The Mitotic Cancer Target Polo-Like Kinase 1: Oncogene or Tumor Suppressor?
Abstract
:1. Introduction
2. Tumor-Prone Mechanisms of PLK1
2.1. Plk1 Transcriptional Regulation Feedback Loops
2.2. Plk1 Promotes the Inactivation of Tumor Suppressors
3. Plk1 Is Able to Play as a Tumor Suppressor
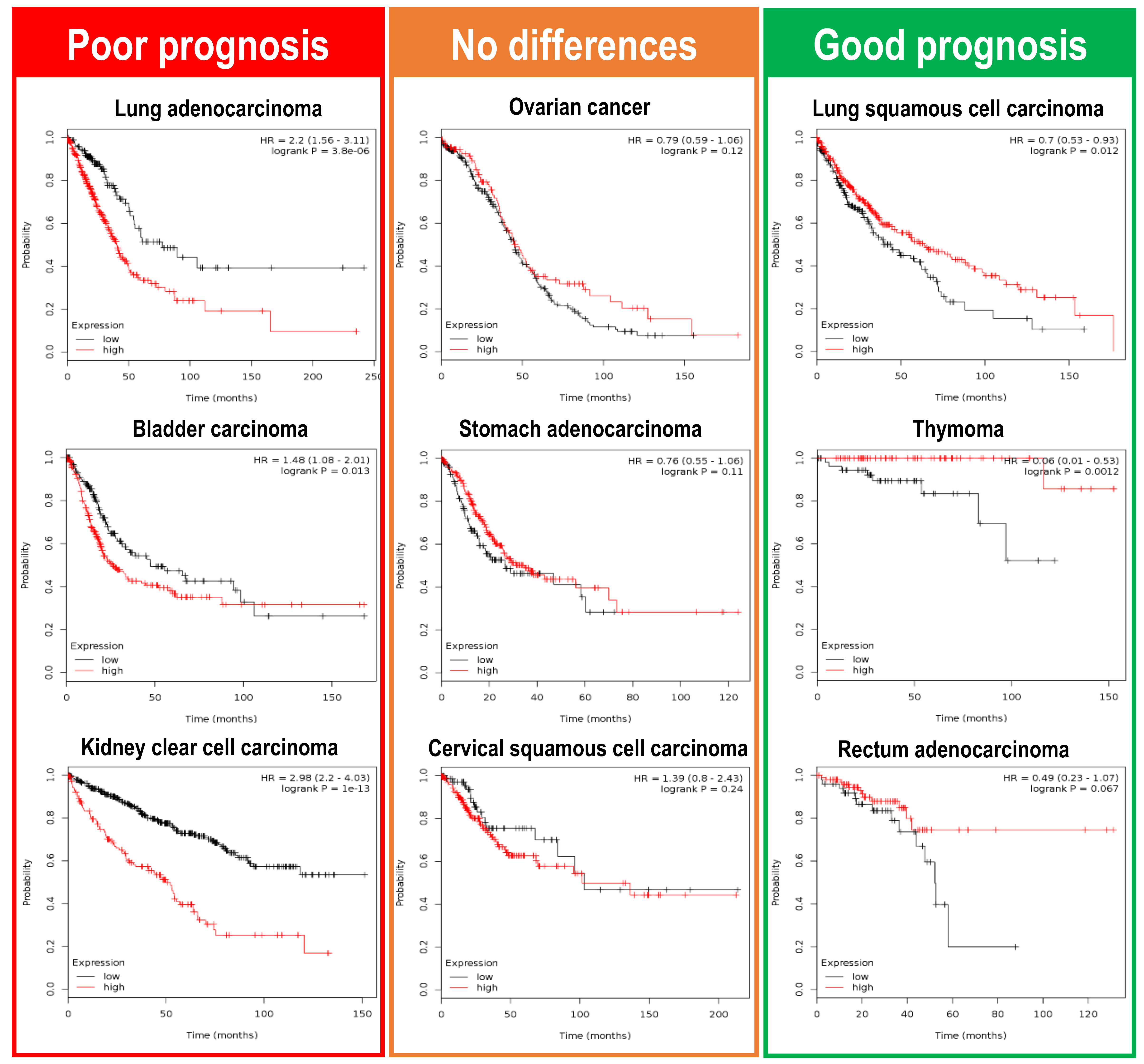
4. Clinical Consequences of Plk1 Upregulation
5. Discussion
6. Future Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Sunkel, C.E.; Glover, D.M. Polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J. Cell Sci. 1988, 89, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Llamazares, S.; Moreira, A.; Tavares, A.; Girdham, C.; Spruce, B.A.; Gonzalez, C.; Karess, R.E.; Glover, D.M.; Sunkel, C.E. Polo encodes a protein kinase homolog required for mitosis in Drosophila. Genes Dev. 1991, 5, 2153–2165. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.S.; Hartwell, L.H. A dependent pathway of gene functions leading to chromosome segregation in Saccharomyces Cerevisiae. J. Cell Biol. 1982, 94, 718–726. [Google Scholar] [CrossRef] [PubMed]
- Hartwell, L.H.; Smith, D. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics 1985, 110, 381–395. [Google Scholar] [PubMed]
- Kitada, K.; Johnson, A.L.; Johnston, L.H.; Suginol, A. A multicopy suppressor gene of the Saccharomyces cerevisiae G1 cell cycle mutant gene dbf4 encodes a protein kinase and is identified as CDC5. Mol. Cell. Biol. 1993, 13, 4445–4457. [Google Scholar] [CrossRef] [PubMed]
- Golsteyn, R.M.; Mundt, K.E.; Fry, A.M.; Nigg, E.A. Cell cycle regulation of the activity and subcellular localization of Plk1, a human protein kinase implicated in mitotic spindle function. J. Cell Biol. 1995, 129, 1617–1628. [Google Scholar] [CrossRef] [PubMed]
- Barr, F.A.; Silljé, H.H.; Nigg, E.A. Polo-like kinases and the orchestration of cell division. Oncogene 2004, 5, 429–440. [Google Scholar] [CrossRef] [PubMed]
- De Carcer, G.; Manning, G.; Malumbres, M. From Plk1 to Plk5: Functional evolution of polo-like kinases. Cell Cycle 2011, 10, 2255–2262. [Google Scholar] [CrossRef] [PubMed]
- Archambault, V.; Lepine, G.; Kachaner, D. Understanding the polo kinase machine. Oncogene 2015, 34, 4799–4807. [Google Scholar] [CrossRef] [PubMed]
- Petronczki, M.; Lenart, P.; Peters, J.M. Polo on the rise-from mitotic entry to cytokinesis with Plk1. Dev. Cell 2008, 14, 646–659. [Google Scholar] [CrossRef] [PubMed]
- Wachowicz, P.; Fernandez-Miranda, G.; Marugan, C.; Escobar, B.; de Cárcer, G. Genetic depletion of polo-like kinase 1 leads to embryonic lethality due to mitotic aberrancies. Bioessays 2016, 38 (Suppl. 1), S96–S106. [Google Scholar] [CrossRef]
