Abstract
Pathogen fitness landscapes change when transmission cycles establish in non-native environments or spill over into new vectors and hosts. The introduction of Leishmania infantum in the Americas into the Neotropics during European colonization represents a unique case study to investigate the mechanisms of ecological adaptation of this important parasite. Defining the evolutionary trajectories that drive L. infantum fitness in this new environment are of great public health importance as they will allow unique insight into pathways of host/pathogen co-evolution and their consequences for region-specific changes in disease manifestation. This review summarizes current knowledge on L. infantum genetic and phenotypic diversity in the Americas and its possible role in the unique epidemiology of visceral leishmaniasis (VL) in the New World. We highlight the importance of appreciating adaptive molecular mechanisms in L. infantum to understand the parasite’s successful establishment on the continent.
1. Introduction
Leishmania infantum, an etiological agent of visceral leishmaniasis (VL), is a member of the globally dispersed Leishmania donovani species complex, prevalent in many regions of the African, American, Asian, and European continents. Despite ongoing debate [1,2,3], it is generally accepted that L. infantum was introduced to the American continent during its colonization from Europe beginning ca. five hundred years ago [4,5,6,7,8]. The imported strains were most likely carried by asymptomatic humans, dogs, or other mammalian hosts. Although equally suitable ecological conditions for L. infantum transmission are observed in the Old and New Worlds (OW and NW), including the common urban reservoir Canis familiaris and the presence of permissive sandfly vector populations, there are important differences that may have uniquely shaped American L. infantum genomes (Figure 1). Domestic dogs in European and Latin American countries generally differ in behavior, nutritional status, vaccination, and treatment [9]. Wild canids (e.g., Lycalopex vetulus and Cerdocyon thous) appear to contribute little to zoonotic transmission in South America whereas the red fox (Vulpes vulpes) may represent an important synanthropic host species in Europe. Lagomorphs (comprising hares and rabbits, e.g., Lepus granatensis and Oryctolagus cuniculus) have also been implicated in suburban outbreaks of disease in Europe [10,11], but infections are seldom reported from South America where marsupials (e.g., the opossums Didelphis albiventris in Brazil and Didelphis marsupialis in Colombia) may be more relevant to disease transmission from sylvatic to urban environments [12]. Sylvatic host diversity (primates, edentates, rodents, etc.) in the New World is not well described but is likely much higher than in the Old World. With respect to the insect vector of L. infantum, the zoonotic transmission cycle in the Mediterranean basin depends on vectors belonging to the Phlebotomus genus [13], while the Lutzomyia genus represents the vector species for L. infantum in Central and South America. These insect genera not only differ in ecological/physiological traits [14] but also, and just as importantly, inter- and intra-species species variances are described [15]. Finally, socio-economic disparities between the New and Old Worlds are relevant for VL, which is associated with poor living conditions and infant malnutrition [12].
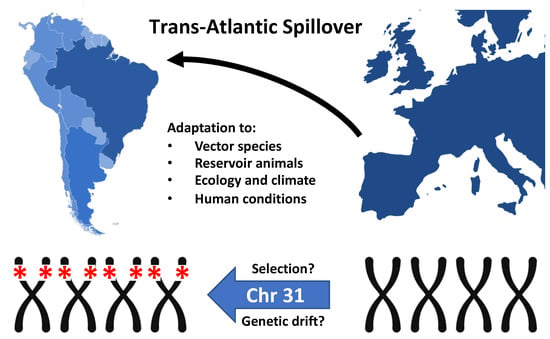
Figure 1.
Trans-Atlantic Spillover of Leishmania infantum from Southern Europe to the Americas during ‘La Conquista’. The homozygous deletion of four genes from tetrasomic chromosome 31 may either result from genetic drift and expansion of a founder population, or natural selection caused by the encountered, region-specific environmental and ecological conditions.
Visceral leishmaniasis in the Americas now extends from southern parts of the USA [16,17] and Mexico to the North of Uruguay, including countries such as Argentina, Brazil, Paraguay, Bolivia, Venezuela, Suriname, Guyana, Colombia, Honduras, Panama, Costa Rica, El Salvador, Guadeloupe, Guatemala, and Nicaragua [12]. Brazil accounts for 90% of cases [12], and changes in the epidemiological profile have been described over the last 30 years [18]. Initially considered rural, the majority of American Visceral Leishmaniasis (AVL) cases are now observed in peri-urban and urban areas of cities in Northeast, Central-West, and Southeast Brazil [19]. The AVL burden remains very high in the Northeast [20], where the parasites’ rapid adaptation to urban niches has been especially prominent. The first major urban outbreaks occurred in the period 1986–1989 [21] and 1991 [22]. A shift in infection demographics has also become evident. Adult infection rates are increasing while those in children have decreased [23], probably due to improvements in nutrition and immunization rates for other diseases. Atypical cutaneous leishmaniasis caused by L. infantum has also been described in the country [24,25]. In such circumstances, the diagnosis based strictly on visual inspection of clinical signs lacking proper Leishmania species identification leads to the misassumption of a low risk of VL in the region, further complicating the management of the spread of the parasite and challenging disease surveillance and control.
VL and dengue are considered the main failures of infectious disease management in Brazil [26]. The observed epidemiological changes and geographic expansion of AVL are associated (but direct causality is yet to be determined) with many variables [27] that compromise the control of this disease [26]. In this review, we describe the underestimated diversity of the neotropical L. infantum parasite and its possible role in generating unique epidemiological patterns and clinical outcomes of VL in the New World. We will further comment on the molecular adaptive mechanisms of Leishmania and the evolutionary trajectories shaped by region-specific host/parasite genotype-genotype and parasite genotype/environment interactions and discuss their potential impact on the epidemiological scenario in the Americas.
