Mitochondrial Protein Quality Control Mechanisms
Abstract
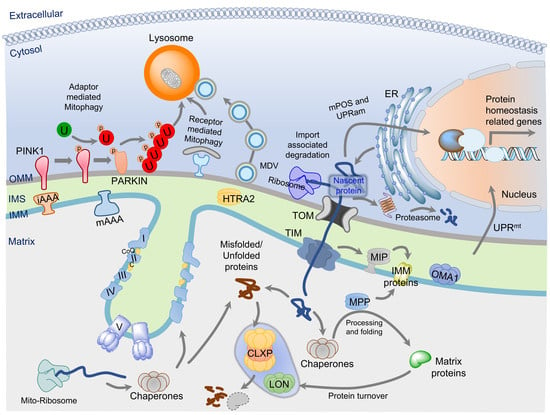
:1. Introduction
2. Mitochondria Proteome
3. Mitochondrial Protein Quality Control System
3.1. Mitochondrial Protein Import and Mitochondria-Associated Degradation Systems
3.2. Chaperone-Mediated Quality Control
3.3. Proteases Involved in MPQC
3.4. Mitophagy and Mitochondria-Derived Vesicles in MPQC
3.5. Mitochondrial Unfolded Protein Response (UPRmt)
4. Conclusions and Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nunnari, J.; Suomalainen, A. Mitochondria: In sickness and in health. Cell 2012, 148, 1145–1159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DiMauro, S.; Schon, E.A. Mitochondrial respiratory-chain diseases. N. Engl. J. Med. 2003, 348, 2656–2668. [Google Scholar] [CrossRef] [PubMed]
- Jadiya, P.; Kolmetzky, D.W.; Tomar, D.; Di Meco, A.; Lombardi, A.A.; Lambert, J.P.; Luongo, T.S.; Ludtmann, M.H.; Pratico, D.; Elrod, J.W. Impaired mitochondrial calcium efflux contributes to disease progression in models of Alzheimer’s disease. Nat. Commun. 2019, 10, 3885. [Google Scholar] [CrossRef] [PubMed]
- Lowell, B.B.; Shulman, G.I. Mitochondrial dysfunction and type 2 diabetes. Science 2005, 307, 384–387. [Google Scholar] [CrossRef] [Green Version]
- Vyas, S.; Zaganjor, E.; Haigis, M.C. Mitochondria and Cancer. Cell 2016, 166, 555–566. [Google Scholar] [CrossRef]
- Gray, M.W.; Burger, G.; Lang, B.F. The origin and early evolution of mitochondria. Genome Biol. 2001, 2, REVIEWS1018. [Google Scholar] [CrossRef] [Green Version]
- Frey, T.G.; Mannella, C.A. The internal structure of mitochondria. Trends Biochem. Sci. 2000, 25, 319–324. [Google Scholar] [CrossRef]
- Friedman, J.R.; Nunnari, J. Mitochondrial form and function. Nature 2014, 505, 335–343. [Google Scholar] [CrossRef] [Green Version]
- Comte, J.; Maisterrena, B.; Gautheron, D.C. Lipid composition and protein profiles of outer and inner membranes from pig heart mitochondria. Comparison with microsomes. Biochim. Biophys. Acta 1976, 419, 271–284. [Google Scholar] [CrossRef]
- Tischler, M.E.; Hecht, P.; Williamson, J.R. Determination of mitochondrial/cytosolic metabolite gradients in isolated rat liver cells by cell disruption. Arch. Biochem. Biophys. 1977, 181, 278–293. [Google Scholar] [CrossRef]
- Schwerzmann, K.; Cruz-Orive, L.M.; Eggman, R.; Sanger, A.; Weibel, E.R. Molecular architecture of the inner membrane of mitochondria from rat liver: A combined biochemical and stereological study. J. Cell Biol. 1986, 102, 97–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caplan, A.I.; Greenawalt, J.W. Biochemical and ultrastructural properties of osmotically lysed rat-liver mitochondria. J. Cell Biol. 1966, 31, 455–472. [Google Scholar] [CrossRef] [PubMed]
- Pagliarini, D.J.; Calvo, S.E.; Chang, B.; Sheth, S.A.; Vafai, S.B.; Ong, S.E.; Walford, G.A.; Sugiana, C.; Boneh, A.; Chen, W.K.; et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell 2008, 134, 112–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, O.; Pfanner, N.; Meisinger, C. Mitochondrial protein import: From proteomics to functional mechanisms. Nat. Rev. Mol. Cell Biol. 2010, 11, 655–667. [Google Scholar] [CrossRef]
- Pfeffer, S.; Woellhaf, M.W.; Herrmann, J.M.; Forster, F. Organization of the mitochondrial translation machinery studied in situ by cryoelectron tomography. Nat. Commun. 2015, 6, 6019. [Google Scholar] [CrossRef] [Green Version]
- Chacinska, A.; Koehler, C.M.; Milenkovic, D.; Lithgow, T.; Pfanner, N. Importing mitochondrial proteins: Machineries and mechanisms. Cell 2009, 138, 628–644. [Google Scholar] [CrossRef] [Green Version]
- Anderson, S.; Bankier, A.T.; Barrell, B.G.; de Bruijn, M.H.; Coulson, A.R.; Drouin, J.; Eperon, I.C.; Nierlich, D.P.; Roe, B.A.; Sanger, F.; et al. Sequence and organization of the human mitochondrial genome. Nature 1981, 290, 457–465. [Google Scholar] [CrossRef]
- Lopez, M.F.; Kristal, B.S.; Chernokalskaya, E.; Lazarev, A.; Shestopalov, A.I.; Bogdanova, A.; Robinson, M. High-throughput profiling of the mitochondrial proteome using affinity fractionation and automation. Electrophoresis 2000, 21, 3427–3440. [Google Scholar] [CrossRef]
- Mootha, V.K.; Bunkenborg, J.; Olsen, J.V.; Hjerrild, M.; Wisniewski, J.R.; Stahl, E.; Bolouri, M.S.; Ray, H.N.; Sihag, S.; Kamal, M.; et al. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell 2003, 115, 629–640. [Google Scholar] [CrossRef] [Green Version]
- Gaston, D.; Tsaousis, A.D.; Roger, A.J. Predicting proteomes of mitochondria and related organelles from genomic and expressed sequence tag data. Methods Enzymol. 2009, 457, 21–47. [Google Scholar] [CrossRef]
- Ozawa, T.; Sako, Y.; Sato, M.; Kitamura, T.; Umezawa, Y. A genetic approach to identifying mitochondrial proteins. Nat. Biotechnol. 2003, 21, 287–293. [Google Scholar] [CrossRef]
