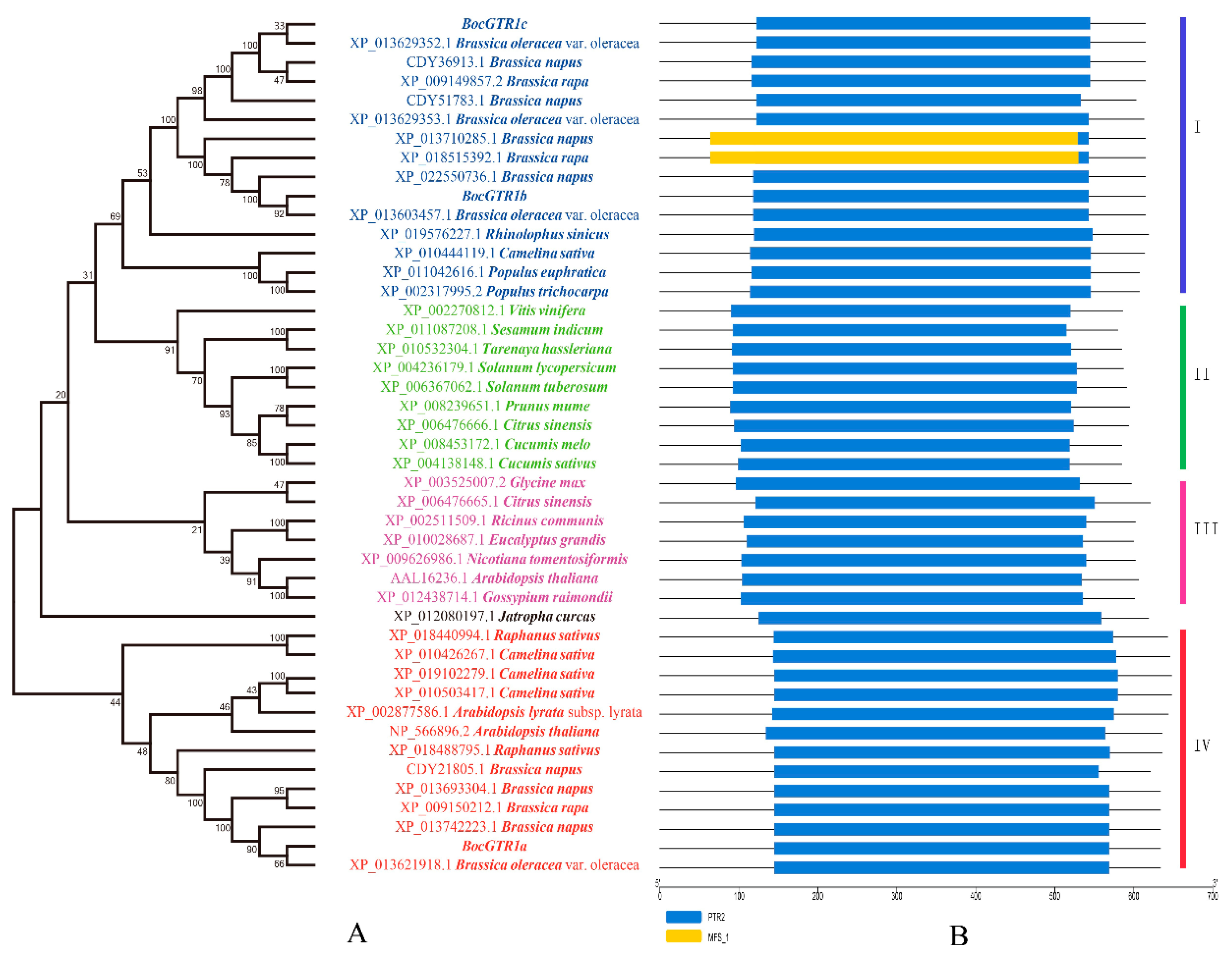
2.3. Phylogenetic Analysis of Three BocGTR1 Proteins
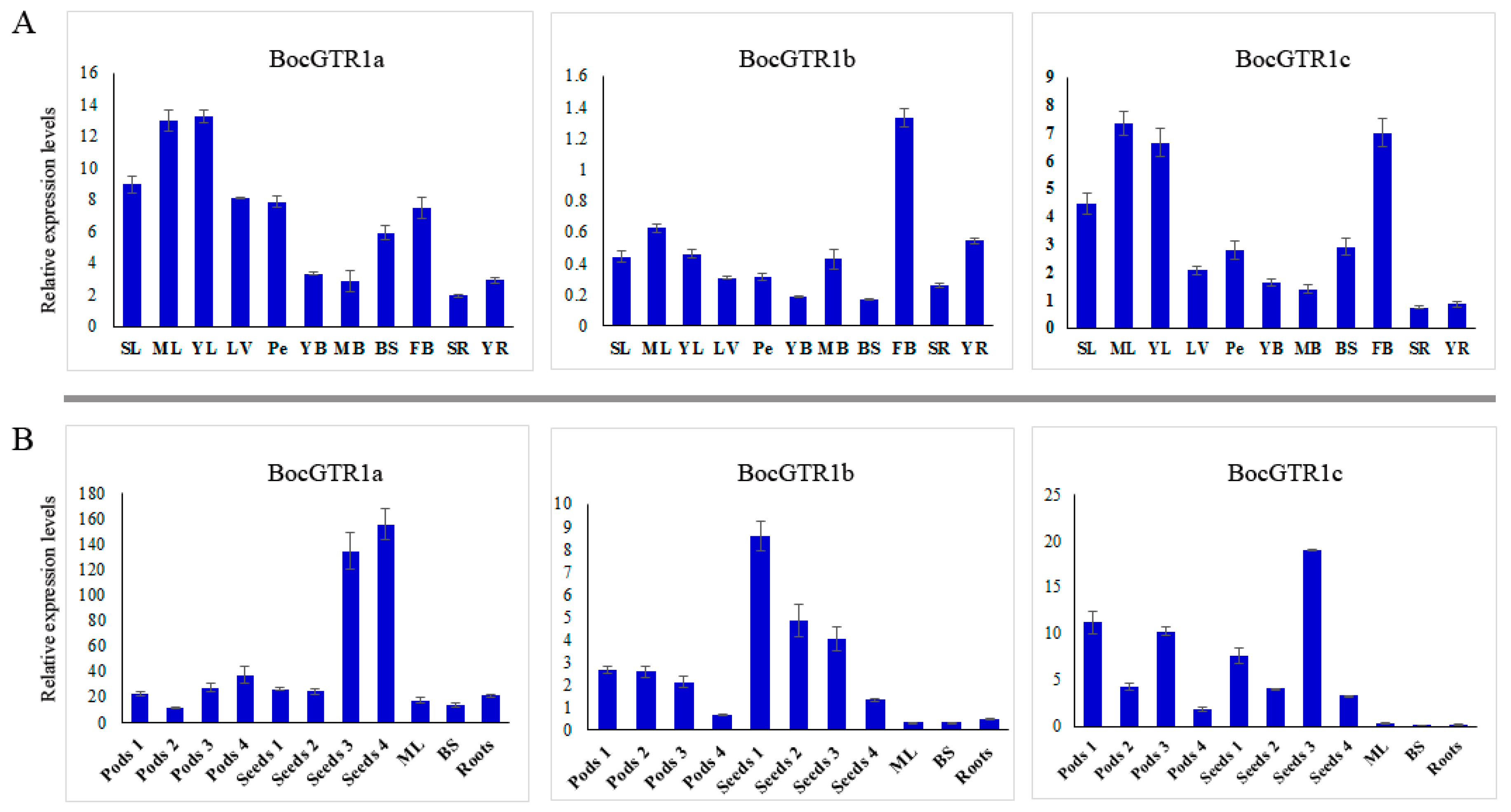
2.4. Expression Profile of BocGTR1s in Different Tissues
2.5. Expression Profile of BocGTR1s under Different Stress Treatments
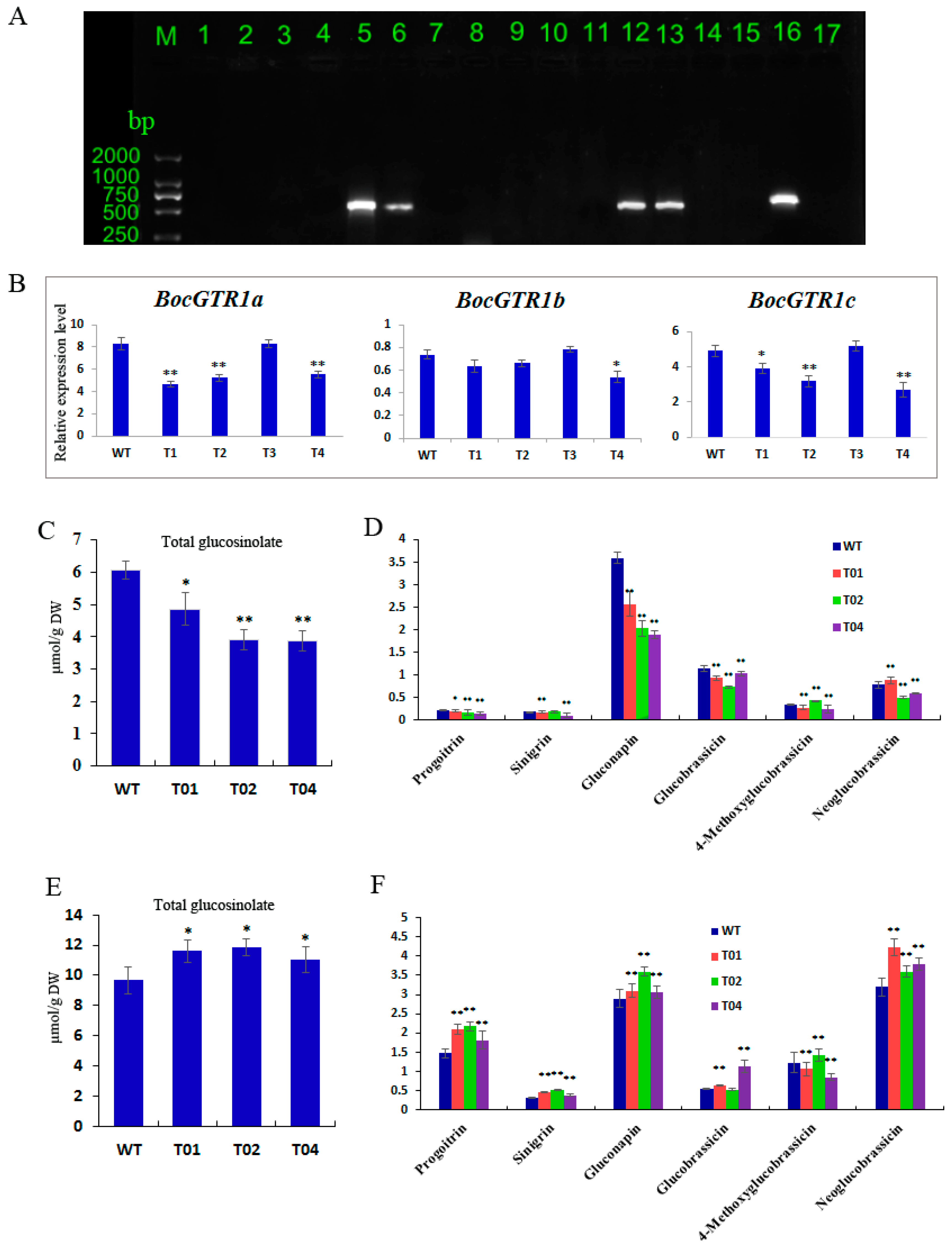
2.6. BocGTR1a RNAi Transgenic Chinese Kale Plants Exhibited Altered Glucosinolate Levels in Roots and Leaves
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Conditions and Stress Treatments
4.2. Molecular Cloning of Three BocGTR1s
4.3. Sequence Analysis
4.4. Expression Analysis by qRT-PCR
4.5. Vector Construction and Transformation of the BoGTR1a RNAi gene
4.6. Extraction and Determination of Glucosinolates
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Fahey, J.W.; Talalay, P. Antioxidant functions of sulforaphane: A potent inducer of phase II detoxication enzymes. Food Chem. Toxicol. 1999, 37, 973–979. [Google Scholar] [CrossRef]
- Halkier, B.A.; Gershenzon, J. Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 2006, 57, 303–333. [Google Scholar] [CrossRef] [PubMed]
- Sønderby, I.E.; Geu-Flores, F.; Halkier, B.A. Biosynthesis of glucosinolates-gene discovery and beyond. Trends Plant Sci. 2010, 15, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Grubb, C.D.; Abel, S. Glucosinolate metabolism and its control. Trends Plant Sci. 2006, 11, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Nour-Eldin, H.H.; Andersen, T.G.; Burow, M.; Madsen, S.R.; Jørgensen, M.E.; Olsen, C.E.; Dreyer, I.; Hedrich, R.; Geiger, D.; Halkier, B.A. NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature 2012, 488, 531–534. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.X.; Petersen, B.L.; Olsen, C.E.; Schulz, A.; Halkier, B.A. Long-distance phloem transport of glucosinolates in Arabidopsis. Plant Physiol. 2001, 127, 194–201. [Google Scholar] [CrossRef] [PubMed]