2. Underestimated Genetic Diversity of Neotropical L. infantum
The relatively recent arrival of L. infantum in the New World is expected to leave a signature of loss of diversity among the neotropical strains. The low mean microsatellite allelic richness of L. infantum compared to European L. infantum populations indicate such a founder effect [4,8,28]. Lower genetic diversity led to the assumption that any clinical/epidemiological variance observed in the New World should be attributed to host or vector [29] and not to parasite traits. For instance, genetically similar L. infantum strains were described, causing dermal (CL) and visceral (VL) leishmaniasis in Honduras [29,30,31], in contrast to descriptions from the Old World. In the Mediterranean region, L. infantum parasites also cause cutaneous and visceral disease [32,33,34,35] with significant correlations detected between zymodeme and pathology, as well as genetic differentiation between viscerotropic and dermotropic zymodemes by random amplified polymorphic DNA (RAPD)–PCR fingerprinting [36,37]. However, the low genetic resolution of these approaches compared to those available today likely underestimated the role of parasite diversity in the epidemiology and clinical outcomes of American VL. Moreover, New World sample sets were generally very limited in size (e.g., only four Honduran samples [31]) and spatial extent (representing three Honduran islands [31]), likely further contributing to an assumption of extreme genetic homogeneity among neotropical L. infantum populations. In contrast, a more recent study with representative sampling from different Brazilian regions analyzed by random amplified polymorphic DNA (RAPD) showed that, despite the relative genetic homogeneity, most isolates clustered according to geographical origin [28]. Likewise, multilocus microsatellite typing (MLMT) by Ferreira et al. [38] detected substantial population structure in the New World, distinguishing three divergent L. infantum populations within Brazil. Such findings call for the application of more resolutive, next generation sequencing (NGS)-based studies to better understand New World L. infantum diversity, including genomic structural variation, which is considered a major driver of adaptation in the Leishmania genus, as detailed in the following section.
3. Mechanisms of Leishmania Genomic Adaptation In Vitro and In Vivo
A hallmark of Leishmania biology is represented by the parasite’s capacity to adapt to changing environments despite the largely constitutive expression of its genetic material and the absence of classical transcriptional regulation [39]. The primary mechanisms are little understood even though they may play important roles in driving parasite fitness gain in new reservoirs and host tissues, or in response to drug pressure. Some of Leishmania’s adaptative strategies have been described, including the ability to modulate the gene dosage of therapeutic targets (or other determinants of resistance) [40] by chromosomal amplification or the generation of episomal amplicons [41]. A recent series of publications has indeed associated Leishmania adaptation to its remarkable genome plasticity, with frequent changes in chromosome and gene copy number generating important intra-strain heterogeneity and complex mosaic populations that are substrates for environmental selection.
In 2011, Sterkers et al. demonstrated by fluorescence in situ hybridization (FISH) that, in Leishmania, mosaic aneuploidy in culture is a common feature [42]. Such karyotypic fluctuations in Leishmania have been subsequently confirmed in situ by Barja et al. in amastigotes from infected hamster spleen and liver, where they can drive tissue-specific haplotype selection [43]. These karyotypic variations modulate transcript abundance [39,41,43,44], and, thus, likely generate selectable phenotypes. Longitudinal analyses in vitro further demonstrated that the karyoptypic mosaic structure of infecting parasite populations allows for polyclonal adaptation in response to the culture environment, revealing trisomy of chromosome 26 as the major driver of fitness gain in this experimental evolution system [43].
In addition to karyotype variation, read depth analysis of newly established field isolates early after culture adaptation revealed that gene copy number variation (CNV) is a major factor in intra-species genetic heterogeneity and likely drives geographic adaptation. Bussotti et al. [45] evaluated CNVs from the genomes of L. infantum field isolates from three continents: South America (Brazil), Europe (Spain), and North Africa (Tunisia). Although evolved in different regions, the genomes were highly homogenous as judged by SNP analysis, suggesting that this kind of genetic variation may have only a little impact on parasite evolution in the field. In contrast, aside from chromosome amplification, the L. infantum isolates significant differences in gene CNV, with both gene amplification and gene loss observed for genes previously linked to parasite infectivity, such as GP63 or amastins [45]. These differences could be the result of positive/purifying selection that adapts parasite fitness to a given ecology or transmission cycle [45].
Together, these studies reveal highly dynamic fluctuations in chromosome and gene copy number as the major mechanisms allowing for adaptation in the field and in culture. As a result, significant differences exist between tissue-resident amastigotes in patients and derived isolates that are propagated as promastigotes in culture and are subjected to epidemiological association studies. Indeed, significant changes in read depth for 21 chromosomes between in vitro passages two and five were demonstrated in fresh field isolates of L. infantum from Brazil [45]. The same study showed that strain-specific genomic adaptation is not only governed by the environment, but also by intrinsic genetic factors, as judged by the karyotypic differences observed during in vitro adaptation of highly related Spanish L. infantum isolates that were cultured under the same conditions. Furthermore, the tissue-origin of parasites may have a similar impact on karyotypic adaptation, as demonstrated by Zackay et al. [46]. The authors reported that strains simultaneously isolated from spleen and skin from the same patient presented different karyotypes and clustered in a tissue-specific way based on aneuploidy profile [46]. These results suggest that genetically different sub-populations of a given isolate are colonizing different organs, with the genetic differences being revealed in culture by distinct adaptive trajectories selected during in vitro adaptation.
The available data suggest that chromosome amplification is associated with short-term environmental adaptation to in vitro culture, while gain and loss of gene copies might affect long-term environmental adaptation in the field [45]. The detection of alleles under selection in a given region could thus provide relevant epidemiological information. Regarding any field-related inferences, it is important to consider polyclonality with respect to karyotype and gene CNV as an intrinsic feature of any Leishmania population. Polyclonal infections are expected to be more difficult for the host’s immune system to control, thus increasing the success of infection. In this way, poly-clonal infections are expected to contribute to parasite fitness gain and virulence in subsequent hosts. [43].
Until direct sequencing from tissue is not routinely applied, a useful strategy to infer about the epidemiologically relevant genetic traits of Leishmania might be to target CNV signals (amplification, deletion, fixed indels) of fresh field isolates from short-term culture, or purified amastigotes from infected tissues (e.g., from infected dogs). Studying the association of these genotypes with phenotypic traits identified in epidemiological or in vitro studies can reveal genetic loci that are under geographic selection. This approach will shed new light on the mechanisms underlying region-specific parasite evolution and clinical manifestations; moreover, it opens new venues for the discovery of biomarkers with diagnostic and prognostic value.
