Cytosolic/Plastid Glyceraldehyde-3-Phosphate Dehydrogenase Is a Negative Regulator of Strawberry Fruit Ripening †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. ABA and Sucrose Treatment
2.3. Cloning of the FaGAPC2 and FaGAPCp1 Gene
2.4. Phylogenetic Analysis
2.5. Plasmid Construction
2.6. Transient Gene Expression in Strawberry Fruit
2.7. Quantitative RT–PCR
2.8. Determination of Total Anthocyanins, ABA, Sucrose, H2O2, Reduced Glutathione (GSH), Glutathione Disulfide (GSSG) Level, and GAPDH Enzyme Content
2.9. Treatment of H2O2 and Dithiothreitol (DTT)
2.10. Metabolomic Analysis of FaGAPCp1 Overexpressing Fruits
2.11. HPLC Conditions
2.12. ESI-Q TRAP-MS/MS
2.13. Metabolite Profiling
2.14. Statistical Analyses
3. Results
3.1. Phylogenetic Tree and Distribution of GAPDH Family
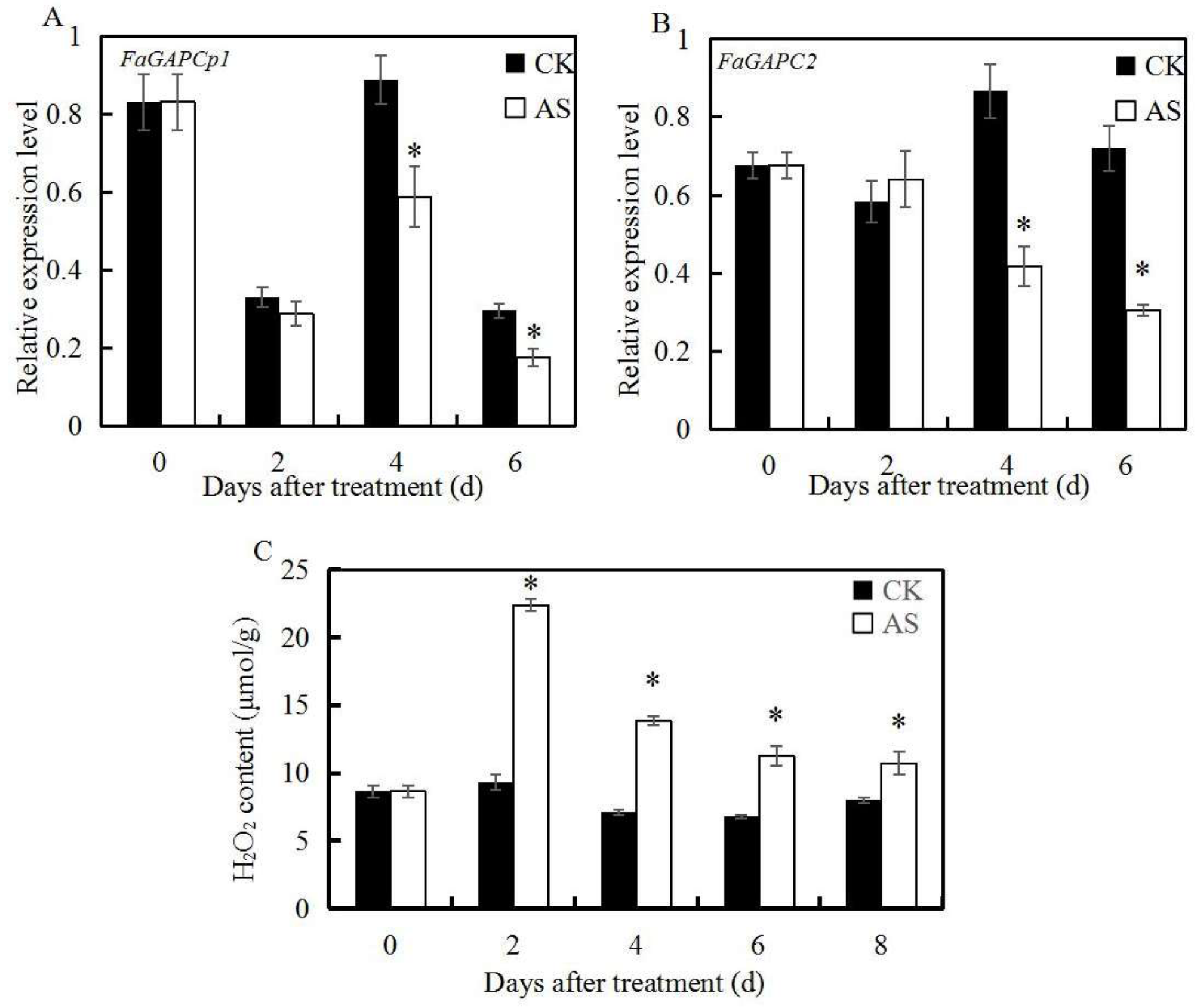
3.2. ABA and Sucrose Induce H2O2 Production and Inhibit the Expression of FaGAPC2 and FaGAPCp1
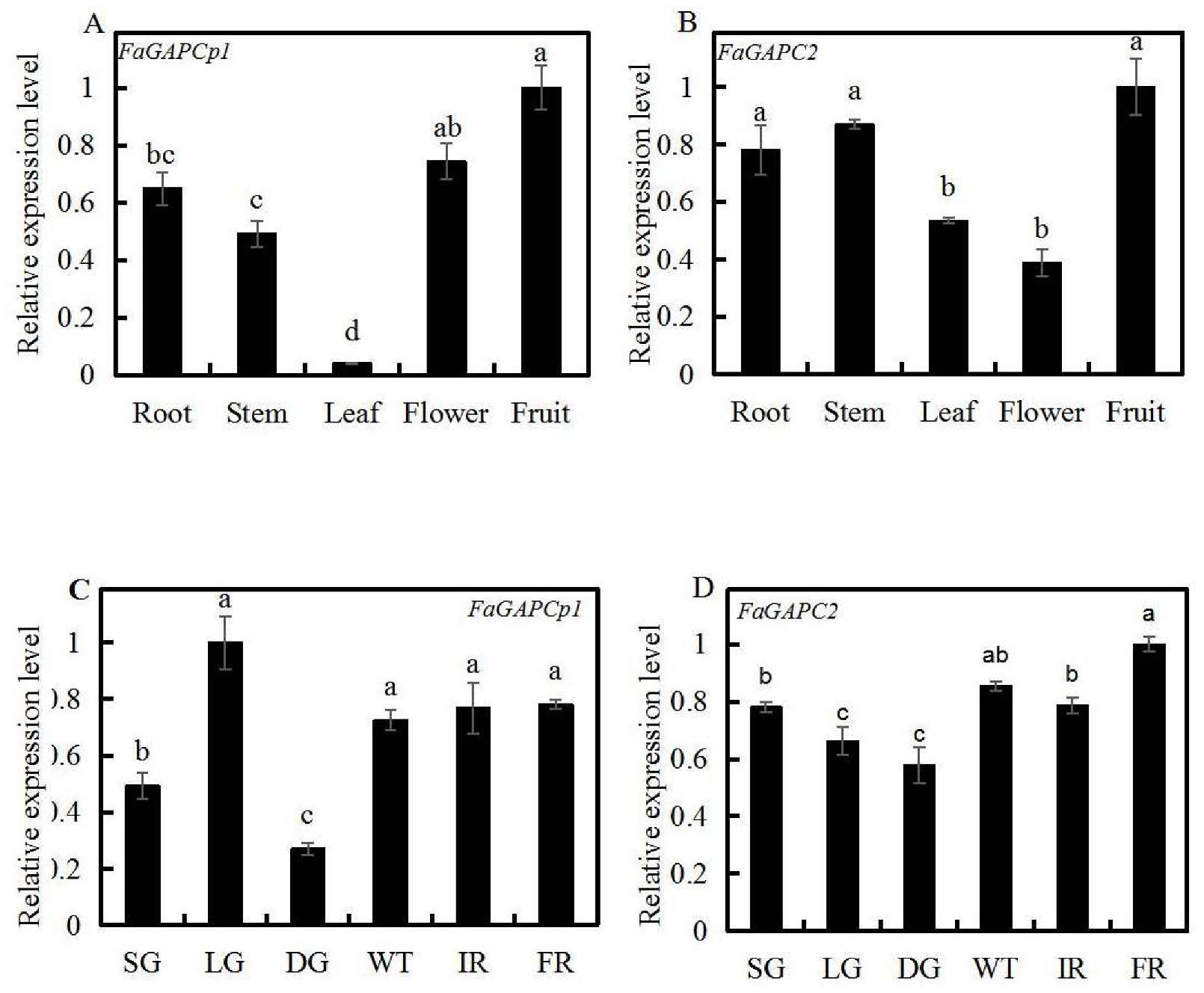
3.3. Spatial and Temporal Expression Profiles of Strawberry FaGAPC2 and FaGAPCp1
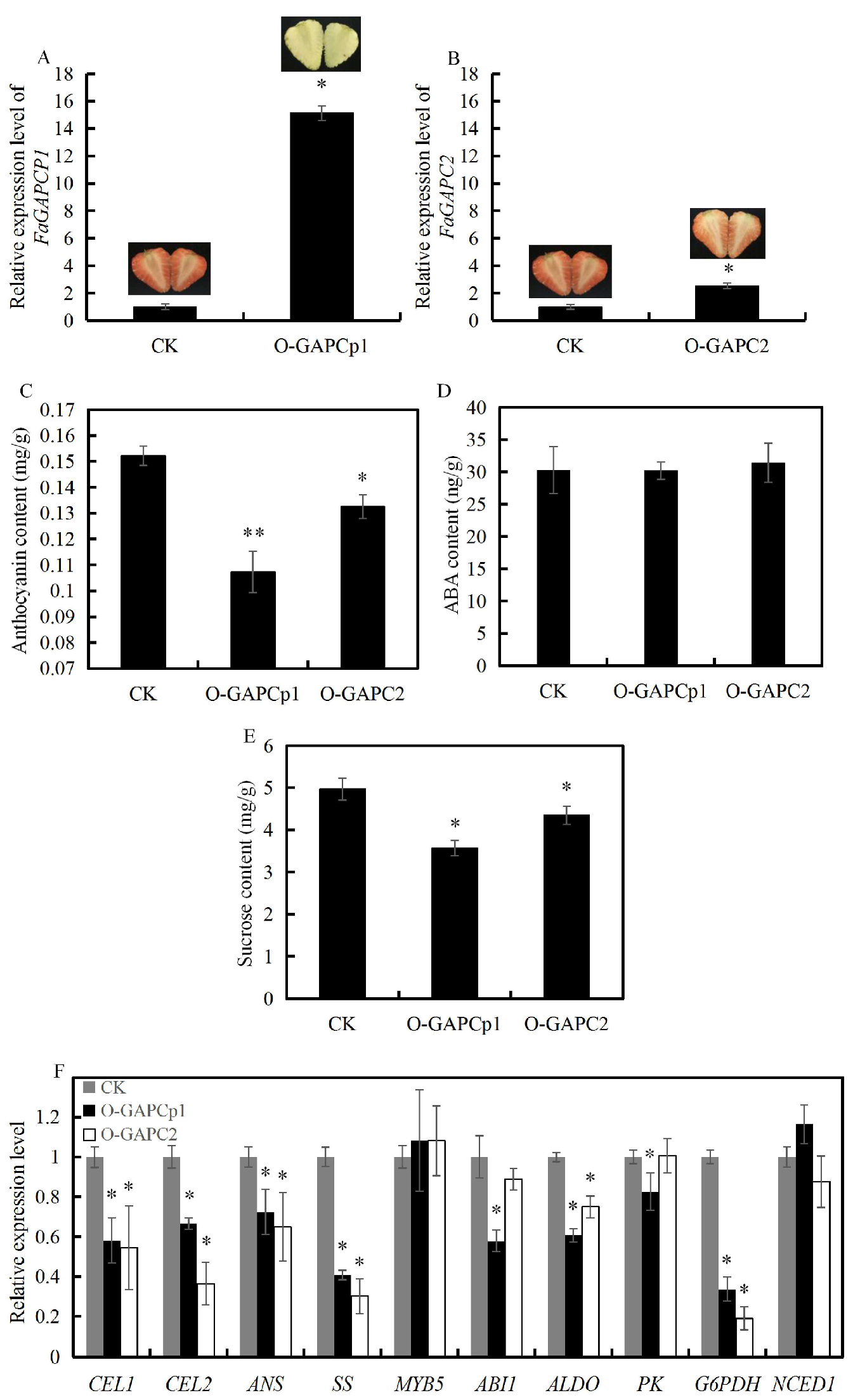
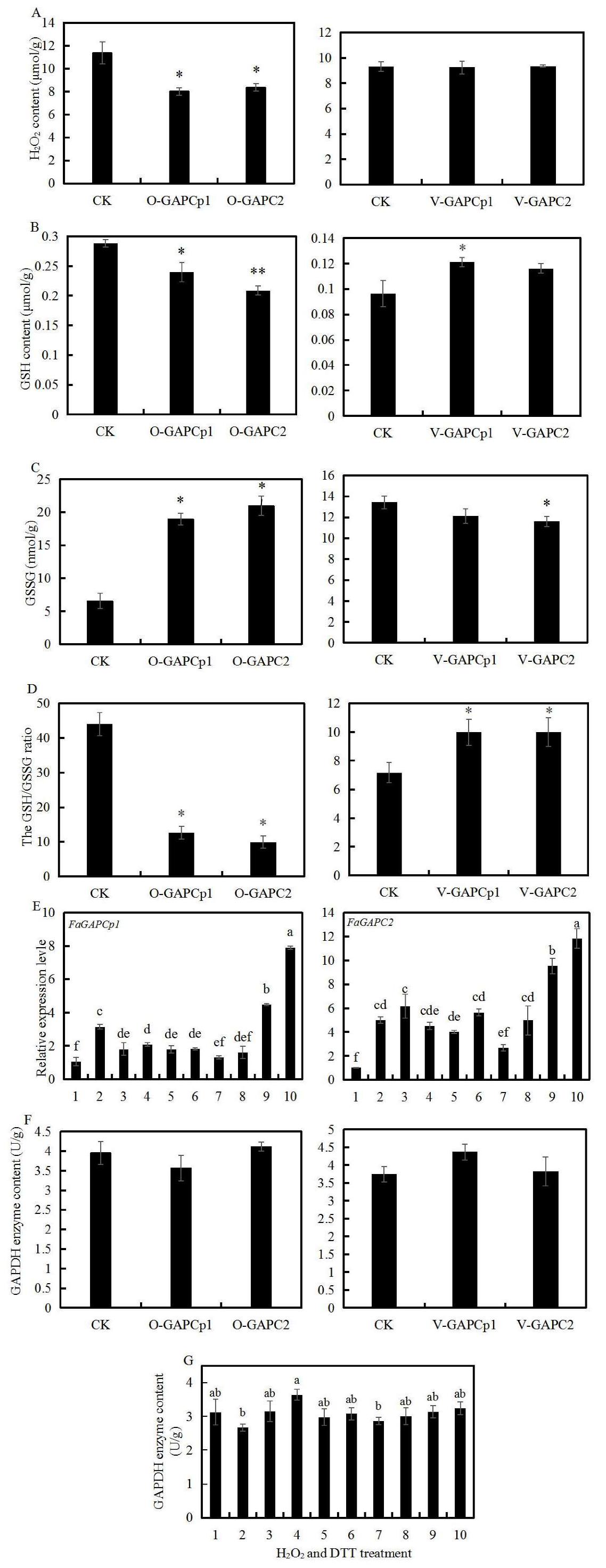
3.4. Overexpression of FaGAPC2 and FaGAPCp1 Inhibits Strawberry Fruit Ripening
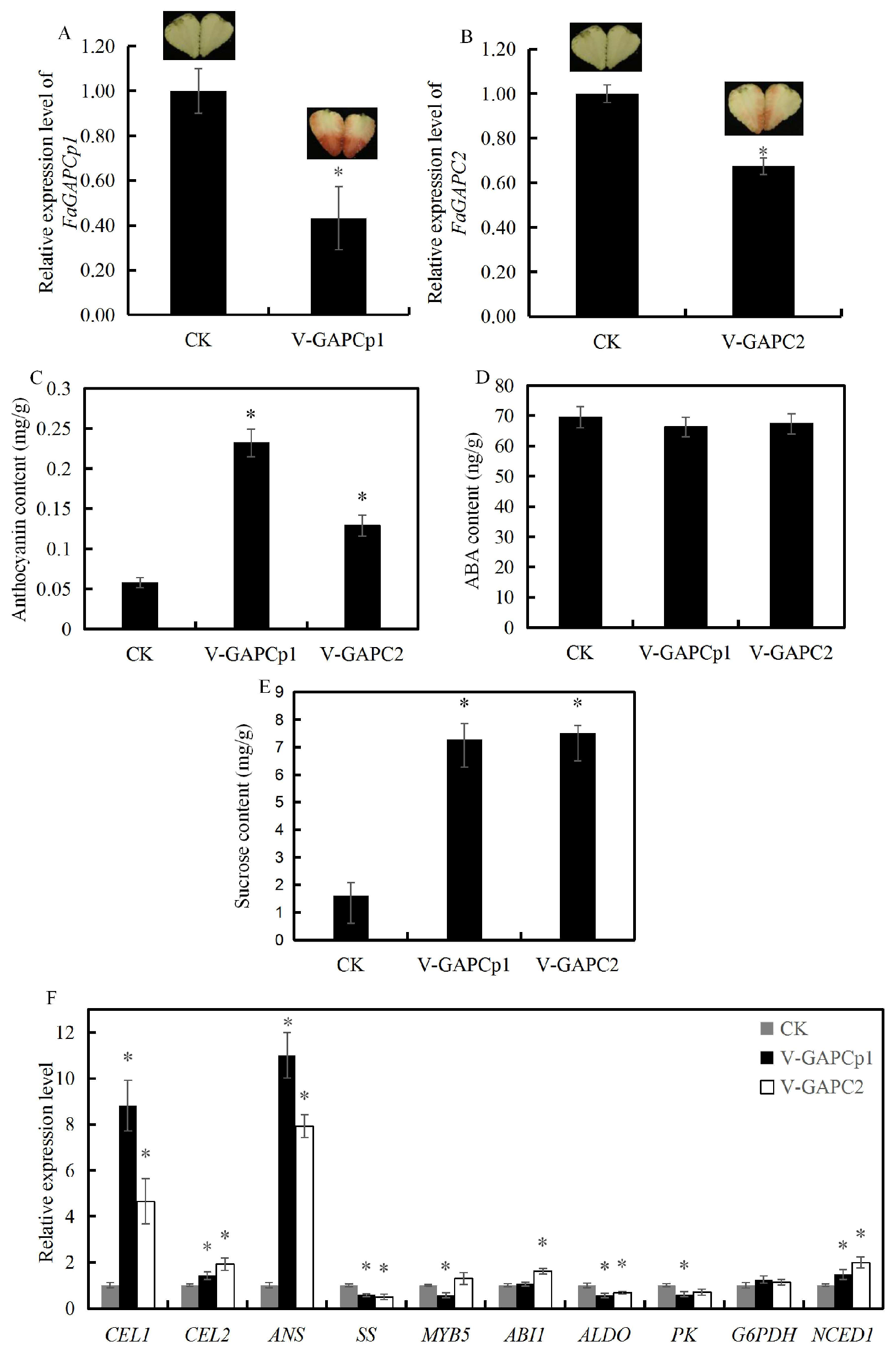
3.5. Transient Knock-Down of FaGAPC2 and FaGAPCp1 Promotes Strawberry Fruit Ripening
3.6. Transient FaGAPC2 and FaGAPCp1 Expression and Oxidative Stress
3.7. Overexpression of FaGAPCp1 Inhibits Ripening-Related Metabolism
4. Discussion
4.1. FaGAPC2 and FaGAPCp1 as a Negative Regulator in the Regulation of Strawberry Fruit Ripening
4.2. The Function of FaGAPC2 and FaGAPCp1 Is Closely Related to the Change in Oxidative Stress
4.3. FaGAPCp1 Changes Ripening-Related Metabolite Synthesis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sun, M.L.; Wang, Y.S.; Yang, D.Q.; Wei, C.L.; Gao, L.P.; Xia, T.; Dan, Y.; Luo, Y. Selection of internal reference genes in real-time fluorescence quantitative PCR qnalysis of tea trees. J. Bot. 2010, 45, 579–587. [Google Scholar] [CrossRef]