- Lykkesfeldt, J.; Møller, B.L. Synthesis of Benzylglucosinolate in Tropaeolum majus L. (Isothiocyanates as Potent Enzyme Inhibitors). Plant Physiol. 1993, 102, 609–613. [Google Scholar] [CrossRef]
- Kliebenstein, D.J. Genetic Control of Natural Variation in Arabidopsis Glucosinolate Accumulation. Plant Physiol. 2001, 126, 811–825. [Google Scholar] [CrossRef] [Green Version]
- Thangstad, O.P.; Bones, A.M.; Holtan, S.; Moen, L.; Rossiter, J.T. Microautoradiographic localisation of a glucosinolate precursor to specific cells in Brassica napus L. embryos indicates a separate transport pathway into myrosin cells. Planta 2001, 213, 207–213. [Google Scholar] [CrossRef]
- Du, L.; Halkier, B.A. Biosynthesis of glucosinolates in the developing silique walls and seeds of Sinapis alba. Phytochemistry 1998, 48, 1145–1150. [Google Scholar]
- Andersen, T.G.; Nour-Eldin, H.H.; Fuller, V.L.; Olsen, C.E.; Burow, M.; Halkier, B.A. Integration of biosynthesis and long-distance transport establish organ-specific glucosinolate profiles in vegetative Arabidopsis. Plant Cell 2013, 25, 3133–3145. [Google Scholar] [CrossRef]
- Nour-Eldin, H.H.; Madsen, S.R.; Engelen, S.; Jørgensen, M.E.; Olsen, C.E.; Andersen, J.S.; David Seynnaeve, D.; Verhoye, T.; Rudy Fulawka, R.; Peter Denolf, P.; et al. Reduction of antinutritional glucosinolates in Brassica oilseeds by mutation of genes encoding transporters. Nat. Biotechnol. 2017, 35, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Fang, L.; Liu, N.; Yan, H.; Zhang, Y.; Shi, Q.; Wang, Q. Studies on main nutritional components of Chinese kale among different organs. Acta Hortic. Sin. 2011, 38, 541–548. (In Chinese) [Google Scholar] [CrossRef]