- Cheng, M.W.; Wang, B.C.; Weng, Z.Q.; Zhu, X.W. Clinicopathological significance of polo-like kinase 1 (PLK1) expression in human malignant glioma. Acta Histochem. 2012, 114, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Donizy, P.; Halon, A.; Surowiak, P.; Kaczorowski, M.; Kozyra, C.; Matkowski, R. Augmented expression of polo-like kinase 1 is a strong predictor of shorter cancer-specific overall survival in early stage breast cancer at 15-year follow-up. Oncol. Lett. 2016, 12, 1667–1674. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Wen, D.Y.; Dang, Y.W.; He, Y.; Yang, H.; Chen, G. Comprehensive and integrative analysis reveals the diagnostic, clinicopathological and prognostic significance of polo-Like kinase 1 in hepatocellular carcinoma. Cell. Physiol. Biochem. 2018, 47, 925–947. [Google Scholar] [CrossRef] [PubMed]
- Weichert, W.; Denkert, C.; Schmidt, M.; Gekeler, V.; Wolf, G.; Kobel, M.; Dietel, M.; Hauptmann, S. Polo-like kinase isoform expression is a prognostic factor in ovarian carcinoma. Br. J. Cancer 2004, 90, 815–821. [Google Scholar] [CrossRef] [PubMed]
- Weichert, W.; Kristiansen, G.; Schmidt, M.; Gekeler, V.; Noske, A.; Niesporek, S.; Dietel, M.; Denkert, C. Polo-like kinase 1 expression is a prognostic factor in human colon cancer. World J. Gastroenterol. 2005, 11, 5644–5650. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Ohira, M.; Horie, H.; Ando, K.; Takayasu, H.; Suzuki, Y.; Sugano, S.; Hirata, T.; Goto, T.; Matsunaga, T.; et al. Expression profiling and differential screening between hepatoblastomas and the corresponding normal livers: Identification of high expression of the PLK1 oncogene as a poor-prognostic indicator of hepatoblastomas. Oncogene 2004, 23, 5901–5911. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.R.; Wilson, M.L.; Hamanaka, R.; Chase, D.; Kung, H.; Longo, D.L.; Ferris, D.K. Malignant transformation of mammalian cells initiated by constitutive expression of the polo-like kinase. Biochem. Biophys. Res. Commun. 1997, 234, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Eckerdt, F.; Yuan, J.; Strebhardt, K. Polo-like kinases and oncogenesis. Oncogene 2005, 24, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Weng Ng, W.T.; Shin, J.S.; Roberts, T.L.; Wang, B.; Lee, C.S. Molecular interactions of polo-like kinase 1 in human cancers. J. Clin. Pathol. 2016, 69, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Takai, N.; Hamanaka, R.; Yoshimatsu, J.; Miyakawa, I. Polo-like kinases (Plks) and cancer. Oncogene 2005, 24, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Cholewa, B.D.; Liu, X.; Ahmad, N. The role of polo-like kinase 1 in carcinogenesis: Cause or consequence? Cancer Res. 2013, 73, 6848–6855. [Google Scholar] [CrossRef] [PubMed]
- Gutteridge, R.E.; Ndiaye, M.A.; Liu, X.; Ahmad, N. Plk1 inhibitors in cancer therapy: From laboratory to clinics. Mol. Cancer Ther. 2016, 15, 1427–1435. [Google Scholar] [CrossRef] [PubMed]
- Van den Bossche, J.; Lardon, F.; Deschoolmeester, V.; De Pauw, I.; Vermorken, J.B.; Specenier, P.; Pauwels, P.; Peeters, M.; Wouters, A. Spotlight on volasertib: Preclinical and clinical evaluation of a promising Plk1 inhibitor. Med. Res. Rev. 2016, 36, 749–786. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.Y.; Wood, J.L.; Minter-Dykhouse, K.; Ye, L.; Saunders, T.L.; Yu, X.; Chen, J. Polo-like kinase 1 is essential for early embryonic development and tumor suppression. Mol. Cell. Biol. 2008, 28, 6870–6876. [Google Scholar] [CrossRef] [PubMed]