- Petersen, J.; Brinkmann, H.; Cerff, R. Origin, evolution, and metabolic role of a novel glycolytic GAPDH enzyme recruited by land plant plastids. J. Mol. Evol. 2003, 57, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Iddar, A.; Valverde, F.; Assobhei, O.; Serrano, A.; Soukri, A. Widespread occurrence of non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase among gram-positive bacteria. Int. Microbiol. 2005, 8, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Cerff, R.; Chambers, S.E. Subunit structure of higher plant glyceraldehyde-3-phosphate dehydrogenases. J. Biol. Chem. 1979, 254, 6094–6098. [Google Scholar] [PubMed]
- Spadaro, D.; Yun, B.W.; Spoel, S.H.; Chu, C.; Wang, Y.Q.; Loake, G.J. The redox switch: Dynamic regulation of protein function by cysteine modifications. Physiol. Plant 2010, 138, 360–371. [Google Scholar] [CrossRef] [PubMed]
- Vescovi, M.; Zaffagnini, M.; Festa, M.; Trost, P.; Lo Schiavo, F.; Costa, A. Nuclear accumulation of cytosolic glyceraldehyde-3-phosphate dehydrogenase in cadmium-stressed Arabidopsis roots. Plant Physiol. 2013, 162, 333–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hancock, J.T.; Henson, D.; Nyirenda, M.; Desikan, R.; Harrison, J.; Lewis, M.; Hughes, J.; Neill, S.J. Proteomic identification of glyceraldehyde 3-phosphate dehydrogenase as an inhibitory target of hydrogen peroxide in Arabidopsis. Plant. Physiol. Biochem. 2005, 43, 828–835. [Google Scholar] [CrossRef]
- Holtgrefe, S.; Gohlke, J.; Starmann, J.; Druce, S.; Klocke, S.; Altmann, B.; Wojtera, J.; Lindermayr, C.; Scheibe, R. Regulation of plant cytosolic glyceraldehyde 3-phosphate dehydrogenase isoforms by thiol modifications. Physiol. Plant 2010, 133, 211–228. [Google Scholar] [CrossRef]
