Hybrid of Restricted and Penalized Maximum Likelihood Method for Efficient Genome-Wide Association Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Arabidopsis thaliana Dataset
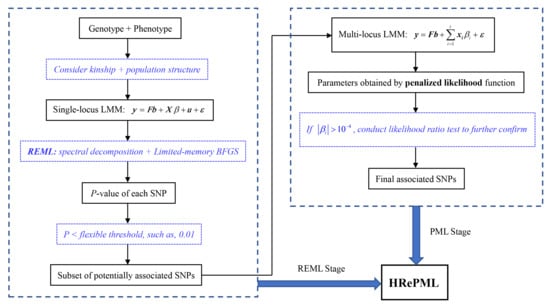
2.2. Restricted Maximum Likelihood (REML) Method in Single-Locus Screening Stage
2.2.1. Single-Locus Linear Mixed Model
2.2.2. Equation Transformation and Update Covariance
2.2.3. Optimal Solution via Efficient REML
2.3. Penalized Maximum Likelihood (PML) Method in Multi-Locus Screening Stage
2.3.1. Multi-Locus Linear Mixed Model and Penalized Likelihood Function
2.3.2. Iterative Method for Parameter Estimation
2.4. Design of Simulation Experiments
3. Results
3.1. Statistical Properties
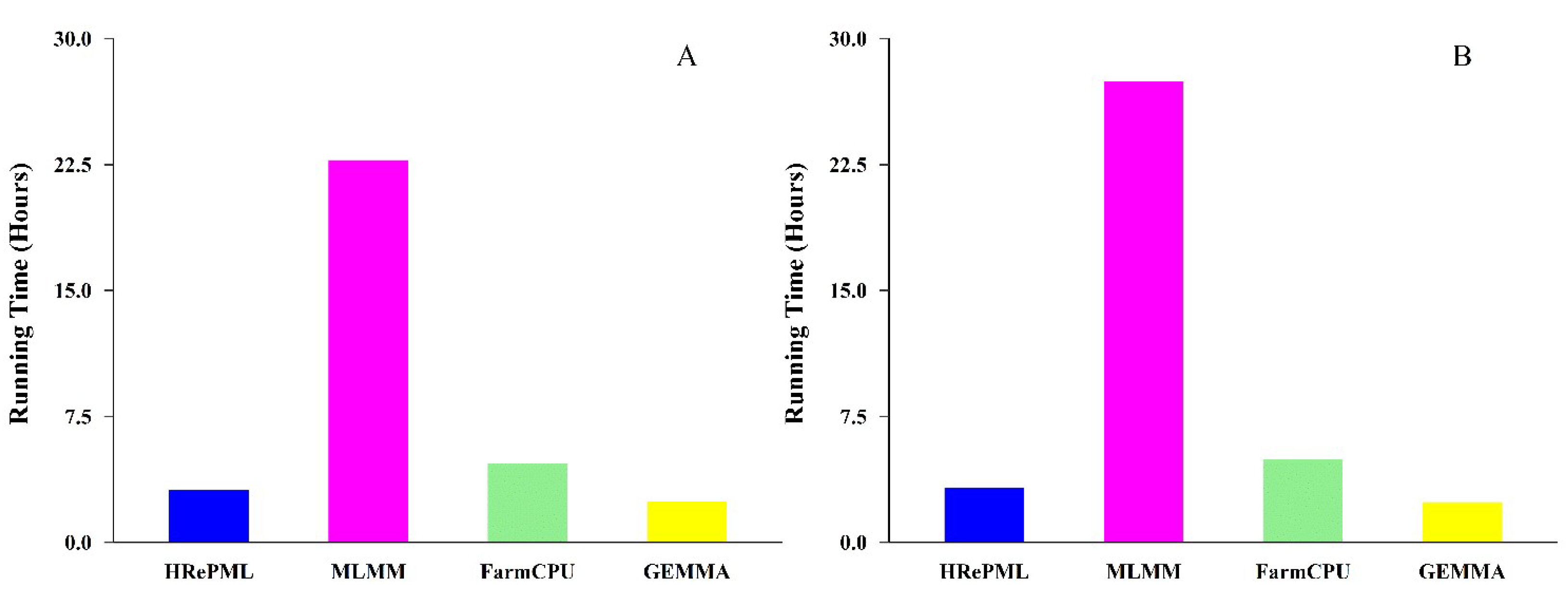
3.2. Running Time
3.3. Association Analysis of Real Data in Arabidopsis
3.4. An Example for the Use of HRePML
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Buniello, A.; MacArthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E.; et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kichaev, G.; Bhatia, G.; Loh, P.R.; Gazal, S.; Burch, K.; Freund, M.K.; Schoech, A.; Pasaniuc, B.; Price, A.L. Leveraging Polygenic Functional Enrichment to Improve GWAS Power. Am. J. Hum. Genet. 2019, 104, 65–75. [Google Scholar] [CrossRef] [Green Version]
- Porcu, E.; Rueger, S.; Lepik, K.; eQTLGen Consortium; BIOS Consortium; Santoni, F.A.; Reymond, A.; Kutalik, Z. Mendelian randomization integrating GWAS and eQTL data reveals genetic determinants of complex and clinical traits. Nat. Commun. 2019, 10, 3300. [Google Scholar] [CrossRef] [Green Version]
- Ganjgahi, H.; Winkler, A.M.; Glahn, D.C.; Blangero, J.; Donohue, B.; Kochunov, P.; Nichols, T.E. Fast and powerful genome wide association of dense genetic data with high dimensional imaging phenotypes. Nat. Commun. 2018, 9, 3254. [Google Scholar] [CrossRef]
- Xu, Y.; Xing, L.; Su, J.; Zhang, X.; Qiu, W. Model-based clustering for identifying disease-associated SNPs in case-control genome-wide association studies. Sci. Rep. 2019, 9, 13686. [Google Scholar] [CrossRef]
- Lee, T.; Lee, I. araGWAB: Network-based boosting of genome-wide association studies in Arabidopsis thaliana. Sci. Rep. 2018, 8, 2925. [Google Scholar] [CrossRef]
- Yang, J.; Zaitlen, N.A.; Goddard, M.E.; Visscher, P.M.; Price, A.L. Advantages and pitfalls in the application of mixed-model association methods. Nat. Genet. 2014, 46, 100–106. [Google Scholar] [CrossRef] [Green Version]