- Smith, A.C.; Blackshaw, J.A.; Robinson, A.J. MitoMiner: A data warehouse for mitochondrial proteomics data. Nucleic Acids Res. 2012, 40, D1160–D1167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rhee, H.W.; Zou, P.; Udeshi, N.D.; Martell, J.D.; Mootha, V.K.; Carr, S.A.; Ting, A.Y. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 2013, 339, 1328–1331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calvo, S.E.; Clauser, K.R.; Mootha, V.K. MitoCarta2.0: An updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016, 44, D1251–D1257. [Google Scholar] [CrossRef] [PubMed]
- Elstner, M.; Andreoli, C.; Klopstock, T.; Meitinger, T.; Prokisch, H. The mitochondrial proteome database: MitoP2. Methods Enzymol. 2009, 457, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Ingman, M.; Gyllensten, U. mtDB: Human Mitochondrial Genome Database, a resource for population genetics and medical sciences. Nucleic Acids Res. 2006, 34, D749–D751. [Google Scholar] [CrossRef]
- Vogtle, F.N.; Burkhart, J.M.; Gonczarowska-Jorge, H.; Kucukkose, C.; Taskin, A.A.; Kopczynski, D.; Ahrends, R.; Mossmann, D.; Sickmann, A.; Zahedi, R.P.; et al. Landscape of submitochondrial protein distribution. Nat. Commun. 2017, 8, 290. [Google Scholar] [CrossRef] [Green Version]
- Hung, V.; Lam, S.S.; Udeshi, N.D.; Svinkina, T.; Guzman, G.; Mootha, V.K.; Carr, S.A.; Ting, A.Y. Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. Elife 2017, 6, e24463. [Google Scholar] [CrossRef] [Green Version]
- Zahedi, R.P.; Sickmann, A.; Boehm, A.M.; Winkler, C.; Zufall, N.; Schonfisch, B.; Guiard, B.; Pfanner, N.; Meisinger, C. Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Mol. Biol. Cell 2006, 17, 1436–1450. [Google Scholar] [CrossRef] [Green Version]
- Hung, V.; Zou, P.; Rhee, H.W.; Udeshi, N.D.; Cracan, V.; Svinkina, T.; Carr, S.A.; Mootha, V.K.; Ting, A.Y. Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging. Mol. Cell 2014, 55, 332–341. [Google Scholar] [CrossRef] [Green Version]
- Vogtle, F.N.; Burkhart, J.M.; Rao, S.; Gerbeth, C.; Hinrichs, J.; Martinou, J.C.; Chacinska, A.; Sickmann, A.; Zahedi, R.P.; Meisinger, C. Intermembrane space proteome of yeast mitochondria. Mol. Cell Proteom. 2012, 11, 1840–1852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- John, G.B.; Shang, Y.; Li, L.; Renken, C.; Mannella, C.A.; Selker, J.M.; Rangell, L.; Bennett, M.J.; Zha, J. The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol. Biol. Cell 2005, 16, 1543–1554. [Google Scholar] [CrossRef] [PubMed]
- Baughman, J.M.; Perocchi, F.; Girgis, H.S.; Plovanich, M.; Belcher-Timme, C.A.; Sancak, Y.; Bao, X.R.; Strittmatter, L.; Goldberger, O.; Bogorad, R.L.; et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature 2011, 476, 341–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Stefani, D.; Raffaello, A.; Teardo, E.; Szabo, I.; Rizzuto, R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature 2011, 476, 336–340. [Google Scholar] [CrossRef]
- Harner, M.; Korner, C.; Walther, D.; Mokranjac, D.; Kaesmacher, J.; Welsch, U.; Griffith, J.; Mann, M.; Reggiori, F.; Neupert, W. The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J. 2011, 30, 4356–4370. [Google Scholar] [CrossRef] [Green Version]
- Haworth, R.A.; Hunter, D.R. The Ca2+-induced membrane transition in mitochondria. II. Nature of the Ca2+ trigger site. Arch. Biochem. Biophys. 1979, 195, 460–467. [Google Scholar] [CrossRef]
- Crompton, M.; Costi, A.; Hayat, L. Evidence for the presence of a reversible Ca2+-dependent pore activated by oxidative stress in heart mitochondria. Biochem. J. 1987, 245, 915–918. [Google Scholar] [CrossRef] [Green Version]
- Hirst, J.; Carroll, J.; Fearnley, I.M.; Shannon, R.J.; Walker, J.E. The nuclear encoded subunits of complex I from bovine heart mitochondria. Biochim. Biophys. Acta 2003, 1604, 135–150. [Google Scholar] [CrossRef] [Green Version]
- Bourges, I.; Ramus, C.; Mousson de Camaret, B.; Beugnot, R.; Remacle, C.; Cardol, P.; Hofhaus, G.; Issartel, J.P. Structural organization of mitochondrial human complex I: Role of the ND4 and ND5 mitochondria-encoded subunits and interaction with prohibitin. Biochem. J. 2004, 383, 491–499. [Google Scholar] [CrossRef] [Green Version]
- Schagger, H.; Pfeiffer, K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000, 19, 1777–1783. [Google Scholar] [CrossRef] [Green Version]
- Rathore, S.; Berndtsson, J.; Marin-Buera, L.; Conrad, J.; Carroni, M.; Brzezinski, P.; Ott, M. Cryo-EM structure of the yeast respiratory supercomplex. Nat. Struct. Mol. Biol. 2019, 26, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Cruciat, C.M.; Brunner, S.; Baumann, F.; Neupert, W.; Stuart, R.A. The cytochrome bc1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria. J. Biol. Chem. 2000, 275, 18093–18098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bianchi, C.; Genova, M.L.; Parenti Castelli, G.; Lenaz, G. The mitochondrial respiratory chain is partially organized in a supercomplex assembly: Kinetic evidence using flux control analysis. J. Biol. Chem. 2004, 279, 36562–36569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lapuente-Brun, E.; Moreno-Loshuertos, R.; Acin-Perez, R.; Latorre-Pellicer, A.; Colas, C.; Balsa, E.; Perales-Clemente, E.; Quiros, P.M.; Calvo, E.; Rodriguez-Hernandez, M.A.; et al. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science 2013, 340, 1567–1570. [Google Scholar] [CrossRef]