- Peralta, D.A.; Araya, A.; Busi, M.V.; Gomez-Casati, D.F. The E3 ubiquitin-ligase SEVEN IN ABSENTIA like 7 mono-ubiquitinates glyceraldehyde-3-phosphate dehydrogenase 1 isoform in vitro and is required for its nuclear localization in Arabidopsis thaliana. Int. J. Biochem. Cell Biol. 2016, 70, 48–56. [Google Scholar] [CrossRef]
- Baek, D.; Jin, Y.; Jeong, J.C.; Lee, H.J.; Moon, H.; Lee, J.; Shin, D.; Kang, C.H.; Kim, D.H.; Nam, J.; et al. Suppression of reactive oxygen species by glyceraldehyde-3-phosphate dehydrogenase. Phytochemistry 2008, 69, 333–338. [Google Scholar] [CrossRef]
- Munoz-Bertomeu, J.; Cascales-Minana, B.; Mulet, J.M.; Baroja-Fernandez, E.; Pozueta-Romero, J.; Kuhn, J.M.; Segura, J.; Ros, R. Plastidial glyceraldehyde-3-phosphate dehydrogenase deficiency leads to altered root development and affects the sugar and amino acid balance in Arabidopsis. Plant Physiol. 2009, 151, 541–558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munoz-Bertomeu, J.; Cascales-Minana, B.; Irles-Segura, A.; Mateu, I.; Nunes-Nesi, A.; Fernie, A.R.; Segura, J.; Ros, R. The plastidial glyceraldehyde-3-phosphate dehydrogenase is critical for viable pollen development in Arabidopsis. Plant Physiol. 2010, 152, 1830–1841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Backhausen, J.E.; Vetter, S.; Baalmann, E.; Kitzmann, C.; Scheibe, R. NAD-dependent malate dehydrogenase and glyceraldehyde 3-phosphate dehydrogenase isoenzymes play an important role in dark metabolism of various plastid types. Planta 1998, 205, 359–366. [Google Scholar] [CrossRef]
- Gu, T.; Jia, S.; Huang, X.; Wang, L.; Fu, W.; Huo, G.; Gan, L.; Ding, J.; Li, Y. Transcriptome and hormone analyses provide insights into hormonal regulation in strawberry ripening. Planta 2019, 250, 1–18. [Google Scholar] [CrossRef]