4. Inferences on the Transmission Dynamics of Neotropical L. infantum Strains and the Shaping of AVL Epidemiology
As commented above, until quite recently, L. infantum strains from the Neotropics were considered genetically homogeneous, a consequence of either low resolutive typing methods employed and/or poor/biased sampling. Then, in 2012, Ferreira et al. applied microsatellite markers to analyze the population structure of 162 L. infantum strains isolated from humans and dogs from most of the Brazilian states endemic for VL and from Paraguay [38]. Samples grouped into three genetically distinct populations: POP1 (relatively more frequent) was observed in all but one endemic area; POP2 was also well-dispersed, but was predominant in Mato Grosso (MT), characterized by the presence of distinct biomes harboring intense biodiversity (Amazon, Cerrado, and Pantanal); POP3 was less dispersed and was observed primarily in Mato Grosso do Sul (MS), likely composed by variable biomes (Cerrado, Atlantic forest, and Pantanal). Several associations with the eco-epidemiological aspects of VL could be identified: First, the distribution of VL in MS seems to follow the west-east construction of the Bolivia-Brazil pipeline from the Corumbá municipality, an inference corroborated by Motoie et al. [47]. Second, regarding vectors, the distribution of POP3 may have been the result of a strong association of this L. infantum population and Lutzomyia cruzi, which is an important VL vector in the region (Corumbá–MS) [48]. In addition, Lutzomyia cruzi also occurs in MT and may influence the structure of POP2 [38,48] as well. The study of Ribolla et al. [49] corroborates that the distinct genetic structure of the sandfly vector plays an important role in shaping the genetic structure of L. infantum in Brazil. Quintana et.al. raised the hypothesis of an expanding urban population of Lutzomyia longipalpis at the Argentina-Bolivia border, with introgressive hybridisation of older haplogroups found in their path in natural forest or rural environments, acquiring a new adaptability to urban environments, and the possibility of changes in vector capacity [50]. The genetic diversity of Lu. longipalpis in Brazil has been recently explored by Casaril et al., who demonstrated the occurrence of three Lu. longipalpis populations in endemic municipalities [51]. Such processes of divergence and speciation of vectors represent mechanisms generating the heterogeneity of vector capacity and competence, as well as vector susceptibility to infectious agents or insecticides.
These studies explored ecological features that possibly shape specific transmission cycles. The question of how the vector/parasite interplay could shape the distribution of Leishmania genotypes implies the selection of parasite populations with higher fitness. Such a process involves (among other factors) the variable ability of Leishmania strains to attach to the gut and survive the sandfly immune response. The complexities of parasite and sandfly interactions, and their molecular background, have been partially characterized (and revised), but mainly consider only parasites and sandflies species, not genotypes [52,53,54]. For instance, defensin of Phlebotomus duboscqi is induced by L. major challenge [52], but not by Leishmania mexicana in Lu. longipalpis [53]. In a more resolutive approach, Mahoney et al. [55,56] assessed intra-species variance, showing polymorphisms in lipophosphoglycan structure controlling Leishmania donovani-sandfly interactions. Thus, the question posed for (now recognized heterogeneous) neotropical parasites is: Could L. infantum genotype variants represent a fitness gain for survival within the population of vectors from a certain area favoring its spread? Could such successful parasite populations harbor biological differences affecting the interaction with the mammal host, thus also influencing disease phenotypes and transmission patterns?
These questions were recently addressed by Courtenay et al. [57] in the context of VL in the Indian subcontinent. The authors combined epidemiology with the basic biology of sandflies, parasite, and hosts to infer transmission dynamics. Two modes of sandfly transmission were proposed based on infection dose and its impact on transmission and disease. It was assumed that uninfected insects feeding on asymptomatic vertebrate hosts with low parasitic burden acquire a low number of parasites. This would result in few metacyclic promastigotes to be transmitted during the next blood meal, resulting in a less severe disease. This mode of transmission could explain the maintenance of the parasite in a given population, without severe clinical disease. The alternative possibility would be the feeding of sandflies on a severely infected individual, with a high parasitic load. These insects likely acquire a higher number of parasites during the blood meal and, consequently, many metacyclic promastigotes would be generated. The blood meal on a second host could result in a larger number of metacyclic parasites being transmitted, and this may lead to more acute and severe disease. In both proposed models, the variable of sandfly density also impacts on the dynamics of Leishmania transmission [58]. The complexity of this issue was boosted by the recent study of Giraud et al. [59]. The authors have demonstrated that experimental transmission of L. infantum by Lu. longipalpis is heterogenous (i.e., sandflies can deliver mixed doses of metacyclic and non-metacyclic promastigotes), and this composition is an important determinant of disease outcome and onward transmission. It is plausible, therefore, that genotypic variants of L. infantum may differ in their competencies for transmission, which may result in changes in both the dose number and quality, and are worthy of future phenotypic analysis.
The transmission models presented by Courtenay et al., could be templates for projects to explore the specificity of host-parasite-vector relationships and the consequent epidemiological variances in neotropical ecological conditions that shaped the transmission cycles. Genetically and biologically distinct neotropical populations of L. infantum could be structured by both the sandfly population distribution and the interplay with mammal hosts, ultimately shaping epidemiological variances of AVL.