- Wierer, M.; Verde, G.; Pisano, P.; Molina, H.; Font-Mateu, J.; Di Croce, L.; Beato, M. PLK1 signaling in breast cancer cells cooperates with estrogen receptor-dependent gene transcription. Cell Rep. 2013, 3, 2021–2032. [Google Scholar] [CrossRef] [PubMed]
- De Carcer, G.; Venkateswaran, S.V.; Salgueiro, L.; El Bakkali, A.; Somogyi, K.; Rowald, K.; Montanes, P.; Sanclemente, M.; Escobar, B.; de Martino, A.; et al. Plk1 overexpression induces chromosomal instability and suppresses tumor development. Nat. Commun. 2018, 9, 3012. [Google Scholar] [CrossRef]
- Raab, M.; Sanhaji, M.; Matthess, Y.; Horlin, A.; Lorenz, I.; Dotsch, C.; Habbe, N.; Waidmann, O.; Kurunci-Csacsko, E.; Firestein, R.; et al. PLK1 has tumor-suppressive potential in APC-truncated colon cancer cells. Nat. Commun. 2018, 9, 1106. [Google Scholar] [CrossRef] [PubMed]
- Laoukili, J.; Kooistra, M.R.; Bras, A.; Kauw, J.; Kerkhoven, R.M.; Morrison, A.; Clevers, H.; Medema, R.H. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat. Cell Biol. 2005, 7, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Olsson, A.Y.; Feber, A.; Edwards, S.; Te Poele, R.; Giddings, I.; Merson, S.; Cooper, C.S. Role of E2F3 expression in modulating cellular proliferation rate in human bladder and prostate cancer cells. Oncogene 2007, 26, 1028–1037. [Google Scholar] [CrossRef] [PubMed]
- Tategu, M.; Nakagawa, H.; Sasaki, K.; Yamauchi, R.; Sekimachi, S.; Suita, Y.; Watanabe, N.; Yoshid, K. Transcriptional regulation of human polo-like kinases and early mitotic inhibitor. J. Genet. Genom. 2008, 35, 215–224. [Google Scholar] [CrossRef]
- Zhu, H.; Chang, B.D.; Uchiumi, T.; Roninson, I.B. Identification of promoter elements responsible for transcriptional inhibition of polo-like kinase 1 and topoisomerase IIalpha genes by p21(WAF1/CIP1/SDI1). Cell Cycle 2002, 1, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Vousden, K.H.; Prives, C. Blinded by the light: The growing complexity of p53. Cell 2009, 137, 413–431. [Google Scholar] [CrossRef] [PubMed]
- Bykov, V.J.; Wiman, K.G. Novel cancer therapy by reactivation of the p53 apoptosis pathway. Ann. Med. 2003, 35, 458–465. [Google Scholar] [CrossRef] [PubMed]
- Hainaut, P.; Hollstein, M. p53 and human cancer: The first ten thousand mutations. Adv. Cancer Res. 2000, 77, 81–137. [Google Scholar] [PubMed]
- McKenzie, L.; King, S.; Marcar, L.; Nicol, S.; Dias, S.S.; Schumm, K.; Robertson, P.; Bourdon, J.C.; Perkins, N.; Fuller-Pace, F.; et al. p53-dependent repression of polo-like kinase-1 (PLK1). Cell Cycle 2010, 9, 4200–4212. [Google Scholar] [CrossRef] [PubMed]
- Uchiumi, T.; Longo, D.L.; Ferris, D.K. Cell cycle regulation of the human polo-like kinase (PLK) promoter. J. Biol. Chem. 1997, 272, 9166–9174. [Google Scholar] [CrossRef] [PubMed]