- Luo, Y.; Ge, C.; Ling, Y.J.; Mo, F.; Yang, M.; Jiang, L.Y.; Chen, Q.; Lin, Y.X.; Sun, B.; Zhang, Y.; et al. ABA and sucrose co-regulate strawberry fruit ripening and show inhibition of glycolysis. Mol. Genet. Genom. 2019, 294, 1–18. [Google Scholar] [CrossRef]
- Luo, Y.; Lin, Y.X.; Mo, F.; Ge, C.; Jiang, L.Y.; Zhang, Y.; Chen, Q.; Sun, B.; Wang, Y.; Wang, X.R.; et al. Sucrose Promotes Strawberry Fruit Ripening and Affects Ripening-Related Processes. Int. J. Genom. 2019, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Jin, P.; Zhu, H.; Wang, J.; Chen, J.; Wang, X.; Zheng, Y. Effect of methyl jasmonate on energy metabolism in peach fruit during chilling stress. J. Sci. Food Agric. 2013, 93, 1827–1832. [Google Scholar] [CrossRef]
- Kan, J.; Wang, H.M.; Jin, C.H.; Xie, H.Y. Changes of reactive oxygen species and related enzymes in mitochondria respiratory metabolism during the ripening of peach fruit. Agric. Sci. China 2010, 9, 138–146. [Google Scholar] [CrossRef]
- Zhang, J.J.; Wang, X.; Yu, O.; Tang, J.J.; Gu, X.G.; Wan, X.C.; Fang, C.B. Metabolic profiling of strawberry (Fragaria × ananassa Duch.) during fruit development and maturation. J. Exp. Bot. 2011, 62, 1103–1118. [Google Scholar] [CrossRef]
- Wang, Q.H.; Zhao, C.; Zhang, M.; Li, Y.Z.; Shen, Y.Y.; Guo, J.X. Transcriptome analysis around the onset of strawberry fruit ripening uncovers an important role of oxidative phosphorylation in ripening. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Jia, H.F.; Zhu, X.Q.; Jin, X.S.; Shen, Y.Y. An effective method and its modifications for isolation of high-quality total RNA from fruit pulps. J. Agric. Sci. Technol. 2008, 2, 58–62. [Google Scholar]
- Jia, H.F.; Chai, Y.M.; Li, C.L.; Lu, D.; Luo, J.J.; Qin, L.; Shen, Y.Y. Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol. 2011, 157, 188–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, G.W.; Breen, P.J. Activity of phenylalanine ammonia-lyase (PAL) and concentrations of anthocyanins and phenolics in developing strawberry fruit. J. Am. Soc. Hortic. Sci. 1991, 116, 865–869. [Google Scholar] [CrossRef]