As discussed above, the Leishmania “genotype variants” differ not only by the nucleotide sequence-based genotypes (primary sequence) but also, and mainly, in the genomic content, i.e., chromosome and gene copy number variations. To detect this variability, neotropical L. infantum genomic diversity has been recently explored by deep sequencing approaches. Teixeira et al. [60] performed a comparative analysis of whole genome sequences of twenty L. infantum isolates from humans and dogs in northeastern Brazil. Despite high sequence identity, individual differences displayed sufficient variation to allow the isolates to be clustered based on the primary sequence. A major source of variation detected was in chromosome somy, but the analyses do not suggest that individual sequence variants account for differences in clinical outcome or adaptation to different hosts. The lack of association observed by the authors does not discard the value of somy and gene CNV as informative targets to be coupled with phenotypes. As already mentioned, the plasticity of the Leishmania genome could lead to a loss of signal for those samples kept under lab conditions [45]. The karyotypic fluctuations after culture adaptation [45] impacts transcriptomics leading to phenotypic variations [43]. That means that studies that do not use fresh field Leishmania isolates, or direct sequencing, might lose relevant signals, unless a strong genomic signal—a result of long-term adaptation, such as gene amplification or deletion [45]—is positively selected. In that circumstance, such signals could eventually be associated with phenotypes or ecological traits.
This latter possibility is supported by recent work from Carnielli et al. [61]. The authors described a full deletion comprising four genes in tandem on chromosome 31 (the only stable chromosome within the aneuploid mosaicism) of Brazilian L. infantum strains. The locus was named ‘miltefosine sensitivity locus’ (MSL) based on the observation that the patients infected with deletion parasites had a 9.4-fold increased risk of treatment failure after miltefosine administration. This deletion further correlated with reduced parasite miltefosine susceptibility in vitro, suggesting natural resistance to this drug given that it had never been used in Brazil before this trial was carried out [62]. As a result of the identified correlations, the authors proposed this locus as a potential molecular marker to predict miltefosine treatment outcome in VL. This is of upmost relevance because miltefosine was recently approved in Brazil for treatment of canine visceral leishmanisis (CVL), and little information on the impact of the dog treatment on the epidemiology of AVL is available. Only one study [63], evaluating infected dogs from São Paulo, Brazil, is published. Authors described a remission of clinical signs, reduction in parasitic load by qPCR, and reduction in infectivity to sandflies. However, dogs were evaluated for only a short period, and the L. infantum strains were not typed for the MSL locus. If there are indeed non-susceptible miltefosine strains infecting dogs in Brazil, the current treatment policy might represent a risk of worsening the epidemiological scenario of VL in the country, which represents a major public health threat. Nevertheless, a stronger link between MSL and susceptibility to miltefosine is yet to be further explored and confirmed. “How does MSL loss contribute to treatment failure?” is a question already posed in a comment by Bhattacharya and Ouellette [64], which will require much effort to answer.
The deletion strains described in Carnielli’s work were isolated mainly from patients from northeastern and southeastern Brazil, while all OW strains tested did not present the trait. This pattern indicates fitness gain in response to Brazilian ecological conditions, although more consistent screening of NW and OW strains must be done to better determine frequency and geographic distribution of deleted (Del) and non-deleted (NonDel) samples (Figure 1). The mechanism of loss of the genomic region is possibly mediated by homologous recombination between repetitive elements bordering the locus [61]. Thus, the existence of deleted sub-population among non-deleted Leishmania cells cannot be discarded—especially in OW strains, a hypothesis yet to be tested. The possibility of extinction, or reduced frequency of deletion parasites in the OW as a result of changes in environments and socio-economic patterns [65] cannot be disregarded. It is also important to consider the possibility of recombination between the Del and NonDel genotypes, since both were detected co-circulating, and even corresponding to subpopulation within the same isolate [61].
Lastly, as already pointed out by other authors [64], the dispersion of deletion parasites indicates that the variant is beneficial, but the nature of the selective advantage driving natural selection of this genomic trait and the selective forces acting within the neotropical ecology remain open questions. The genomic deletion may indeed represent a relevant biomarker, but not necessarily for miltefosine resistance.
5. Concluding Remarks
Parasite expansion into new environments and new host/vector populations creates potential for strong natural selection and bottlenecks, and associated genetic drift can provide the opportunity for rapid evolutionary change. The possibility of major evolutionary change in imported L. infantum populations has received little attention. Lack of research in this direction is surprising given the parasite’s extreme medical importance and unique capacity for adaptation based on rapid karyotypic and genetic change. More comprehensive surveys of L. infantum genomic and phenotypic diversity across heterogeneous, biodiverse, neotropical environments are required to understand the extensive proliferation of AVL, especially to disentangle demographic from selection events behind complex L. infantum population structure and the diverse clinical outcomes observed in the New World. Comparisons between different biomes are especially interesting, e.g., in Central West vs. Northestern Brazil, where different vector species circulate and divergence through natural selection may occur. Alternatively, differences may result from random drift or from distinct parasite importation events. Selection for the distinct imported clones possibly occurred differently, by the diverse environments Leishmania parasites were exposed to. Along the years, that possibly led to the founding of specific transmission cycles in the Americas and the respective epidemiological variances observed in the continent. This polyclonal adaptation scenario opens important questions on how the neotropical conditions—with their specific selective pressures (reservoirs, vectors, and other environmental factors) might have shaped New World L. infantum evolution. High sampling effort and demographic history reconstruction is therefore also an essential component of future work. Experimental validations of miltefosine resistance represents another priority in the field, as is the potential for putative resistance markers to spread through populations via genetic exchange. To shed new light on the ecology and epidemiology of VL in the Americas, it will be important to combine phenotypic assays with phylogenomic and CNV approaches to trace patterns of demography and selection.