- Zhang, S.; Reljic, B.; Liang, C.; Kerouanton, B.; Francisco, J.C.; Peh, J.H.; Mary, C.; Jagannathan, N.S.; Olexiouk, V.; Tang, C.; et al. Mitochondrial peptide BRAWNIN is essential for vertebrate respiratory complex III assembly. Nat. Commun. 2020, 11, 1312. [Google Scholar] [CrossRef]
- Chu, Q.; Martinez, T.F.; Novak, S.W.; Donaldson, C.J.; Tan, D.; Vaughan, J.M.; Chang, T.; Diedrich, J.K.; Andrade, L.; Kim, A.; et al. Regulation of the ER stress response by a mitochondrial microprotein. Nat. Commun. 2019, 10, 4883. [Google Scholar] [CrossRef] [Green Version]
- Rathore, A.; Chu, Q.; Tan, D.; Martinez, T.F.; Donaldson, C.J.; Diedrich, J.K.; Yates, J.R., 3rd; Saghatelian, A. MIEF1 Microprotein Regulates Mitochondrial Translation. Biochemistry 2018, 57, 5564–5575. [Google Scholar] [CrossRef]
- Stein, C.S.; Jadiya, P.; Zhang, X.; McLendon, J.M.; Abouassaly, G.M.; Witmer, N.H.; Anderson, E.J.; Elrod, J.W.; Boudreau, R.L. Mitoregulin: A lncRNA-Encoded Microprotein that Supports Mitochondrial Supercomplexes and Respiratory Efficiency. Cell Rep. 2018, 23, 3710–3720.e3718. [Google Scholar] [CrossRef]
- Makarewich, C.A.; Baskin, K.K.; Munir, A.Z.; Bezprozvannaya, S.; Sharma, G.; Khemtong, C.; Shah, A.M.; McAnally, J.R.; Malloy, C.R.; Szweda, L.I.; et al. MOXI Is a Mitochondrial Micropeptide That Enhances Fatty Acid β-Oxidation. Cell Rep. 2018, 23, 3701–3709. [Google Scholar] [CrossRef]
- Akimoto, C.; Sakashita, E.; Kasashima, K.; Kuroiwa, K.; Tominaga, K.; Hamamoto, T.; Endo, H. Translational repression of the McKusick-Kaufman syndrome transcript by unique upstream open reading frames encoding mitochondrial proteins with alternative polyadenylation sites. Biochim. Biophys. Acta 2013, 1830, 2728–2738. [Google Scholar] [CrossRef]
- Van Heesch, S.; Witte, F.; Schneider-Lunitz, V.; Schulz, J.F.; Adami, E.; Faber, A.B.; Kirchner, M.; Maatz, H.; Blachut, S.; Sandmann, C.L.; et al. The Translational Landscape of the Human Heart. Cell 2019, 178, 242–260.e229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiebler, M.; Pfaller, R.; Sollner, T.; Griffiths, G.; Horstmann, H.; Pfanner, N.; Neupert, W. Identification of a mitochondrial receptor complex required for recognition and membrane insertion of precursor proteins. Nature 1990, 348, 610–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiedemann, N.; Pfanner, N. Mitochondrial Machineries for Protein Import and Assembly. Annu. Rev. Biochem. 2017, 86, 685–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mokranjac, D.; Neupert, W. Cell biology: Architecture of a protein entry gate. Nature 2015, 528, 201–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogtle, F.N.; Wortelkamp, S.; Zahedi, R.P.; Becker, D.; Leidhold, C.; Gevaert, K.; Kellermann, J.; Voos, W.; Sickmann, A.; Pfanner, N.; et al. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell 2009, 139, 428–439. [Google Scholar] [CrossRef]
- Araiso, Y.; Tsutsumi, A.; Qiu, J.; Imai, K.; Shiota, T.; Song, J.; Lindau, C.; Wenz, L.S.; Sakaue, H.; Yunoki, K.; et al. Structure of the mitochondrial import gate reveals distinct preprotein paths. Nature 2019, 575, 395–401. [Google Scholar] [CrossRef]
- Wiedemann, N.; Kozjak, V.; Chacinska, A.; Schonfisch, B.; Rospert, S.; Ryan, M.T.; Pfanner, N.; Meisinger, C. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature 2003, 424, 565–571. [Google Scholar] [CrossRef]
- Chacinska, A.; Pfannschmidt, S.; Wiedemann, N.; Kozjak, V.; Sanjuan Szklarz, L.K.; Schulze-Specking, A.; Truscott, K.N.; Guiard, B.; Meisinger, C.; Pfanner, N. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 2004, 23, 3735–3746. [Google Scholar] [CrossRef] [Green Version]
- Allen, S.; Balabanidou, V.; Sideris, D.P.; Lisowsky, T.; Tokatlidis, K. Erv1 mediates the Mia40-dependent protein import pathway and provides a functional link to the respiratory chain by shuttling electrons to cytochrome c. J. Mol. Biol. 2005, 353, 937–944. [Google Scholar] [CrossRef]
- Heo, J.M.; Livnat-Levanon, N.; Taylor, E.B.; Jones, K.T.; Dephoure, N.; Ring, J.; Xie, J.; Brodsky, J.L.; Madeo, F.; Gygi, S.P.; et al. A stress-responsive system for mitochondrial protein degradation. Mol. Cell 2010, 40, 465–480. [Google Scholar] [CrossRef] [Green Version]
- Izawa, T.; Park, S.H.; Zhao, L.; Hartl, F.U.; Neupert, W. Cytosolic Protein Vms1 Links Ribosome Quality Control to Mitochondrial and Cellular Homeostasis. Cell 2017, 171, 890–903.e18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruan, L.; Zhou, C.; Jin, E.; Kucharavy, A.; Zhang, Y.; Wen, Z.; Florens, L.; Li, R. Cytosolic proteostasis through importing of misfolded proteins into mitochondria. Nature 2017, 543, 443–446. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, X.J. A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature 2015, 524, 481–484. [Google Scholar] [CrossRef] [Green Version]
- Wrobel, L.; Topf, U.; Bragoszewski, P.; Wiese, S.; Sztolsztener, M.E.; Oeljeklaus, S.; Varabyova, A.; Lirski, M.; Chroscicki, P.; Mroczek, S.; et al. Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature 2015, 524, 485–488. [Google Scholar] [CrossRef]