- Lei, J.; Chen, G.; Chen, C.; Cao, B. Germplasm diversity of chinese kale in china. Hortic. Plant J. 2017, 3, 101–104. [Google Scholar] [CrossRef]
- Sun, B.; Liu, N.; Zhao, Y.; Yan, H.; Wang, Q. Variation of glucosinolates in three edible parts of Chinese kale (Brassica alboglabra Bailey) varieties. Food Chem. 2011, 124, 941–947. [Google Scholar] [CrossRef]
- Sun, B.; Yan, H.; Zhang, F.; Wang, Q. Effects of plant hormones on main health-promoting compounds and antioxidant capacity of Chinese kale. Food Res. Int. 2012, 48, 359–366. [Google Scholar] [CrossRef]
- Qian, H.; Liu, T.; Deng, M.; Miao, H.; Cai, C.; Shen, W.S.; Wang, Q. Effects of light quality on main health-promoting compounds and antioxidant capacity of Chinese kale sprouts. Food Chem. 2016, 196, 1232–1238. [Google Scholar] [CrossRef]
- Chen, X.; Zhu, Z.; Gerendás, J.; Zimmermann, N. Glucosinolates in Chinese Brassica campestris vegetables: Chinese cabbage, purple cai-tai, choysum, pakchoi and turnip. Hortscience 2008, 43, 571–574. [Google Scholar] [CrossRef]
- Guo, L.; Yang, R.; Zhou, Y.; Gu, Z. Heat and hypoxia stresses enhance the accumulation of aliphatic glucosinolates and sulforaphane in broccoli sprouts. Eur. Food Res. Technol. 2016, 242, 107–116. [Google Scholar] [CrossRef]
- Cheng, F.; Liu, S.; Wu, J.; Fang, L.; Sun, S.; Liu, B.; Li, P.; Hua, W.; Wang, X. BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol. 2011, 11, 136. [Google Scholar] [CrossRef]