Author Contributions
M.C.B. conceptualization and writing—original draft; G.F.S. and E.C. scientific critical overview and constant writing revision; G.F.S. designed the figure; G.B., preliminary data production R.P., F.N.M., M.L., P.S., scientifically debated the conceptualization. All authors contributed to a final critical review of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by: Mariana C. Boite—Inova Fiocruz/VPPCB Fundação Oswaldo Cruz; MCTIC/CNPq No 28/2018—Universal, processo 425347/2018-4; Gerald F. Späth —Seeding grant from the Institut Pasteur International Department to the LeiSHield Consortium; Philipp Schwabl —Division of Microbiology and Infectious Diseases, the National Institute of Allergy and Infectious Diseases and the National Institutes of Health (DMID/NIADID/NIH grants AI077896-01 and AI105749-01A1); and the Scottish Universities Life Sciences Alliance. The Wellcome Trust; Elisa Cupolillo—Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001; CNPq—Researcher Fellow (302622/2017-9) Faperj—CNE-E26-202.569/2019 and Apoio às Instituições Sediadas no estado do Rio de Janeiro - E-26/010.101083/2018; PASTEUR—FIOCRUZ—USP PROGRAM (2018), Programa PRINT FIOCRUZ-CAPES.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Marcili, A.; Sperança, M.A.; Da Costa, A.P.; Madeira, M.D.F.; Soares, H.S.; Sanches, C.D.O.; Acosta, I.D.C.; Girotto, A.; Minervino, A.; Horta, M.C.; et al. Phylogenetic relationships of Leishmania species based on trypanosomatid barcode (SSU rDNA) and gGAPDH genes: Taxonomic revision of Leishmania (L.) infantum chagasi in South America. Infect. Genet. Evol. 2014, 25, 44–51. [Google Scholar] [CrossRef]
- Sereno, D.; Akhoundi, M.; Dorkeld, F.; Oury, B.; Momen, H.; Perrin, P. What pre-Columbian mummies could teach us about South American leishmaniasis? Pathog. Dis. 2017, 75, 3. [Google Scholar] [CrossRef]
- Shaw, J.J. Further thoughts on the use of the name Leishmania (Leishmania) infantum chagasi for the aetiological agent of American visceral leishmaniasis. Memórias Inst. Oswaldo Cruz 2006, 101, 577–579. [Google Scholar] [CrossRef]
- Leblois, R.; Kuhls, K.; François, O.; Schönian, G.; Wirth, T. Guns, germs and dogs: On the origin of Leishmania chagasi. Infect. Genet. Evol. 2011, 11, 1091–1095. [Google Scholar] [CrossRef]
- Momen, H.; Grimaldi, J.G.; Deane, L.M. Leishmania infantum, the aetiological agent of American visceral leishmaniasis (AVL)? Mem. Inst. Oswaldo Cruz. 1987, 82, 447–448. [Google Scholar] [CrossRef] [PubMed]
- Mauricio, I.L.; Stothard, J.R.; Miles, M.A. The Strange Case of Leishmania chagasi. Parasitol. Today 2000, 16, 188–189. [Google Scholar] [CrossRef]
- Mauricio, I.L.; Gaunt, M.W.; Stothard, J.R.; Miles, M.A. Glycoprotein 63 (gp63) genes show gene conversion and reveal the evolution of Old World Leishmania. Int. J. Parasitol. 2007, 37, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Kuhls, K.; Alam, M.Z.; Cupolillo, E.; Ferreira, G.E.M.; Mauricio, I.L.; Oddone, R.; Feliciangeli, M.D.; Wirth, T.; Miles, M.A.; Schönian, G. Comparative Microsatellite Typing of New World Leishmania infantum Reveals Low Heterogeneity among Populations and Its Recent Old World Origin. PLoS Negl. Trop. Dis. 2011, 5, e1155. [Google Scholar] [CrossRef]
- Dantas-Torres, F.; Solano-Gallego, L.; Baneth, G.; Ribeiro, V.M.; De Paiva-Cavalcanti, M.; Otranto, D. Canine leishmaniosis in the Old and New Worlds: unveiled similarities and differences. Trends Parasitol. 2012, 28, 531–538. [Google Scholar] [CrossRef]
- Antoniou, M.; Gramiccia, M.; Molina, R.; Dvorak, V.; Volf, P. The role of indigenous phlebotomine sandflies and mammals in the spreading of leishmaniasis agents in the Mediterranean region. Eurosurveillance 2013, 18, 20540. [Google Scholar] [CrossRef]
- Arce, A.; Estirado, A.; Ordobas, M.; Sevilla, S.; García, N.; Moratilla, L.; De La Fuente, S.; Martínez, A.M.; Pérez, A.M.; Aránguez, E.; et al. Re-emergence of leishmaniasis in Spain: community outbreak in Madrid, Spain, 2009 to 2012. Eurosurveillance 2013, 18, 20546. [Google Scholar] [CrossRef] [PubMed]
- Alvar, J.; Velez, I.D.; Bern, C.; Herrero, M.; Desjeux, P.; Cano, J.; Jannin, J.; Boer, M.D. Leishmaniasis Worldwide and Global Estimates of Its Incidence. PLOS ONE 2012, 7, e35671. [Google Scholar] [CrossRef] [PubMed]
- Manual on Case Management and Surveillance of the Leishmaniases in the WHO European Region (2017) 20 June 2018. Available online: http://www.euro.who.int/en/publications/abstracts/manual-on-case-management-and-surveillance-of-the-leishmaniases-in-the-who-european-region-2017 (accessed on 6 September 2019).
- Akhoundi, M.; Kuhls, K.; Cannet, A.; Votýpka, J.; Marty, P.; Delaunay, P.; Sereno, D. A Historical Overview of the Classification, Evolution, and Dispersion of Leishmania Parasites and Sandflies. PLoS Negl. Trop. Dis. 2016, 10, e0004349. [Google Scholar] [CrossRef] [PubMed]
- Souza, N.A.; Brazil, R.P.; Araki, A.S. The current status of the Lutzomyia longipalpis (Diptera: Psychodidae: Phlebotominae) species complex. Memórias Instituto Oswaldo Cruz 2017, 112, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Rosypal, A.C.; Troy, G.C.; Zajac, A.M.; Duncan, R.B.; Waki, K.; Chang, K.-P.; Lindsay, D.S. Emergence of Zoonotic Canine Leishmaniasis in the United States: Isolation and Immunohistochemical Detection of Leishmania infantum from Foxhounds from Virginia. J. Eukaryot. Microbiol. 2003, 50, 691–693. [Google Scholar] [CrossRef] [PubMed]
- Rosypal, A.C.; Zajac, A.M.; Lindsay, D.S. Canine visceral leishmaniasis and its emergence in the United States. Vete Clin. N. A. Small Anim. Pract. 2003, 33, 921–937. [Google Scholar] [CrossRef]
- Maia-Elkhoury, A.N.S.; Alves, W.A.; De Sousa-Gomes, M.L.; De Sena, J.M.; Luna, E.A. Visceral leishmaniasis in Brazil: Trends and challenges. Cadernos Saúde Pública 2008, 24, 2941–2947. [Google Scholar] [CrossRef]
- Nascimento, E.L.T.; Martins, D.R.; Monteiro, G.R.; Barbosa, J.D.; Ximenes, M.F.F.M.; Maciel, B.L.; Duarte, I.; Jerônimo, S.M.B. Forum: geographic spread and urbanization of visceral leishmaniasis in Brazil. Postscript: New challenges in the epidemiology of Leishmania chagasi infection. Cadernos de Saúde Pública 2008, 24, 2964–2967. [Google Scholar]
- Lima, Á.L.; de Lima, I.D.; Coutinho, J.F.; de Sousa, Ú.P.; Rodrigues, M.A.; Wilson, M.E.; Pearson, R.D.; Queiroz, J.W.; Jerônimo, S.M. Changing Epidemiology of Visceral Leishmaniasis in Northeastern Brazil: A 25-Year Follow-Up of an Urban Outbreak. -PubMed -NCBI. Available online: https://www.ncbi.nlm.nih.gov/pubmed/29394411 (accessed on 14 October 2019).