- Weidberg, H.; Amon, A. MitoCPR-A surveillance pathway that protects mitochondria in response to protein import stress. Science 2018, 360, eaan4146. [Google Scholar] [CrossRef] [Green Version]
- Martensson, C.U.; Priesnitz, C.; Song, J.; Ellenrieder, L.; Doan, K.N.; Boos, F.; Floerchinger, A.; Zufall, N.; Oeljeklaus, S.; Warscheid, B.; et al. Mitochondrial protein translocation-associated degradation. Nature 2019, 569, 679–683. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.M.; Leboucher, G.P.; Livnat-Levanon, N.; Glickman, M.H.; Weissman, A.M. Ubiquitin-proteasome-dependent degradation of a mitofusin, a critical regulator of mitochondrial fusion. Mol. Biol. Cell 2008, 19, 2457–2464. [Google Scholar] [CrossRef] [Green Version]
- Brandman, O.; Hegde, R.S. Ribosome-associated protein quality control. Nat. Struct. Mol. Biol. 2016, 23, 7–15. [Google Scholar] [CrossRef]
- Shao, S.; Brown, A.; Santhanam, B.; Hegde, R.S. Structure and assembly pathway of the ribosome quality control complex. Mol. Cell 2015, 57, 433–444. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.C.; Umanah, G.K.; Dephoure, N.; Andrabi, S.A.; Gygi, S.P.; Dawson, T.M.; Dawson, V.L.; Rutter, J. Msp1/ATAD1 maintains mitochondrial function by facilitating the degradation of mislocalized tail-anchored proteins. EMBO J. 2014, 33, 1548–1564. [Google Scholar] [CrossRef]
- Okreglak, V.; Walter, P. The conserved AAA-ATPase Msp1 confers organelle specificity to tail-anchored proteins. Proc. Natl. Acad. Sci. USA 2014, 111, 8019–8024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehmann, G.; Ziv, T.; Braten, O.; Admon, A.; Udasin, R.G.; Ciechanover, A. Ubiquitination of specific mitochondrial matrix proteins. Biochem. Biophys. Res. Commun. 2016, 475, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Chen, J. The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat. Struct. Mol. Biol. 2012, 19, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef]
- Schwartz, A.L.; Trausch, J.S.; Ciechanover, A.; Slot, J.W.; Geuze, H. Immunoelectron microscopic localization of the ubiquitin-activating enzyme E1 in HepG2 cells. Proc. Natl. Acad. Sci. USA 1992, 89, 5542–5546. [Google Scholar] [CrossRef] [Green Version]
- Patil, H.; Yoon, D.; Bhowmick, R.; Cai, Y.; Cho, K.I.; Ferreira, P.A. Impairments in age-dependent ubiquitin proteostasis and structural integrity of selective neurons by uncoupling Ran GTPase from the Ran-binding domain 3 of Ranbp2 and identification of novel mitochondrial isoforms of ubiquitin-conjugating enzyme E2I (ubc9) and Ranbp2. Small GTPases 2019, 10, 146–161. [Google Scholar] [CrossRef]
- Lavie, J.; De Belvalet, H.; Sonon, S.; Ion, A.M.; Dumon, E.; Melser, S.; Lacombe, D.; Dupuy, J.W.; Lalou, C.; Benard, G. Ubiquitin-Dependent Degradation of Mitochondrial Proteins Regulates Energy Metabolism. Cell Rep. 2018, 23, 2852–2863. [Google Scholar] [CrossRef]
- Jeon, H.B.; Choi, E.S.; Yoon, J.H.; Hwang, J.H.; Chang, J.W.; Lee, E.K.; Choi, H.W.; Park, Z.Y.; Yoo, Y.J. A proteomics approach to identify the ubiquitinated proteins in mouse heart. Biochem. Biophys. Res. Commun. 2007, 357, 731–736. [Google Scholar] [CrossRef]
- Buneeva, O.; Kopylov, A.; Kapitsa, I.; Ivanova, E.; Zgoda, V.; Medvedev, A. The Effect of Neurotoxin MPTP and Neuroprotector Isatin on the Profile of Ubiquitinated Brain Mitochondrial Proteins. Cells 2018, 7, 91. [Google Scholar] [CrossRef] [Green Version]
- Voos, W. Chaperone-protease networks in mitochondrial protein homeostasis. Biochim. Biophys. Acta 2013, 1833, 388–399. [Google Scholar] [CrossRef] [Green Version]
- Deocaris, C.C.; Kaul, S.C.; Wadhwa, R. On the brotherhood of the mitochondrial chaperones mortalin and heat shock protein 60. Cell Stress Chaperones 2006, 11, 116–128. [Google Scholar] [CrossRef] [PubMed]
- Dutkiewicz, R.; Schilke, B.; Knieszner, H.; Walter, W.; Craig, E.A.; Marszalek, J. Ssq1, a mitochondrial Hsp70 involved in iron-sulfur (Fe/S) center biogenesis. Similarities to and differences from its bacterial counterpart. J. Biol. Chem. 2003, 278, 29719–29727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voisine, C.; Craig, E.A.; Zufall, N.; von Ahsen, O.; Pfanner, N.; Voos, W. The protein import motor of mitochondria: Unfolding and trapping of preproteins are distinct and separable functions of matrix Hsp70. Cell 1999, 97, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Gambill, B.D.; Voos, W.; Kang, P.J.; Miao, B.; Langer, T.; Craig, E.A.; Pfanner, N. A dual role for mitochondrial heat shock protein 70 in membrane translocation of preproteins. J. Cell Biol. 1993, 123, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Flaherty, K.M.; DeLuca-Flaherty, C.; McKay, D.B. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature 1990, 346, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Qi, R.; Sarbeng, E.B.; Liu, Q.; Le, K.Q.; Xu, X.; Xu, H.; Yang, J.; Wong, J.L.; Vorvis, C.; Hendrickson, W.A.; et al. Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP. Nat. Struct. Mol. Biol. 2013, 20, 900–907. [Google Scholar] [CrossRef] [Green Version]
- Craig, E.A.; Huang, P.; Aron, R.; Andrew, A. The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev. Physiol. Biochem. Pharmacol. 2006, 156, 1–21. [Google Scholar] [CrossRef]