- Ando, K.; Ozaki, T.; Yamamoto, H.; Furuya, K.; Hosoda, M.; Hayashi, S.; Fukuzawa, M.; Nakagawara, A. Polo-like kinase 1 (Plk1) inhibits p53 function by physical interaction and phosphorylation. J. Biol. Chem. 2004, 279, 25549–25561. [Google Scholar] [CrossRef] [PubMed]
- Dias, S.S.; Hogan, C.; Ochocka, A.M.; Meek, D.W. Polo-like kinase-1 phosphorylates MDM2 at Ser260 and stimulates MDM2-mediated p53 turnover. FEBS Lett. 2009, 583, 3543–3548. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Dai, G.; Wang, Y.Q.; Wang, S.; Pan, F.Y.; Xue, B.; Zhao, D.H.; Li, C.J. Polo-like kinase 1 regulates mitotic arrest after UV irradiation through dephosphorylation of p53 and inducing p53 degradation. FEBS Lett. 2006, 580, 3624–3630. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, H.; Zhou, Z.; Wang, W.H.; Deng, A.; Andrisani, O.; Liu, X. Plk1-mediated phosphorylation of topors regulates p53 stability. J. Biol. Chem. 2009, 284, 18588–18592. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.S.; Li, H.; Song, B.; Liu, X. Polo-like kinase 1 phosphorylation of G2 and S-phase-expressed 1 protein is essential for p53 inactivation during G2 checkpoint recovery. EMBO Rep. 2010, 11, 626–632. [Google Scholar] [CrossRef] [PubMed]
- Xiao, D.; Yue, M.; Su, H.; Ren, P.; Jiang, J.; Li, F.; Hu, Y.; Du, H.; Liu, H.; Qing, G. Polo-like kinase-1 regulates myc stabilization and activates a feedforward circuit promoting tumor cell survival. Mol. Cell 2016, 64, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Bi, C.; Zhao, X.; Lwin, T.; Wang, C.; Yuan, J.; Silva, A.S.; Shah, B.D.; Fang, B.; Li, T.; et al. PLK1 stabilizes a MYC-dependent kinase network in aggressive B cell lymphomas. J. Clin. Investig. 2018, 128, 5517–5530. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, N.; Arai, H.; Iwasaki, J.; Shiina, M.; Ogata, K.; Hunter, T.; Osada, H. Cyclin-dependent kinase (CDK) phosphorylation destabilizes somatic Wee1 via multiple pathways. Proc. Natl. Acad. Sci. USA 2005, 102, 11663–11668. [Google Scholar] [CrossRef] [PubMed]
- Seki, A.; Coppinger, J.A.; Du, H.; Jang, C.Y.; Yates, J.R., 3rd; Fang, G. Plk1- and beta-TrCP-dependent degradation of Bora controls mitotic progression. J. Cell Biol. 2008, 181, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Eckerdt, F.; Strebhardt, K. Polo-like kinase 1: Target and regulator of anaphase-promoting complex/cyclosome-dependent proteolysis. Cancer Res. 2006, 66, 6895–6898. [Google Scholar] [CrossRef] [PubMed]
- Karlin, K.L.; Mondal, G.; Hartman, J.K.; Tyagi, S.; Kurley, S.J.; Bland, C.S.; Hsu, T.Y.; Renwick, A.; Fang, J.E.; Migliaccio, I.; et al. The oncogenic STP axis promotes triple-negative breast cancer via degradation of the REST tumor suppressor. Cell Rep. 2014, 9, 1318–1332. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Diab, A.; Fan, H.; Mani, S.K.; Hullinger, R.; Merle, P.; Andrisani, O. PLK1 and HOTAIR accelerate proteasomal degradation of SUZ12 and ZNF198 during hepatitis B virus-induced liver carcinogenesis. Cancer Res. 2015, 75, 2363–2374. [Google Scholar] [CrossRef] [PubMed]