- Lippert, C.; Listgarten, J.; Liu, Y.; Kadie, C.M.; Davidson, R.I.; Heckerman, D. FaST linear mixed models for genome-wide association studies. Nat. Methods 2011, 8, 833–835. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loh, P.R.; Tucker, G.; Bulik-Sullivan, B.K.; Vilhjalmsson, B.J.; Finucane, H.K.; Salem, R.M.; Chasman, D.I.; Ridker, P.M.; Neale, B.M.; Berger, B.; et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 2015, 47, 284–290. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Zheng, Z.; Qi, T.; Kemper, K.E.; Wray, N.R.; Visscher, P.M.; Yang, J. A resource-efficient tool for mixed model association analysis of large-scale data. Nat. Genet. 2019, 51, 1749–1755. [Google Scholar] [CrossRef]
- Border, R.; Becker, S. Stochastic Lanczos estimation of genomic variance components for linear mixed-effects models. Bmc Bioinform. 2019, 20, 411. [Google Scholar] [CrossRef] [Green Version]
- Hadfield, J.D. MCMC Methods for Multi-Response Generalized Linear Mixed Models: The MCMCglmm R Package. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Usinglme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Lourenco, V.M.; Rodrigues, P.C.; Pires, A.M.; Piepho, H.P. A robust DF-REML framework for variance components estimation in genetic studies. Bioinformatics 2017, 33, 3584–3594. [Google Scholar] [CrossRef]
- Cesarani, A.; Pocrnic, I.; Macciotta, N.P.P.; Fragomeni, B.O.; Misztal, I.; Lourenco, D.A.L. Bias in heritability estimates from genomic restricted maximum likelihood methods under different genotyping strategies. J. Anim Breed. Genet. 2019, 136, 40–50. [Google Scholar] [CrossRef] [Green Version]
- Tibshirani, R. Regression Shrinkage and Selection via the Lasso. J. R. Stat. Soc. Ser. B 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Yuan, M. Model Selection and Estimation in Regression With Grouped Variables. J. R. Stat. Soc. Ser. B 2006, 68, 49–67. [Google Scholar] [CrossRef]
- Zou, H. The Adaptive Lasso and Its Oracle Properties. J. Am. Stat. Assoc. 2006, 101, 1418–1429. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.M.; Xu, S. A penalized maximum likelihood method for estimating epistatic effects of QTL. Heredity 2005, 95, 96–104. [Google Scholar] [CrossRef] [Green Version]
- Hoffman, G.E.; Logsdon, B.A.; Mezey, J.G. PUMA: A unified framework for penalized multiple regression analysis of GWAS data. PLoS Comput. Biol. 2013, 9, e1003101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamuri, A.U.; Goldman, N.; dos Reis, M. A penalized-likelihood method to estimate the distribution of selection coefficients from phylogenetic data. Genetics 2014, 197, 257–271. [Google Scholar] [CrossRef] [Green Version]
- Meyer, K. Simple Penalties on Maximum-Likelihood Estimates of Genetic Parameters to Reduce Sampling Variation. Genetics 2016, 203, 1885–1900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gianola, D. Priors in whole-genome regression: The bayesian alphabet returns. Genetics 2013, 194, 573–596. [Google Scholar] [CrossRef] [Green Version]