- Ohnmeiss, T.E.; Baldwin, I.T. Optimal defense theory predicts the ontogeny of an induced nicotine defense. Ecology 2000, 81, 1765–1783. [Google Scholar] [CrossRef]
- Chen, G.; Si, Y.; Cao, B.; Feng, E.; Lei, J. Analysis of combining ability and heredity parameters of glucosinolates in Chinese kale. Afr. J. Biotechnol. 2010, 9, 9026–9031. [Google Scholar] [CrossRef]
- Park, J.Y.; Koo, D.H.; Hong, C.P.; Lee, S.J.; Jeon, J.W.; Lee, S.H.; Yun, P.Y.; Park, B.S.; Kim, H.R.; Bang, J.W.; et al. Physicalmapping and microsynteny of Brassica rapa ssp. Pekinensis genome corresponding to a 222 kb gene-rich region of Arabidopsis chromosome 4 and partially duplicated on chromosome 5. Mol. Gen. Genom. 2005, 274, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Schranz, M.E.; Lysak, M.A.; Mitchell-Olds, T. The ABC’s of comparative genomics in the Brassicaceae: Building blocks of crucifer genomes. Trends Plant Sci. 2006, 11, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Nour-Eldin, H.H.; Halkier, B.A. Piecing together the transport pathway of aliphatic glucosinolates. Phytochem. Rev. 2009, 8, 53–67. [Google Scholar] [CrossRef]
- Qasim, M.; Ashraf, M.; Ashraf, M.Y.; Rehman, S.U.; Rha, E.S. Salt induced changes in two canola cultivars differing in salt tolerance. Biol. Plant. 2003, 46, 629–632. [Google Scholar] [CrossRef]
- Velasco, P.; Cartea, M.E.; Gonzalez, C.; Vilar, M.; Ordas, A. Factors affecting the glucosinolate content of kale (Brassica oleracea acephala group). J. Agric. Food Chem. 2007, 55, 955–962. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Z.; Pang, Q.Y.; He, Y.; Zhu, N.; Branstroma, I.; Yan, X.F.; Chen, S.X. Proteomics and metabolomics of arabidopsis esponses to perturbation of glucosinolate biosynthesis. Mol. Plant 2012, 5, 1138–1150. [Google Scholar] [CrossRef]