- Costa, C.H.; Pereira, H.F.; Araújo, M.V. Visceral leishmaniasis epidemic in the State of Piauí, Brazil, 1980–1986. Revista Saúde Pública 1990, 24, 361–372. [Google Scholar] [CrossRef]
- Jeronimo, S.M.; Oliveira, R.M.; Mackay, S.; Costa, R.M.; Sweet, J.; Nascimento, E.T.; Luz, K.G.; Fernandes, M.Z.; Jernigan, J.; Pearson, R.D. An urban outbreak of visceral leishmaniasis in Natal, Brazil. Trans. R. Soc. Trop. Med. Hyg. 1994, 88, 386–388. [Google Scholar] [CrossRef]
- Lima, I.D.; Lima, A.L.M.; Mendes-Aguiar, C.D.O.; Coutinho, J.F.V.; Wilson, M.E.; Pearson, R.D.; Queiroz, J.W.; Jeronimo, S.M.B. Changing demographics of visceral leishmaniasis in northeast Brazil: Lessons for the future. PLoS Negl. Trop. Dis. 2018, 12, e0006164. [Google Scholar] [CrossRef] [PubMed]
- Lyra, M.R.; Pimentel, M.I.F.; Madeira, M.D.F.; Antonio, L.D.F.; Lyra, J.P.D.M.; Fagundes, A.; Schubach, A.D.O. First report of cutaneous leishmaniasis caused by Leishmania (Leishmania) infantum chagasi in an urban area of Rio de Janeiro, Brazil. Rev. Inst. Med. Trop. Sao Paulo 2015, 57, 451–454. [Google Scholar] [CrossRef] [PubMed]
- Castro, L.S.; França, A.D.O.; Ferreira, E.D.C.; Filho, G.H.; Júnior, M.G.H.; Gontijo, C.M.F.; Pereira, A.A.S.; Dorval, M.E.M.C.; Hans, G.; Higa, M.G. Leishmania infantum AS A Causative agent of cutaneous leishmaniasis in the state of mato grosso do sul, Brazil. Revista Instituto Medicina Tropical São Paulo 2016, 58, 23. [Google Scholar] [CrossRef] [PubMed]
- Barreto, M.L.; Teixeira, M.G.; Bastos, F.I.; Ximenes, R.A.; Barata, R.B.; Rodrigues, L.C. Successes and failures in the control of infectious diseases in Brazil: social and environmental context, policies, interventions, and research needs. Lancet 2011, 377, 1877–1889. [Google Scholar] [CrossRef]
- Pasquali, A.K.S.; Baggio, R.A.; Boeger, W.A.; González-Britez, N.; Guedes, D.C.; Chaves, E.C.; Thomaz-Soccol, V. Dispersion of Leishmania (Leishmania) infantum in central-southern Brazil: Evidence from an integrative approach. PLoS Negl. Trop. Dis. 2019, 13, e0007639. [Google Scholar] [CrossRef]
- Segatto, M.; Ribeiro, L.S.; Costa, D.L.; Costa, C.H.N.; De Oliveira, M.R.; Carvalho, S.F.G.; Macedo, A.M.; Valadares, H.M.S.; Dietze, R.; De Brito, C.F.A.; et al. Genetic diversity of Leishmania infantum field populations from Brazil. Memórias Instituto Oswaldo Cruz 2012, 107, 39–47. [Google Scholar] [CrossRef][Green Version]
- Campos-Ponce, M.; Ponce, C.; Ponce, E.; Maingon, R. Leishmania chagasi/infantum: further investigations on Leishmania tropisms in atypical cutaneous and visceral leishmaniasis foci in Central America. Exp. Parasitol. 2005, 109, 209–219. [Google Scholar] [CrossRef]
- Noyes, H.; Chance, M.; Ponce, C.; Ponce, E.; Maingon, R. Leishmania chagasi: Genotypically Similar Parasites from Honduras Cause both Visceral and Cutaneous Leishmaniasis in Humans. Exp. Parasitol. 1997, 85, 264–273. [Google Scholar] [CrossRef]
- Ponce, C.; Ponce, E.; Cruz, A.; Kreutzer, R.; Pratt, D.M.; Neva, F. Leishmania donovani chagasi: new clinical variant of cutaneous leishmaniasis in Honduras. Lancet 1991, 337, 67–70. [Google Scholar] [CrossRef]
- Campino, L.; Santos-Gomes, G.M.; Pratlong, F.; Antunes, F.; Maurício, I.; Dedet, J.P.; Abranches, P. HIV/Leishmania co-infections in Portugal: diagnosis and isoenzyme characterization of Leishmania. Ann. Trop. Med. Parasitol. 1997, 91, 433–436. [Google Scholar] [CrossRef]
- Cascio, A.; Giordano, S.; Gramiccia, M.; Gradoni, L.; Titone, L.; Scalone, A.; Camma, C.; Russo, R.; Scarlata, F. Epidemiologic surveillance of visceral leishmaniasis in Sicily, Italy. Am. J. Trop. Med. Hyg. 1997, 57, 75–78. [Google Scholar] [CrossRef]
- Fenech, B.F.F. Leishmaniasis in Malta and the Mediterranean Basin. Ann. Trop. Med. Parasitol. 1997, 91, 747–754. [Google Scholar] [CrossRef]
- Guessous-Idrissi, N.; Hamdani, A.; Rhalem, A.; Riyad, M.; Sahibi, H.; Dehbi, F.; Bichichi, M.; Essari, A.; Berrag, B. Epidemiology of human visceral leishmaniasis in Taounate, a northern province of Morocco. Parasite 1997, 4, 181–185. [Google Scholar] [CrossRef]
- Angelici, M.C.; Gramiccia, M.; Gradoni, L. Study on genetic polymorphism of Leishmania infantum through the analysis of restriction enzyme digestion patterns of kinetoplast DNA. Parasitology 1989, 99, 301–309. [Google Scholar] [CrossRef]