- Cao, J.K.; Jiang, W.B.; Zhao, Y.M. Guidance for Postharvest Physiological and Biochemical Experiments of Fruits and Vegetables; China Light Industry Press: Beijing, China, 2007. [Google Scholar]
- Zou, X. Effects of Abscisic Acid and Protein Reversible Phosphorylation on the Activities of Glucose Metabolizing Enzymes in Grape and Apple Fruits. Doctoral Dissertation, Agricultural University of China, Beijing, China, 2004. [Google Scholar] [CrossRef]
- André, S.; Stefan, W.; Johannes, S.; Michael, M.; Hidgund, S. The gene encoding the cellulase (avicelase) CEL1 from streptomyces reticuli and analysis of protein domains. Mol. Microbiol. 1992, 23, 3611–3621. [Google Scholar] [CrossRef]
- Chen, F.; Ling, W.; Hao-Ru, T.; Jie, X. Cloning and expression analysis of myb transcription factor famyb5 gene from strawberry. Acta Agric. Zhejiangensis 2016, 8, 1351–1359. [Google Scholar] [CrossRef]
- Plaxton, W.C. The organization and regulation of plant glycolysis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 185–214. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.; Devaiah, S.P.; Narasimhan, R.; Pan, X.; Zhang, Y.; Zhang, W.; Wang, X. Cytosolic glyceraldehyde-3-phosphate dehydrogenases interact with phospholipase Dδ to transduce hydrogen peroxide signals in the arabidopsis response to stress. Plant Cell 2012, 24, 2200–2212. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Yang, B.; Harris, N.S.; Deyholos, M.K. Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J. Exp. Bot. 2007, 5, 3591–3607. [Google Scholar] [CrossRef] [Green Version]
- Wawer, I.; Bucholc, M.; Astier, J.; Anielska-Mazur, A.; Dahan, J.; Kulik, A.; Wysłouch-Cieszynska, A.; Zaręba-Kozioł, M.; Krzywinska, E.; Dadlez, M.; et al. Regulation of Nicotiana tabacum osmotic stress-activated protein kinase and its cellular partner GAPDH by nitric oxide in response to salinity. Biochem. J. 2010, 429, 73–83. [Google Scholar] [CrossRef] [Green Version]