- Richly, H.; Aloia, L.; Di Croce, L. Roles of the polycomb group proteins in stem cells and cancer. Cell Death Dis. 2011, 2, e204. [Google Scholar] [CrossRef] [PubMed]
- Madden, C.R.; Finegold, M.J.; Slagle, B.L. Hepatitis B virus X protein acts as a tumor promoter in development of diethylnitrosamine-induced preneoplastic lesions. J. Virol. 2001, 75, 3851–3858. [Google Scholar] [CrossRef] [PubMed]
- Terradillos, O.; Billet, O.; Renard, C.A.; Levy, R.; Molina, T.; Briand, P.; Buendia, M.A. The hepatitis B virus X gene potentiates c-myc-induced liver oncogenesis in transgenic mice. Oncogene 1997, 14, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Studach, L.; Wang, W.H.; Weber, G.; Tang, J.; Hullinger, R.L.; Malbrue, R.; Liu, X.; Andrisani, O. Polo-like kinase 1 activated by the hepatitis B virus X protein attenuates both the DNA damage checkpoint and DNA repair resulting in partial polyploidy. J. Biol. Chem. 2010, 285, 30282–30293. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Abdelmohsen, K.; Kim, J.; Yang, X.; Martindale, J.L.; Tominaga-Yamanaka, K.; White, E.J.; Orjalo, A.V.; Rinn, J.L.; Kreft, S.G.; et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat. Commun. 2013, 4, 2939. [Google Scholar] [CrossRef] [PubMed]
- Hou, S.Q.; Ouyang, M.; Brandmaier, A.; Hao, H.; Shen, W.H. PTEN in the maintenance of genome integrity: From DNA replication to chromosome segregation. Bioessays 2017, 39. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Carracedo, A.; Pandolfi, P.P. Tenets of PTEN tumor suppression. Cell 2008, 133, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, J.; Bi, P.; Lu, Y.; Burcham, G.; Elzey, B.D.; Ratliff, T.; Konieczny, S.F.; Ahmad, N.; Kuang, S.; et al. Plk1 phosphorylation of PTEN causes a tumor-promoting metabolic state. Mol. Cell. Biol. 2014, 34, 3642–3661. [Google Scholar] [CrossRef] [PubMed]
- Maehama, T.; Dixon, J.E. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 1998, 273, 13375–13378. [Google Scholar] [CrossRef] [PubMed]
- Simizu, S.; Osada, H. Mutations in the Plk gene lead to instability of Plk protein in human tumour cell lines. Nat. Cell. Biol. 2000, 2, 852–854. [Google Scholar] [CrossRef] [PubMed]
- Ng, W.; Shin, J.-S.; Wang, B.; Lee, C.S. Exploration of mutation and DNA methylation of polo-like kinase 1 (PLK1) in colorectal cancer. Open J. Pathol. 2017, 7, 13. [Google Scholar] [CrossRef]
- Raab, M.; Kappel, S.; Kramer, A.; Sanhaji, M.; Matthess, Y.; Kurunci-Csacsko, E.; Calzada-Wack, J.; Rathkolb, B.; Rozman, J.; Adler, T.; et al. Toxicity modelling of Plk1-targeted therapies in genetically engineered mice and cultured primary mammalian cells. Nat. Commun. 2011, 2, 395. [Google Scholar] [CrossRef] [PubMed]
- De Carcer, G.; Wachowicz, P.; Martinez-Martinez, S.; Oller, J.; Mendez-Barbero, N.; Escobar, B.; Gonzalez-Loyola, A.; Takaki, T.; El Bakkali, A.; Camara, J.A.; et al. Plk1 regulates contraction of postmitotic smooth muscle cells and is required for vascular homeostasis. Nat. Med. 2017, 23, 964–974. [Google Scholar] [CrossRef] [PubMed]