- Gramiccia, M.; Gradoni, L. Comparison between leishmanins. Trans. R. Soc. Trop. Med. Hyg. 1994, 88, 255. [Google Scholar] [CrossRef]
- Ferreira, G.E.M.; Dos Santos, B.N.; Dorval, M.E.C.; Ramos, T.P.B.; Porrozzi, R.; Peixoto, A.A.; Cupolillo, E. The Genetic Structure of Leishmania infantum Populations in Brazil and Its Possible Association with the Transmission Cycle of Visceral Leishmaniasis. PLoS ONE 2012, 7, e36242. [Google Scholar] [CrossRef]
- Iantorno, S.A.; Durrant, C.; Khan, A.; Sanders, M.J.; Beverley, S.M.; Warren, W.C.; Berriman, M.; Sacks, D.L.; Cotton, J.A.; Grigg, M.E. Gene Expression in Leishmania Is Regulated Predominantly by Gene Dosage. mBio 2017, 8, e01393-17. [Google Scholar] [CrossRef]
- Patino, L.H.; Imamura, H.; Cruz-Saavedra, L.; Pavia, P.; Muskus, C.; Méndez, C.; Dujardin, J.C.; Ramírez, J.D. Major changes in chromosomal somy, gene expression and gene dosage driven by SbIII in Leishmania braziliensis and Leishmania panamensis. Sci. Rep. 2019, 9, 9485. [Google Scholar] [CrossRef]
- Rogers, M.B.; Hilley, J.D.; Dickens, N.J.; Wilkes, J.; Bates, P.A.; Depledge, D.P.; Harris, D.; Her, Y.; Herzyk, P.; Imamura, H.; et al. Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Res. 2011, 21, 2129–2142. [Google Scholar] [CrossRef]
- Sterkers, Y.; Lachaud, L.; Crobu, L.; Bastien, P.; Pagès, M. FISH analysis reveals aneuploidy and continual generation of chromosomal mosaicism in Leishmania major. Cell. Microbiol. 2010, 13, 274–283. [Google Scholar] [CrossRef]
- Barja, P.P.; Pescher, P.; Bussotti, G.; Dumetz, F.; Imamura, H.; Kedra, D.; Domagalska, M.; Chaumeau, V.; Himmelbauer, H.; Pages, M.; et al. Haplotype selection as an adaptive mechanism in the protozoan pathogen Leishmania donovani. Nat. Ecol. Evol. 2017, 1, 1961–1969. [Google Scholar] [CrossRef]
- Dumetz, F.; Imamura, H.; Sanders, M.; Seblova, V.; Myšková, J.; Pescher, P.; Vanaerschot, M.; Meehan, C.J.; Cuypers, B.; De Muylder, G.; et al. Modulation of Aneuploidy in Leishmania donovani during Adaptation to Different In Vitro and In Vivo Environments and Its Impact on Gene Expression. mBio 2017, 8, e00599-17. [Google Scholar] [CrossRef]
- Bussotti, G.; Gouzelou, E.; Boité, M.C.; Kherachi, I.; Harrat, Z.; Eddaikra, N.; Mottram, J.C.; Antoniou, M.; Christodoulou, V.; Bali, A.; et al. Leishmania Genome Dynamics during Environmental Adaptation Reveal Strain-Specific Differences in Gene Copy Number Variation, Karyotype Instability, and Telomeric Amplification. mBio 2018, 9, e01399-18. [Google Scholar] [CrossRef]
- Zackay, A.; Cotton, J.A.; Sanders, M.; Hailu, A.; Nasereddin, A.; Warburg, A.; Jaffe, C.L. Genome wide comparison of Ethiopian Leishmania donovani strains reveals differences potentially related to parasite survival. PLoS Genet. 2018, 14, e1007133. [Google Scholar] [CrossRef]
- Motoie, G.; Ferreira, G.E.M.; Cupolillo, E.; Canavez, F.; Pereira-Chioccola, V.L. Spatial distribution and population genetics of Leishmania infantum genotypes in São Paulo State, Brazil, employing multilocus microsatellite typing directly in dog infected tissues. Infect. Genet. Evol. 2013, 18, 48–59. [Google Scholar] [CrossRef][Green Version]
- De Oliveira, E.F.; Oshiro, E.T.; Fernandes, W.S.; Ferreira, A.M.T.; De Oliveira, A.G.; Galati, E.A.B. Vector Competence of Lutzomyia cruzi Naturally Demonstrated for Leishmania infantum and Suspected for Leishmania amazonensis. Am. J. Trop. Med. Hyg. 2016, 96, 178–181. [Google Scholar] [CrossRef]
- Ribolla, P.E.M.; Gushi, L.T.; Pires e Cruz, M.D.S.; Costa, C.H.N.; Costa, D.L.; Junior, M.S.D.C.L.; Dorval, M.E.M.C.; De Oliveira, A.G.; Santos, M.F.D.C.; Camargo-Neves, V.L.F.; et al. Leishmania infantumGenetic Diversity andLutzomyia longipalpisMitochondrial Haplotypes in Brazil. BioMed Res. Int. 2016, 2016, 9249217. [Google Scholar] [CrossRef]
- Quintana, M.G.; Pech-May, A.; Fuenzalida, A.D.; Mancini, J.M.D.; Barroso, P.A.; Yadón, Z.E.; Zaidenberg, M.; Salomón, O.D. Lutzomyia longipalpis (Diptera: Psychodidae) Argentina-Bolivia border: new report and genetic diversity. Memórias Instituto Oswaldo Cruz 2019, 114, e190184. [Google Scholar] [CrossRef]