- Rassow, J.; Voos, W.; Pfanner, N. Partner proteins determine multiple functions of Hsp70. Trends Cell Biol. 1995, 5, 207–212. [Google Scholar] [CrossRef]
- Lu, B.; Garrido, N.; Spelbrink, J.N.; Suzuki, C.K. Tid1 isoforms are mitochondrial DnaJ-like chaperones with unique carboxyl termini that determine cytosolic fate. J. Biol. Chem. 2006, 281, 13150–13158. [Google Scholar] [CrossRef] [Green Version]
- Cheng, M.Y.; Hartl, F.U.; Horwich, A.L. The mitochondrial chaperonin hsp60 is required for its own assembly. Nature 1990, 348, 455–458. [Google Scholar] [CrossRef]
- Cheng, M.Y.; Hartl, F.U.; Martin, J.; Pollock, R.A.; Kalousek, F.; Neupert, W.; Hallberg, E.M.; Hallberg, R.L.; Horwich, A.L. Mitochondrial heat-shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria. Nature 1989, 337, 620–625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rospert, S.; Looser, R.; Dubaquie, Y.; Matouschek, A.; Glick, B.S.; Schatz, G. Hsp60-independent protein folding in the matrix of yeast mitochondria. EMBO J. 1996, 15, 764–774. [Google Scholar] [CrossRef] [PubMed]
- Cechetto, J.D.; Gupta, R.S. Immunoelectron microscopy provides evidence that tumor necrosis factor receptor-associated protein 1 (TRAP-1) is a mitochondrial protein which also localizes at specific extramitochondrial sites. Exp. Cell Res. 2000, 260, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Felts, S.J.; Owen, B.A.; Nguyen, P.; Trepel, J.; Donner, D.B.; Toft, D.O. The hsp90-related protein TRAP1 is a mitochondrial protein with distinct functional properties. J. Biol. Chem. 2000, 275, 3305–3312. [Google Scholar] [CrossRef] [Green Version]
- Altieri, D.C.; Stein, G.S.; Lian, J.B.; Languino, L.R. TRAP-1, the mitochondrial Hsp90. Biochim. Biophys. Acta 2012, 1823, 767–773. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Sowa, M.E.; Choi, J.M.; Tsai, F.T. The ClpB/Hsp104 molecular chaperone-a protein disaggregating machine. J. Struct. Biol. 2004, 146, 99–105. [Google Scholar] [CrossRef]
- Wortmann, S.B.; Zietkiewicz, S.; Kousi, M.; Szklarczyk, R.; Haack, T.B.; Gersting, S.W.; Muntau, A.C.; Rakovic, A.; Renkema, G.H.; Rodenburg, R.J.; et al. CLPB mutations cause 3-methylglutaconic aciduria, progressive brain atrophy, intellectual disability, congenital neutropenia, cataracts, movement disorder. Am. J. Hum. Genet. 2015, 96, 245–257. [Google Scholar] [CrossRef] [Green Version]
- Quiros, P.M.; Langer, T.; Lopez-Otin, C. New roles for mitochondrial proteases in health, ageing and disease. Nat. Rev. Mol. Cell Biol. 2015, 16, 345–359. [Google Scholar] [CrossRef]
- Deshwal, S.; Fiedler, K.U.; Langer, T. Mitochondrial Proteases: Multifaceted Regulators of Mitochondrial Plasticity. Annu. Rev. Biochem. 2020, 89. [Google Scholar] [CrossRef] [Green Version]
- Serero, A.; Giglione, C.; Sardini, A.; Martinez-Sanz, J.; Meinnel, T. An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway. J. Biol. Chem. 2003, 278, 52953–52963. [Google Scholar] [CrossRef] [Green Version]
- Tsai, C.W.; Wu, Y.; Pao, P.C.; Phillips, C.B.; Williams, C.; Miller, C.; Ranaghan, M.; Tsai, M.F. Proteolytic control of the mitochondrial calcium uniporter complex. Proc. Natl. Acad. Sci. USA 2017, 114, 4388–4393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konig, T.; Troder, S.E.; Bakka, K.; Korwitz, A.; Richter-Dennerlein, R.; Lampe, P.A.; Patron, M.; Muhlmeister, M.; Guerrero-Castillo, S.; Brandt, U.; et al. The m-AAA Protease Associated with Neurodegeneration Limits MCU Activity in Mitochondria. Mol. Cell 2016, 64, 148–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, X.; Neupert, W.; Tzagoloff, A. The metalloprotease encoded by ATP23 has a dual function in processing and assembly of subunit 6 of mitochondrial ATPase. Mol. Biol. Cell 2007, 18, 617–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anand, R.; Wai, T.; Baker, M.J.; Kladt, N.; Schauss, A.C.; Rugarli, E.; Langer, T. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J. Cell Biol. 2014, 204, 919–929. [Google Scholar] [CrossRef] [PubMed]
- Kaser, M.; Kambacheld, M.; Kisters-Woike, B.; Langer, T. Oma1, a novel membrane-bound metallopeptidase in mitochondria with activities overlapping with the m-AAA protease. J. Biol. Chem. 2003, 278, 46414–46423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, Y.; Imai, Y.; Nakayama, H.; Takahashi, K.; Takio, K.; Takahashi, R. A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol. Cell 2001, 8, 613–621. [Google Scholar] [CrossRef]
- Chao, J.R.; Parganas, E.; Boyd, K.; Hong, C.Y.; Opferman, J.T.; Ihle, J.N. Hax1-mediated processing of HtrA2 by Parl allows survival of lymphocytes and neurons. Nature 2008, 452, 98–102. [Google Scholar] [CrossRef]
- Bingol, B.; Tea, J.S.; Phu, L.; Reichelt, M.; Bakalarski, C.E.; Song, Q.; Foreman, O.; Kirkpatrick, D.S.; Sheng, M. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature 2014, 510, 370–375. [Google Scholar] [CrossRef]
- Klionsky, D.J. Autophagy: From phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 2007, 8, 931–937. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Acevedo Arozena, A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef] [Green Version]