- Weichert, W.; Kristiansen, G.; Winzer, K.J.; Schmidt, M.; Gekeler, V.; Noske, A.; Muller, B.M.; Niesporek, S.; Dietel, M.; Denkert, C. Polo-like kinase isoforms in breast cancer: Expression patterns and prognostic implications. Virchows Arch. Int. J. Pathol. 2005, 446, 442–450. [Google Scholar] [CrossRef] [PubMed]
- King, S.I.; Purdie, C.A.; Bray, S.E.; Quinlan, P.R.; Jordan, L.B.; Thompson, A.M.; Meek, D.W. Immunohistochemical detection of Polo-like kinase-1 (PLK1) in primary breast cancer is associated with TP53 mutation and poor clinical outcom. Breast Cancer Res. BCR 2012, 14, R40. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, J.; Li, J.; Kong, Y.; Sandusky, G.; Rao, X.; Liu, Y.; Wan, J.; Liu, X. Polo-like kinase 1 (Plk1) overexpression enhances ionizing radiation-induced cancer formation in mice. J. Biol. Chem. 2017, 292, 17461–17472. [Google Scholar] [CrossRef] [PubMed]
- Moser, A.R.; Luongo, C.; Gould, K.A.; McNeley, M.K.; Shoemaker, A.R.; Dove, W.F. ApcMin: A mouse model for intestinal and mammary tumorigenesis. Eur. J. Cancer 1995, 31A, 1061–1064. [Google Scholar] [CrossRef]
- Strebhardt, K.; Ullrich, A. Targeting polo-like kinase 1 for cancer therapy. Nat. Rev. Cancer 2006, 6, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Gyorffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res. Treat. 2010, 123, 725–731. [Google Scholar] [CrossRef] [PubMed]
- De Carcer, G.; Malumbres, M. A centrosomal route for cancer genome instability. Nat. Cell Biol. 2014, 16, 504–506. [Google Scholar] [CrossRef] [PubMed]
- Carter, S.L.; Eklund, A.C.; Kohane, I.S.; Harris, L.N.; Szallasi, Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet. 2006, 38, 1043–1048. [Google Scholar] [CrossRef] [PubMed]
- Santaguida, S.; Amon, A. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat. Rev. Mol. Cell Biol. 2015, 16, 473–485. [Google Scholar] [CrossRef] [PubMed]
- Weaver, B.A.; Silk, A.D.; Montagna, C.; Verdier-Pinard, P.; Cleveland, D.W. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell 2007, 11, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Birkbak, N.J.; Eklund, A.C.; Li, Q.; McClelland, S.E.; Endesfelder, D.; Tan, P.; Tan, I.B.; Richardson, A.L.; Szallasi, Z.; Swanton, C. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 2011, 71, 3447–3452. [Google Scholar] [CrossRef] [PubMed]



© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Cárcer, G. The Mitotic Cancer Target Polo-Like Kinase 1: Oncogene or Tumor Suppressor? Genes 2019, 10, 208. https://doi.org/10.3390/genes10030208
de Cárcer G. The Mitotic Cancer Target Polo-Like Kinase 1: Oncogene or Tumor Suppressor? Genes. 2019; 10(3):208. https://doi.org/10.3390/genes10030208
Chicago/Turabian Stylede Cárcer, Guillermo. 2019. "The Mitotic Cancer Target Polo-Like Kinase 1: Oncogene or Tumor Suppressor?" Genes 10, no. 3: 208. https://doi.org/10.3390/genes10030208