- Casaril, A.E.; Alonso, D.P.; Franco, K.G.; Alvarez, M.V.N.; Barrios, S.P.G.; Fernandes, W.D.S.; Infran, J.D.O.M.; Rodrigues, A.C.M.; Ribolla, P.E.M.; De Oliveira, A.G. Macrogeographic genetic structure of Lutzomyia longipalpis complex populations using Next Generation Sequencing. PLoS ONE 2019, 14, e0223277. [Google Scholar] [CrossRef]
- Boulanger, N.; Lowenberger, C.; Volf, P.; Ursic, R.; Sigutova, L.; Sabatier, L.; Svobodova, M.; Beverley, S.M.; Späth, G.; Brun, R.; et al. Characterization of a Defensin from the Sand Fly Phlebotomus duboscqi Induced by Challenge with Bacteria or the Protozoan Parasite Leishmania major. Infect. Immun. 2004, 72, 7140–7146. [Google Scholar] [CrossRef]
- Telleria, E.L.; Sant’Anna, M.R.V.; Alkurbi, M.O.; Pitaluga, A.N.; Dillon, R.J.; Traub-Csekö, Y.M. Bacterial feeding, Leishmania infection and distinct infection routes induce differential defensin expression in Lutzomyia longipalpis. Parasites Vectors 2013, 6, 12. [Google Scholar] [CrossRef]
- Dostalova, A.; Volf, P. Leishmania development in sand flies: parasite-vector interactions overview. Parasites Vectors 2012, 5, 276. [Google Scholar] [CrossRef]
- Mahoney, A.B.; Turco, S.J. Characterization of the Glucosyltransferases That Assemble the Side Chains of the Indian Leishmania donovani Lipophosphoglycan. Arch. Biochem. Biophys. 1999, 372, 367–374. [Google Scholar] [CrossRef]
- Mahoney, A.B.; Sacks, D.L.; Saraiva, E.; Modi, G.; Turco, S.J. Intra-Species and Stage-Specific Polymorphisms in Lipophosphoglycan Structure ControlLeishmania donovani−Sand Fly Interactions†. Biochemistry 1999, 38, 9813–9823. [Google Scholar] [CrossRef]
- Courtenay, O.; Peters, N.C.; Rogers, M.E.; Bern, C. Combining epidemiology with basic biology of sand flies, parasites, and hosts to inform leishmaniasis transmission dynamics and control. PLoS Pathog. 2017, 13, e1006571. [Google Scholar] [CrossRef]
- França-Silva, J.C.; Barata, R.A.; Da Costa, R.T.; Monteiro, E.M.; Machado-Coelho, G.L.L.; Vieira, E.P.; Prata, A.; Mayrink, W.; Nascimento, E.; Fortes-Dias, C.L.; et al. Importance of Lutzomyia longipalpis in the dynamics of transmission of canine visceral leishmaniasis in the endemic area of Porteirinha Municipality, Minas Gerais, Brazil. Veter Parasitol. 2005, 131, 213–220. [Google Scholar]
- Giraud, E.; Martin, O.; Yakob, L.; Rogers, M. Quantifying Leishmania Metacyclic Promastigotes from Individual Sandfly Bites Reveals the Efficiency of Vector Transmission. Commun. Boil. 2019, 2, 84. [Google Scholar] [CrossRef]
- Teixeira, D.; Monteiro, G.; Martins, D.; Fernandes, M.; Macedo-Silva, V.; Ansaldi, M.; Nascimento, P.; Kurtz, M.; Streit, J.; Ximenes, M.; et al. Comparative analyses of whole genome sequences of Leishmania infantum isolates from humans and dogs in northeastern Brazil. Int. J. Parasitol. 2017, 47, 655–665. [Google Scholar] [CrossRef]
- Carnielli, J.B.; Crouch, K.; Forrester, S.; Silva, V.C.; Carvalho, S.F.; Damasceno, J.D.; Brown, E.; Dickens, N.J.; Costa, D.L.; Costa, C.H.; et al. A Leishmania infantum genetic marker associated with miltefosine treatment failure for visceral leishmaniasis. EBioMedicine 2018, 36, 83–91. [Google Scholar] [CrossRef]
- Carnielli, J.B.T.; Monti-Rocha, R.; Costa, D.L.; Sesana, A.M.; Pansini, L.N.N.; Segatto, M.; Mottram, J.C.; Costa, C.H.N.; Carvalho, S.F.G.; Dietze, R. Natural Resistance of Leishmania infantum to Miltefosine Contributes to the Low Efficacy in the Treatment of Visceral Leishmaniasis in Brazil. Am. J. Trop. Med. Hyg. 2019, 101, 789–794. [Google Scholar] [CrossRef]
- Nogueira, F.D.S.; Avino, V.C.; Galvis-Ovallos, F.; Pereira-Chioccola, V.L.; Moreira, M.A.B.; Romariz, A.P.P.L.; Molla, L.M.; Menz, I. Use of miltefosine to treat canine visceral leishmaniasis caused by Leishmania infantum in Brazil. Parasites Vectors 2019, 12, 79. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Ouellette, M. New insights with miltefosine unresponsiveness in Brazilian Leishmania infantum isolates. EBioMedicine 2018, 37, 13–14. [Google Scholar] [CrossRef]
- Ready, P.D. Leishmaniasis emergence in Europe. Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. 2010, 15, 19505. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).