- Hale, A.N.; Ledbetter, D.J.; Gawriluk, T.R.; Rucker, E.B., 3rd. Autophagy: Regulation and role in development. Autophagy 2013, 9, 951–972. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of cells and tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, L.; Chen, Y.; Tooze, S.A. Autophagy pathway: Cellular and molecular mechanisms. Autophagy 2018, 14, 207–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, S.L., Jr. Cellular differentiation in the kidneys of newborn mice studies with the electron microscope. J. Biophys. Biochem. Cytol. 1957, 3, 349–362. [Google Scholar] [CrossRef] [Green Version]
- Pickles, S.; Vigie, P.; Youle, R.J. Mitophagy and Quality Control Mechanisms in Mitochondrial Maintenance. Curr. Biol. 2018, 28, R170–R185. [Google Scholar] [CrossRef]
- Zhang, Y.; Qi, H.; Taylor, R.; Xu, W.; Liu, L.F.; Jin, S. The role of autophagy in mitochondria maintenance: Characterization of mitochondrial functions in autophagy-deficient S. cerevisiae strains. Autophagy 2007, 3, 337–346. [Google Scholar] [CrossRef] [Green Version]
- Mijaljica, D.; Prescott, M.; Devenish, R.J. Different fates of mitochondria: Alternative ways for degradation? Autophagy 2007, 3, 4–9. [Google Scholar] [CrossRef] [Green Version]
- Okamoto, K.; Kondo-Okamoto, N.; Ohsumi, Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell 2009, 17, 87–97. [Google Scholar] [CrossRef] [Green Version]
- Castenholz, A. Functional microanatomy of initial lymphatics with special consideration of the extracellular matrix. Lymphology 1998, 31, 101–118. [Google Scholar]
- Murakawa, T.; Yamaguchi, O.; Hashimoto, A.; Hikoso, S.; Takeda, T.; Oka, T.; Yasui, H.; Ueda, H.; Akazawa, Y.; Nakayama, H.; et al. Bcl-2-like protein 13 is a mammalian Atg32 homologue that mediates mitophagy and mitochondrial fragmentation. Nat. Commun. 2015, 6, 7527. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Feng, D.; Chen, G.; Chen, M.; Zheng, Q.; Song, P.; Ma, Q.; Zhu, C.; Wang, R.; Qi, W.; et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat. Cell Biol. 2012, 14, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Novak, I.; Kirkin, V.; McEwan, D.G.; Zhang, J.; Wild, P.; Rozenknop, A.; Rogov, V.; Lohr, F.; Popovic, D.; Occhipinti, A.; et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010, 11, 45–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tracy, K.; Dibling, B.C.; Spike, B.T.; Knabb, J.R.; Schumacker, P.; Macleod, K.F. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol. Cell Biol. 2007, 27, 6229–6242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rikka, S.; Quinsay, M.N.; Thomas, R.L.; Kubli, D.A.; Zhang, X.; Murphy, A.N.; Gustafsson, A.B. Bnip3 impairs mitochondrial bioenergetics and stimulates mitochondrial turnover. Cell Death Differ. 2011, 18, 721–731. [Google Scholar] [CrossRef] [Green Version]
- Kitada, T.; Asakawa, S.; Hattori, N.; Matsumine, H.; Yamamura, Y.; Minoshima, S.; Yokochi, M.; Mizuno, Y.; Shimizu, N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 1998, 392, 605–608. [Google Scholar] [CrossRef]
- Valente, E.M.; Abou-Sleiman, P.M.; Caputo, V.; Muqit, M.M.; Harvey, K.; Gispert, S.; Ali, Z.; Del Turco, D.; Bentivoglio, A.R.; Healy, D.G.; et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science 2004, 304, 1158–1160. [Google Scholar] [CrossRef] [Green Version]
- Greene, J.C.; Whitworth, A.J.; Kuo, I.; Andrews, L.A.; Feany, M.B.; Pallanck, L.J. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc. Natl. Acad. Sci. USA 2003, 100, 4078–4083. [Google Scholar] [CrossRef] [Green Version]
- Clark, I.E.; Dodson, M.W.; Jiang, C.; Cao, J.H.; Huh, J.R.; Seol, J.H.; Yoo, S.J.; Hay, B.A.; Guo, M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 2006, 441, 1162–1166. [Google Scholar] [CrossRef]
- Park, J.; Lee, S.B.; Lee, S.; Kim, Y.; Song, S.; Kim, S.; Bae, E.; Kim, J.; Shong, M.; Kim, J.M.; et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 2006, 441, 1157–1161. [Google Scholar] [CrossRef]
- Wang, X.; Winter, D.; Ashrafi, G.; Schlehe, J.; Wong, Y.L.; Selkoe, D.; Rice, S.; Steen, J.; LaVoie, M.J.; Schwarz, T.L. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell 2011, 147, 893–906. [Google Scholar] [CrossRef] [Green Version]
- Morais, V.A.; Haddad, D.; Craessaerts, K.; De Bock, P.J.; Swerts, J.; Vilain, S.; Aerts, L.; Overbergh, L.; Grunewald, A.; Seibler, P.; et al. PINK1 loss-of-function mutations affect mitochondrial complex I activity via NdufA10 ubiquinone uncoupling. Science 2014, 344, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Narendra, D.; Tanaka, A.; Suen, D.F.; Youle, R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008, 183, 795–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, S.M.; Lazarou, M.; Wang, C.; Kane, L.A.; Narendra, D.P.; Youle, R.J. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J. Cell Biol. 2010, 191, 933–942. [Google Scholar] [CrossRef] [Green Version]