- Rius, S.P.; Paula, C.; Iglesias, A.A.; Gomez-Casati, D.F. Characterization of Arabidopsis lines deficient in GAPC-1, a cytosolic NAD-dependent glyceraldehyde-3-phosphate dehydrogenase. Plant Physiol. 2008, 148, 1655–1667. [Google Scholar] [CrossRef] [Green Version]
- Jia, H.F.; Lu, D.; Sun, J.H.; Li, C.L.; Xing, Y.; Qin, L.; Shen, Y.Y. Type 2C protein phosphatase ABI1 is a negative regulator of strawberry fruit ripening. J. Exp. Bot. 2013, 64, 1677–1687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, H.F.; Jiu, S.T.; Zhang, C.; Wang, C.; Tariq, P.; Liu, Z.; Wang, B.; Cui, L.; Fang, J. Abscisic acid and sucrose regulate tomato and strawberry fruit ripening through the abscisic acid-stress ripening transcription factor. Plant Biotechnol. J. 2016, 14, 2045–2065. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Bertomeu, J.; Anoman, A.D.; Toujani, W.; Cascales-MinAna, B.; Flores-Tornero, M.; Ros, R. Interactions between abscisic acid and plastidial glycolysis in Arabidopsis. Plant Signal. Behav. 2011, 6, 157–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chuankhayan, P.; Hsieh, C.Y.; Huang, Y.C.; Hsieh, Y.Y.; Guan, H.H.; Hsieh, Y.C.; Tien, Y.C.; Chen, C.D.; Chiang, C.M.; Chen, C.J. Crystal structures of Aspergillus japonicus fructosyltransferase complex with donor/acceptor substrates reveal complete subsites in the active site for catalysis. J. Biol. Chem. 2010, 285, 23251–23264. [Google Scholar] [CrossRef] [Green Version]
- Bright, J.; Desikan, R.; Hancock, J.T.; Weir, I.S.; Neill, S.J. ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. Plant J. 2006, 45, 113–122. [Google Scholar] [CrossRef]
- Jones, D.P. Redox potential of GSH/GSSG couple: Assay and biological significance. Methods Enzymol. 2002, 348, 93–112. [Google Scholar] [CrossRef]