- Villarreal-García, D.; Nair, V.; Cisneros-Zevallos, L.; Jacobo-Velázquez, D.A. Plants as biofactories: Postharvest stress-induced accumulation of phenolic compounds and glucosinolates in broccoli subjected to wounding stress and exogenous phytohormones. Front. Plant Sci. 2016, 7, 45. [Google Scholar] [CrossRef]
- Torres-Contreras, A.M.; Senes-Guerrero, C.; Pacheco, A.; Gonzalez-Aguero, M.; Ramos-Parra, P.A.; Cisneros-Zevallos, L.; Jacobo-Velázquez, D.A. Genes differentially expressed in broccoli as an early and late response to wounding stress. Postharvest Biol. Technol. 2017, 145, 172–182. [Google Scholar] [CrossRef]
- Madsen, S.R.; Olsen, C.E.; Nour-Eldin, H.H.; Halkier, B.A. Elucidating the role of transport processes in leaf glucosinolate distribution. Plant Physiol. 2014, 166, 1450–1462. [Google Scholar] [CrossRef] [PubMed]
- Jiang., D.; Chen, G.; Lei, J.; Cao, B.; Chen, C. Advances in the physiological, biochemical and molecular mechanisms of glucosinolate transport. Plant Physiol. J. 2017, 53, 29–37. (In Chinese) [Google Scholar] [CrossRef]
- Yin, L.; Chen, C.; Chen, G.; Cao, B.; Lei, J. Molecular Cloning, Expression Pattern and Genotypic Effects on Glucoraphanin Biosynthetic Related Genes in Chinese Kale (Brassica oleracea var. alboglabra Bailey). Molecules 2015, 20, 20254–20267. [Google Scholar] [CrossRef] [PubMed]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Lei, J.; Chen, G.; Chen, H.; Cao, B.; Chen, C. De novo Transcriptome Assembly of Chinese Kale and Global Expression Analysis of Genes Involved in Glucosinolate Metabolism in Multiple Tissues. Front. Plant Sci. 2017, 8, 92. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Battistuzzi, F.U.; Billing-Ross, P.; Murillo, O.; Filipski, A.; Kumar, S. Estimating Divergence times in large molecular phylogenies. Proc. Natl. Acad. Sci. USA 2012, 109, 19333–19338. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Cao, B.; Lu, Y.; Chen, G.; Lei, J. Functional characterization of the translationally controlled tumor protein (TCTP) gene associated with growth and defense response in cabbage. Plant Cell Tiss Organ Cult. 2010, 103, 217. [Google Scholar] [CrossRef]
- Chen, C.; Zhao, X.; Lei, J.; Cao, B.; Chen, G. Pyramiding of Bt Gene into Brassica alboglabra L.H.Bailey by Biolistic Bombardment and Agrobacterium-mediated Transformation Methods. China Veg. 2016, 1, 21–28. (In Chinese) [Google Scholar]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, D.; Lei, J.; Cao, B.; Wu, S.; Chen, G.; Chen, C. Molecular Cloning and Characterization of Three Glucosinolate Transporter (GTR) Genes from Chinese Kale. Genes 2019, 10, 202. https://doi.org/10.3390/genes10030202
Jiang D, Lei J, Cao B, Wu S, Chen G, Chen C. Molecular Cloning and Characterization of Three Glucosinolate Transporter (GTR) Genes from Chinese Kale. Genes. 2019; 10(3):202. https://doi.org/10.3390/genes10030202
Chicago/Turabian StyleJiang, Ding, Jianjun Lei, Bihao Cao, Siyuan Wu, Guoju Chen, and Changming Chen. 2019. "Molecular Cloning and Characterization of Three Glucosinolate Transporter (GTR) Genes from Chinese Kale" Genes 10, no. 3: 202. https://doi.org/10.3390/genes10030202