- Matsuda, N.; Sato, S.; Shiba, K.; Okatsu, K.; Saisho, K.; Gautier, C.A.; Sou, Y.S.; Saiki, S.; Kawajiri, S.; Sato, F.; et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 2010, 189, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Yamano, K.; Youle, R.J. PINK1 is degraded through the N-end rule pathway. Autophagy 2013, 9, 1758–1769. [Google Scholar] [CrossRef] [Green Version]
- Koyano, F.; Okatsu, K.; Kosako, H.; Tamura, Y.; Go, E.; Kimura, M.; Kimura, Y.; Tsuchiya, H.; Yoshihara, H.; Hirokawa, T.; et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 2014, 510, 162–166. [Google Scholar] [CrossRef]
- Kane, L.A.; Lazarou, M.; Fogel, A.I.; Li, Y.; Yamano, K.; Sarraf, S.A.; Banerjee, S.; Youle, R.J. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 2014, 205, 143–153. [Google Scholar] [CrossRef]
- Lazarou, M.; Sliter, D.A.; Kane, L.A.; Sarraf, S.A.; Wang, C.; Burman, J.L.; Sideris, D.P.; Fogel, A.I.; Youle, R.J. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 2015, 524, 309–314. [Google Scholar] [CrossRef] [Green Version]
- Jin, S.M.; Youle, R.J. The accumulation of misfolded proteins in the mitochondrial matrix is sensed by PINK1 to induce PARK2/Parkin-mediated mitophagy of polarized mitochondria. Autophagy 2013, 9, 1750–1757. [Google Scholar] [CrossRef] [Green Version]
- Sugiura, A.; McLelland, G.L.; Fon, E.A.; McBride, H.M. A new pathway for mitochondrial quality control: Mitochondrial-derived vesicles. EMBO J. 2014, 33, 2142–2156. [Google Scholar] [CrossRef] [Green Version]
- Soubannier, V.; Rippstein, P.; Kaufman, B.A.; Shoubridge, E.A.; McBride, H.M. Reconstitution of mitochondria derived vesicle formation demonstrates selective enrichment of oxidized cargo. PLoS ONE 2012, 7, e52830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neuspiel, M.; Schauss, A.C.; Braschi, E.; Zunino, R.; Rippstein, P.; Rachubinski, R.A.; Andrade-Navarro, M.A.; McBride, H.M. Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr. Biol. 2008, 18, 102–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soubannier, V.; McLelland, G.L.; Zunino, R.; Braschi, E.; Rippstein, P.; Fon, E.A.; McBride, H.M. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr. Biol. 2012, 22, 135–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinus, R.D.; Garth, G.P.; Webster, T.L.; Cartwright, P.; Naylor, D.J.; Hoj, P.B.; Hoogenraad, N.J. Selective induction of mitochondrial chaperones in response to loss of the mitochondrial genome. Eur. J. Biochem. 1996, 240, 98–103. [Google Scholar] [CrossRef]
- Yoneda, T.; Benedetti, C.; Urano, F.; Clark, S.G.; Harding, H.P.; Ron, D. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J. Cell Sci. 2004, 117, 4055–4066. [Google Scholar] [CrossRef] [Green Version]
- King, M.P.; Attardi, G. Human cells lacking mtDNA: Repopulation with exogenous mitochondria by complementation. Science 1989, 246, 500–503. [Google Scholar] [CrossRef]
- Benedetti, C.; Haynes, C.M.; Yang, Y.; Harding, H.P.; Ron, D. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics 2006, 174, 229–239. [Google Scholar] [CrossRef] [Green Version]
- Haynes, C.M.; Yang, Y.; Blais, S.P.; Neubert, T.A.; Ron, D. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol. Cell 2010, 37, 529–540. [Google Scholar] [CrossRef] [Green Version]
- Nargund, A.M.; Pellegrino, M.W.; Fiorese, C.J.; Baker, B.M.; Haynes, C.M. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 2012, 337, 587–590. [Google Scholar] [CrossRef] [Green Version]
- Baker, B.M.; Nargund, A.M.; Sun, T.; Haynes, C.M. Protective coupling of mitochondrial function and protein synthesis via the eIF2alpha kinase GCN-2. PLoS Genet. 2012, 8, e1002760. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Q.; Wang, J.; Levichkin, I.V.; Stasinopoulos, S.; Ryan, M.T.; Hoogenraad, N.J. A mitochondrial specific stress response in mammalian cells. EMBO J. 2002, 21, 4411–4419. [Google Scholar] [CrossRef] [PubMed]
- Aldridge, J.E.; Horibe, T.; Hoogenraad, N.J. Discovery of genes activated by the mitochondrial unfolded protein response (mtUPR) and cognate promoter elements. PLoS ONE 2007, 2, e874. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.K.; Ryu, D.; Kim, K.S.; Chang, J.Y.; Kim, Y.K.; Yi, H.S.; Kang, S.G.; Choi, M.J.; Lee, S.E.; Jung, S.B.; et al. Growth differentiation factor 15 is a myomitokine governing systemic energy homeostasis. J. Cell Biol. 2017, 216, 149–165. [Google Scholar] [CrossRef] [PubMed]
- Munch, C.; Harper, J.W. Mitochondrial unfolded protein response controls matrix pre-RNA processing and translation. Nature 2016, 534, 710–713. [Google Scholar] [CrossRef] [Green Version]
- Dogan, S.A.; Pujol, C.; Maiti, P.; Kukat, A.; Wang, S.; Hermans, S.; Senft, K.; Wibom, R.; Rugarli, E.I.; Trifunovic, A. Tissue-specific loss of DARS2 activates stress responses independently of respiratory chain deficiency in the heart. Cell Metab. 2014, 19, 458–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCullough, K.D.; Martindale, J.L.; Klotz, L.O.; Aw, T.Y.; Holbrook, N.J. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol. Cell Biol. 2001, 21, 1249–1259. [Google Scholar] [CrossRef] [Green Version]
- Horibe, T.; Hoogenraad, N.J. The chop gene contains an element for the positive regulation of the mitochondrial unfolded protein response. PLoS ONE 2007, 2, e835. [Google Scholar] [CrossRef] [Green Version]