- Hossain, M.A.; Pukclai, P.; da Silva, J.A.T.; Fujita, M. Molecular mechanism of heavy metal toxicity and tolerance in plants: Central role of glutathione in detoxification of reactive oxygen species and methylglyoxal and in heavy metal chelation. J. Bot. 2012, 4, 37–47. [Google Scholar] [CrossRef]
- Cascales-Minana, B.; Munoz-Bertomeu, J.; Flores-Tornero, M.; Anoman, A.D.; Pertusa, J.; Alaiz, M.; Osorio, S.; Fernie, A.R.; Segura, J.; Ros, R. The phosphorylated pathway of serine biosynthesis is essential both for male gametophyte and embryo development and for root growth in Arabidopsis. Plant Cell 2013, 25, 2084–2101. [Google Scholar] [CrossRef] [Green Version]
- Tarola, A.M.; Van de Velde, F.; Salvagni, L.; Preti, R. Determination of phenolic compounds in strawberries (Fragaria ananassa Duch) by high performance liquid chromatography with diode array detection. Food Anal. Methods 2013, 6, 227–237. [Google Scholar] [CrossRef]
- Xia, M.; Ling, W.; Zhu, H.L.; Ma, J.; Wang, Q.; Hou, M.J.; Tang, Z.H.; Guo, H.H.; Liu, C.; Ye, Q.Y. Anthocyanin attenuates cd40-mediated endothelial cell activation and apoptosis by inhibiting cd40-induced mapk activation. Atherosclerosis 2009, 202, 41–47. [Google Scholar] [CrossRef]
- Lopes-da-silva, F.; Pascual-Teresa, S.D.; Rivas-Gonzalo, J.; Santos-Buelga, C. Identification of anthocyanin pigments in strawberry (cv Camarosa) by LC using DAD and EST-MS detection. Eur. Food Res. Technol. 2002, 214, 248–253. [Google Scholar] [CrossRef]
- Pourcel, L.; Routaboul, J.M.; Cheynier, V.; Lepiniec, L.; Debeaujon, I. Flavonoid oxidation in plants: From biochemical properties to physiological functions. Trends Plant Sci. 2007, 12, 29–36. [Google Scholar] [CrossRef]