- Perez, P.; de los Campos, G. Genome-wide regression and prediction with the BGLR statistical package. Genetics 2014, 198, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Segura, V.; Vilhjalmsson, B.J.; Platt, A.; Korte, A.; Seren, U.; Long, Q.; Nordborg, M. An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat. Genet. 2012, 44, 825–830. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative Usage of Fixed and Random Effect Models for Powerful and Efficient Genome-Wide Association Studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, N.; Lo, M.T.; Kauppi, K.; Djurovic, S.; Andreassen, O.A.; Johnson, V.E.; Chen, C.H. GWASinlps: Non-local prior based iterative SNP selection tool for genome-wide association studies. Bioinformatics 2019, 35, 1–11. [Google Scholar] [CrossRef]
- Sinoquet, C. A method combining a random forest-based technique with the modeling of linkage disequilibrium through latent variables, to run multilocus genome-wide association studies. BMC Bioinform. 2018, 19, 106. [Google Scholar] [CrossRef] [Green Version]
- Sun, R.; Hui, S.; Bader, G.D.; Lin, X.; Kraft, P. Powerful gene set analysis in GWAS with the Generalized Berk-Jones statistic. PLoS Genet. 2019, 15, e1007530. [Google Scholar] [CrossRef] [Green Version]
- Hamazaki, K.; Iwata, H. RAINBOW: Haplotype-based genome-wide association study using a novel SNP-set method. PLoS Comput. Biol. 2020, 16, e1007663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.B.; Feng, J.Y.; Ren, W.L.; Huang, B.; Zhou, L.; Wen, Y.J.; Zhang, J.; Dunwell, J.M.; Xu, S.; Zhang, Y.M. Improving power and accuracy of genome-wide association studies via a multi-locus mixed linear model methodology. Sci. Rep. 2016, 6, 19444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, S. An expectation-maximization algorithm for the Lasso estimation of quantitative trait locus effects. Heredity 2010, 105, 483–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigue, N. On the statistical interpretation of site-specific variables in phylogeny-based substitution models. Genetics 2013, 193, 557–564. [Google Scholar] [CrossRef] [Green Version]
- Zhu, C.; Byrd, R.H.; Lu, P.; Nocedal, J. Algorithm 778: L-BFGS-B: Fortran subroutines for large-scale bound-constrained optimization. ACM Trans. Math. Softw. 1997, 23, 550–560. [Google Scholar] [CrossRef]
- Atwell, S.; Huang, Y.S.; Vilhjalmsson, B.J.; Willems, G.; Horton, M.; Li, Y.; Meng, D.; Platt, A.; Tarone, A.M.; Hu, T.T.; et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 2010, 465, 627–631. [Google Scholar] [CrossRef]