- Jaeschke, A.; Karasarides, M.; Ventura, J.J.; Ehrhardt, A.; Zhang, C.; Flavell, R.A.; Shokat, K.M.; Davis, R.J. JNK2 is a positive regulator of the cJun transcription factor. Mol. Cell 2006, 23, 899–911. [Google Scholar] [CrossRef]
- Lakdja, F.; Lobera, A.; Faucher, A.; Renaud Salis, J.L.; Becouarn, Y.; Ravaud, A. Symptomatic treatment of pain in cervico-facial cancers. Rev. Laryngol. Otol. Rhinol. Bord. 1992, 113, 165–171. [Google Scholar]
- Fiorese, C.J.; Schulz, A.M.; Lin, Y.F.; Rosin, N.; Pellegrino, M.W.; Haynes, C.M. The Transcription Factor ATF5 Mediates a Mammalian Mitochondrial UPR. Curr. Biol. 2016, 26, 2037–2043. [Google Scholar] [CrossRef] [Green Version]
- Taniuchi, S.; Miyake, M.; Tsugawa, K.; Oyadomari, M.; Oyadomari, S. Integrated stress response of vertebrates is regulated by four eIF2alpha kinases. Sci. Rep. 2016, 6, 32886. [Google Scholar] [CrossRef] [PubMed]
- Harding, H.P.; Zhang, Y.; Zeng, H.; Novoa, I.; Lu, P.D.; Calfon, M.; Sadri, N.; Yun, C.; Popko, B.; Paules, R.; et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 2003, 11, 619–633. [Google Scholar] [CrossRef]
- Teske, B.F.; Fusakio, M.E.; Zhou, D.; Shan, J.; McClintick, J.N.; Kilberg, M.S.; Wek, R.C. CHOP induces activating transcription factor 5 (ATF5) to trigger apoptosis in response to perturbations in protein homeostasis. Mol. Biol. Cell 2013, 24, 2477–2490. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Aviles, G.; Liu, Y.; Tian, R.; Unger, B.A.; Lin, Y.T.; Wiita, A.P.; Xu, K.; Correia, M.A.; Kampmann, M. Mitochondrial stress is relayed to the cytosol by an OMA1-DELE1-HRI pathway. Nature 2020, 579, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Fessler, E.; Eckl, E.M.; Schmitt, S.; Mancilla, I.A.; Meyer-Bender, M.F.; Hanf, M.; Philippou-Massier, J.; Krebs, S.; Zischka, H.; Jae, L.T. A pathway coordinated by DELE1 relays mitochondrial stress to the cytosol. Nature 2020, 579, 433–437. [Google Scholar] [CrossRef] [PubMed]

| Mitochondrial Compartment | Number of Proteins | Number of Microproteins |
|---|---|---|
| OMM | ~140 | 1 |
| IMS | ~130 | unknown |
| IMM | ~800 | 3 |
| Matrix | ~500 | unknown |
| Total | ~1570 | ~25 (mostly uncharacterized) |
| MAD | Components |
|---|---|
| VMS1/VCP/MDM30 | VMS1, Cdc48/VCP/p97, MDM30 (SCF ubiquitin ligases) |
| mitoRQC | VMS1, Rqc2/NEMF, Ltn1/Listerin |
| MAGIC | Cytosolic HSP70s, LONP |
| mPOS | Ribosome associated proteins Gis2, Nog2 |
| UPRam | Downregulated ribosomal and upregulated proteasome protein expression |
| mitoCPR | Cis1, Msp1/ATAD1 |
| mitoTAD | Ubx2, Cdc48/VCP/p97 |
| Protease | Localization | Catalytic Class | Yeast Orthologue |
|---|---|---|---|
| Mitochondrial intermediate peptidase (MIP) | Matrix | Metallo | Oct1 |
| Mitochondrial-processing peptidase subunit α (PMPCA) | Matrix | Metallo | Mas2 (MPP) |
| Mitochondrial-processing peptidase subunit β (PMPCB) | Matrix | Metallo | Mas1 (MPP) |
| Presequence protease 1 (PITRM1) | Matrix | Metallo | Mop112 |
| O-sialoglycoprotein endopeptidase-like protein 1 (OSGEPL1) | Matrix | Metallo | Qri7 |
| Methionine aminopeptidase 1D (METAP1D) | Matrix | Metallo | Map1 |
| Xaa-Pro aminopeptidase 3 (XPNPEP3) | Matrix | Metallo | Icp55 |
| Parkinson disease protein 7 (PARK7) | Matrix | Cysteine | Unknown |
| ATP-dependent Clp protease proteolytic subunit (CLPP) | Matrix | Serine | Unknown |
| Lon protease-like protein (LONP) | Matrix | Serine | Pim1 (Lon) |
| YME1-like protein 1 (YME1L1) | IMM | Metallo | Yme1 (i-AAA) |
| Spastic paraplegia 7 protein (SPG7) | IMM | Metallo | Yta10 (m-AAA) |
| AFG3-like protein 2 (AFG3L2) | IMM | Metallo | Yta12 (m-AAA) |
| Overlapping with the m-AAA protease 1 homolog (OMA1) | IMM | Metallo | Oma1 |
| Ubiquinol-cytochrome-c reductase complex core protein 1 (UQCRC1) | IMM | Metallo | Cor1 |
| Ubiquinol-cytochrome-c reductase complex core protein 2 (UQCRC2) | IMM | Metallo | Qcr2 |
| Mitochondrial inner membrane protease subunit 1 (IMMP1L) | IMM | Serine | Imp1 (IMP) |
| Mitochondrial inner membrane protease subunit 2 (IMMP2L) | IMM | Serine | Imp2 (IMP) |
| Presenilins-associated rhomboid-like protein (PARL) | IMM | Serine | Pcp1 (Rhomboid) |
| Mitochondrial inner membrane protease ATP23 homolog (ATP23) | IMS | Metallo | Atp23 |
| Microsomal endopeptidase (MEP) or Neurolysin | IMS | Metallo | Prd1 |
| High-temperature requirement protein A2 (HTRA2) or OMI | IMS | Serine | Unknown |
| Serine β-lactamase-like protein LACTB (LACTB) | IMS | Serine | Unknown |
| Ubiquitin-specific-processing protease 30 (USP30) | OMM | Cysteine | Ubp16 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jadiya, P.; Tomar, D. Mitochondrial Protein Quality Control Mechanisms. Genes 2020, 11, 563. https://doi.org/10.3390/genes11050563
Jadiya P, Tomar D. Mitochondrial Protein Quality Control Mechanisms. Genes. 2020; 11(5):563. https://doi.org/10.3390/genes11050563
Chicago/Turabian StyleJadiya, Pooja, and Dhanendra Tomar. 2020. "Mitochondrial Protein Quality Control Mechanisms" Genes 11, no. 5: 563. https://doi.org/10.3390/genes11050563