- Griesser, M.; Hoffmann, T.; Bellido, M.L.; Rosati, C.; Fink, B.; Kurtzer, R.; Aharoni, A.; Muñoz-Blanco, J.; Schwab, W. Redirection of flavonoid biosynthesis through the down-regulation of an anthocyanidin glucosyltransferase in ripening strawberry fruit. Plant Physiol. 2008, 146, 1528–1539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, J.Y.; Tang, C.Y. Determination of total phenolic and flavonoid contents in selected fruits and vegetables, as well as their stimulatory effects on mouse splenocyte proliferation. Food Chem. 2007, 101, 140–147. [Google Scholar] [CrossRef]
- Xie, L.; Cheng, X.H.; Feng, X.X.; Yang, T.; Zhang, Z.P.; Wang, L.J. Effects of an amino acid fertilizer on the leaf photosynthesis and fruit quality of ‘Summer Black’ grape. J. Nanjing Agric. Univ. 2013, 2, 31–37. [Google Scholar] [CrossRef]
- Zhang, J.J.; Yang, H.; Wang, X.; Hu, Y.H.; Fang, C.B. Determination of the major amino acid components by high performance liquid chromatography coupled with fluorescence detection during the development of strawberry fruits. J. Anhui Agric. Univ. 2009, 3, 377–381. [Google Scholar]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, Y.; Ge, C.; Yang, M.; Long, Y.; Li, M.; Zhang, Y.; Chen, Q.; Sun, B.; Wang, Y.; Wang, X.; et al. Cytosolic/Plastid Glyceraldehyde-3-Phosphate Dehydrogenase Is a Negative Regulator of Strawberry Fruit Ripening. Genes 2020, 11, 580. https://doi.org/10.3390/genes11050580
Luo Y, Ge C, Yang M, Long Y, Li M, Zhang Y, Chen Q, Sun B, Wang Y, Wang X, et al. Cytosolic/Plastid Glyceraldehyde-3-Phosphate Dehydrogenase Is a Negative Regulator of Strawberry Fruit Ripening. Genes. 2020; 11(5):580. https://doi.org/10.3390/genes11050580
Chicago/Turabian StyleLuo, Ya, Cong Ge, Min Yang, Yu Long, Mengyao Li, Yong Zhang, Qing Chen, Bo Sun, Yan Wang, Xiaorong Wang, and et al. 2020. "Cytosolic/Plastid Glyceraldehyde-3-Phosphate Dehydrogenase Is a Negative Regulator of Strawberry Fruit Ripening" Genes 11, no. 5: 580. https://doi.org/10.3390/genes11050580