- Kang, H.M.; Zaitlen, N.A.; Wade, C.M.; Kirby, A.; Heckerman, D.; Daly, M.J.; Eskin, E. Efficient control of population structure in model organism association mapping. Genetics 2008, 178, 1709–1723. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Ersoz, E.; Lai, C.Q.; Todhunter, R.J.; Tiwari, H.K.; Gore, M.A.; Bradbury, P.J.; Yu, J.; Arnett, D.K.; Ordovas, J.M.; et al. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 2010, 42, 355–360. [Google Scholar] [CrossRef] [Green Version]
- Schraudolph, N.N.; Yu, J.; Günter, S. A stochastic quasi-Newton method for online convex optimization. AISTATS 2007, 2, 436–443. [Google Scholar]
- Nocedal, J. Updating quasi-Newton matrices with limited storage. Math. Comput. 1980, 35, 773–782. [Google Scholar] [CrossRef]
- Schäling, B. The Boost C++ Libraries, 2nd ed.; XML Press: Laguna Niguel, CA, USA, 2014. [Google Scholar]
- Cox, D.D.; O’Sullivan, F. Asymptotic analysis of penalized likelihood and related estimators. Ann. Stat. 1990, 18, 1676–1695. [Google Scholar] [CrossRef]
- Ren, W.L.; Wen, Y.J.; Dunwell, J.M.; Zhang, Y.M. pKWmEB: Integration of Kruskal-Wallis test with empirical Bayes under polygenic background control for multi-locus genome-wide association study. Heredity 2018, 120, 208–218. [Google Scholar] [CrossRef]
- Tamba, C.L.; Ni, Y.L.; Zhang, Y.M. Iterative sure independence screening EM-Bayesian LASSO algorithm for multi-locus genome-wide association studies. PLoS Comput. Biol. 2017, 13, e1005357. [Google Scholar] [CrossRef] [PubMed]
- The Arabidopsis Information Resource. Available online: https://www.arabidopsis.org/index.jsp. (accessed on 29 October 2020).
- Platt, A.; Vilhjalmsson, B.J.; Nordborg, M. Conditions under which genome-wide association studies will be positively misleading. Genetics 2010, 186, 1045–1052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.T.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; O’Connell, J.; et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018, 562, 203–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| QTN | Chr. | Position(bp) | R2 | Effect | Power (%) | Mean Squared Errors (MSE) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HRePML | MLMM | FarmCPU | GEMMA | HRePML | MLMM | FarmCPU | GEMMA | |||||
| 1 | 1 | 404108 | 0.01 | 0.4328 | 9.9 | 2.4 | 1.6 | 0.0 | 0.0509 | 0.1334 | 0.1224 | na # |
| 2 | 1 | 636788 | 0.03 | 0.7497 | 45.1 | 39.9 | 53.7 | 0.2 | 0.0193 | 0.0440 | 0.0241 | 0.3112 |
| 3 | 3 | 507976 | 0.03 | 0.7497 | 66.7 | 40.1 | 13.9 | 8.1 | 0.1443 | 0.0992 | 0.2756 | 0.1597 |
| 4 | 3 | 931437 | 0.05 | 0.9679 | 89.5 | 69.5 | 58.8 | 55.3 | 0.0321 | 0.0276 | 0.0434 | 0.0770 |
| 5 | 4 | 75898 | 0.08 | 1.2243 | 100.0 | 99.8 | 100.0 | 97.5 | 0.0407 | 0.0375 | 0.0527 | 0.0283 |
| 6 | 4 | 461978 | 0.01 | 0.4328 | 12.7 | 5.0 | 8.9 | 0.7 | 0.2488 | 0.3808 | 0.3429 | 0.5502 |
| 7 | 4 | 607026 | 0.05 | 0.9679 | 69.6 | 80.9 | 98.5 | 73.8 | 0.0421 | 0.0988 | 0.0367 | 0.1544 |
| 8 | 5 | 282008 | 0.05 | 0.9679 | 89.6 | 87.6 | 90.3 | 55.1 | 0.0397 | 0.0334 | 0.0345 | 0.0725 |
| Statistical Properties | HRePML | MLMM | FarmCPU | GEMMA |
|---|---|---|---|---|
| Average power (%) | 60.39 | 53.15 | 53.21 | 36.34 |
| Average MSE | 0.0772 | 0.1068 | 0.1165 | 0.1933 |
| Running time (Hour) | 3.1419 | 22.7274 | 4.6653 | 2.4186 |
| QTN | R2 | Sample Size: Power (%) | |||
|---|---|---|---|---|---|
| 500 | 1000 | 2000 | 4000 | ||
| 1 | 0.01 | 12 | 13 | 30 | 61 |
| 2 | 0.03 | 48 | 84 | 94 | 99 |
| 3 | 0.03 | 61 | 91 | 93 | 96 |
| 4 | 0.05 | 91 | 99 | 97 | 99 |
| 5 | 0.08 | 100 | 100 | 100 | 100 |
| 6 | 0.01 | 9 | 23 | 46 | 77 |
| 7 | 0.05 | 71 | 98 | 98 | 99 |
| 8 | 0.05 | 87 | 98 | 98 | 98 |
| Average power (%) | 59.88 | 75.75 | 82.00 | 91.13 | |
| Running time (Hour) | 0.3142 | 1.1244 | 3.9969 | 39.5439 | |
| Detected Genes | Associated Trait | Chr. | Position | Effect Estimate | LOD/p-Value | Methods |
|---|---|---|---|---|---|---|
| AT2G16440 | LFS GH | 2 | 7140030 | −7.461, −9.107, −5.16 | FarmCPU, MLMM, GEMMA | |
| AT3G07160 | LFS GH | 3 | 2280271 | −5.934, −8.845 | FarmCPU, MLMM | |
| AT3G54280 | MT GH | 3 | 20090780 | 1.002, 1.762 | FarmCPU, MLMM | |
| AT4G09960 | FT Duration GH | 4 | 6228754 | 0.822, 1.136 | 3.74, | HRePML, FarmCPU |
| AT4G33620 | LC Duration GH | 4 | 16140068 | 2.996, 2.540 | 4.78, | HRePML, MLMM |
| AT5G45900, AT5G45940 | LC Duration GH | 5 | 18625634, 18625726 | −3.707, −6.051 | 4.78, | HRePML, FarmCPU |
| AT5G45900, AT5G45940 | LFS GH | 5 | 18625634, 18625726, 18625726 | −4.318, −5.147, −5.616 | 5.23, | HRePML, FarmCPU, GEMMA |
| AT5G53360 | MT GH | 5 | 21646741 | 0.236, 0.267 | FarmCPU, GEMMA |
Data Availability: The real datasets for this study can be found in the Arabidopsis Information Resource http://www.arabidopsis.org/. The C++ code implement of HRePML is available on https://github.com/wenlongren/HRePML. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, W.; Liang, Z.; He, S.; Xiao, J. Hybrid of Restricted and Penalized Maximum Likelihood Method for Efficient Genome-Wide Association Study. Genes 2020, 11, 1286. https://doi.org/10.3390/genes11111286
Ren W, Liang Z, He S, Xiao J. Hybrid of Restricted and Penalized Maximum Likelihood Method for Efficient Genome-Wide Association Study. Genes. 2020; 11(11):1286. https://doi.org/10.3390/genes11111286
Chicago/Turabian StyleRen, Wenlong, Zhikai Liang, Shu He, and Jing Xiao. 2020. "Hybrid of Restricted and Penalized Maximum Likelihood Method for Efficient Genome-Wide Association Study" Genes 11, no. 11: 1286. https://doi.org/10.3390/genes11111286