Transcriptomic Analysis Provides Insights into the Differential Effects of Aluminum on Peanut (Arachis hypogaea L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatment Conditions
2.2. Raw Sequencing Data
2.3. Enrichment Analysis
2.4. Real-Time Quantitative RT-PCR
2.5. Determination of Soluble Sugar Content
2.6. Determination of ABA Content
3. Results
3.1. Effects of Different Concentrations of Al3+ on the Morphological Characteristics of Peanut and the Expression of AhALMT9 and AhFRDL1 Genes
3.2. Sequencing Statistics
3.3. Mapping of Reads to the Reference Genome
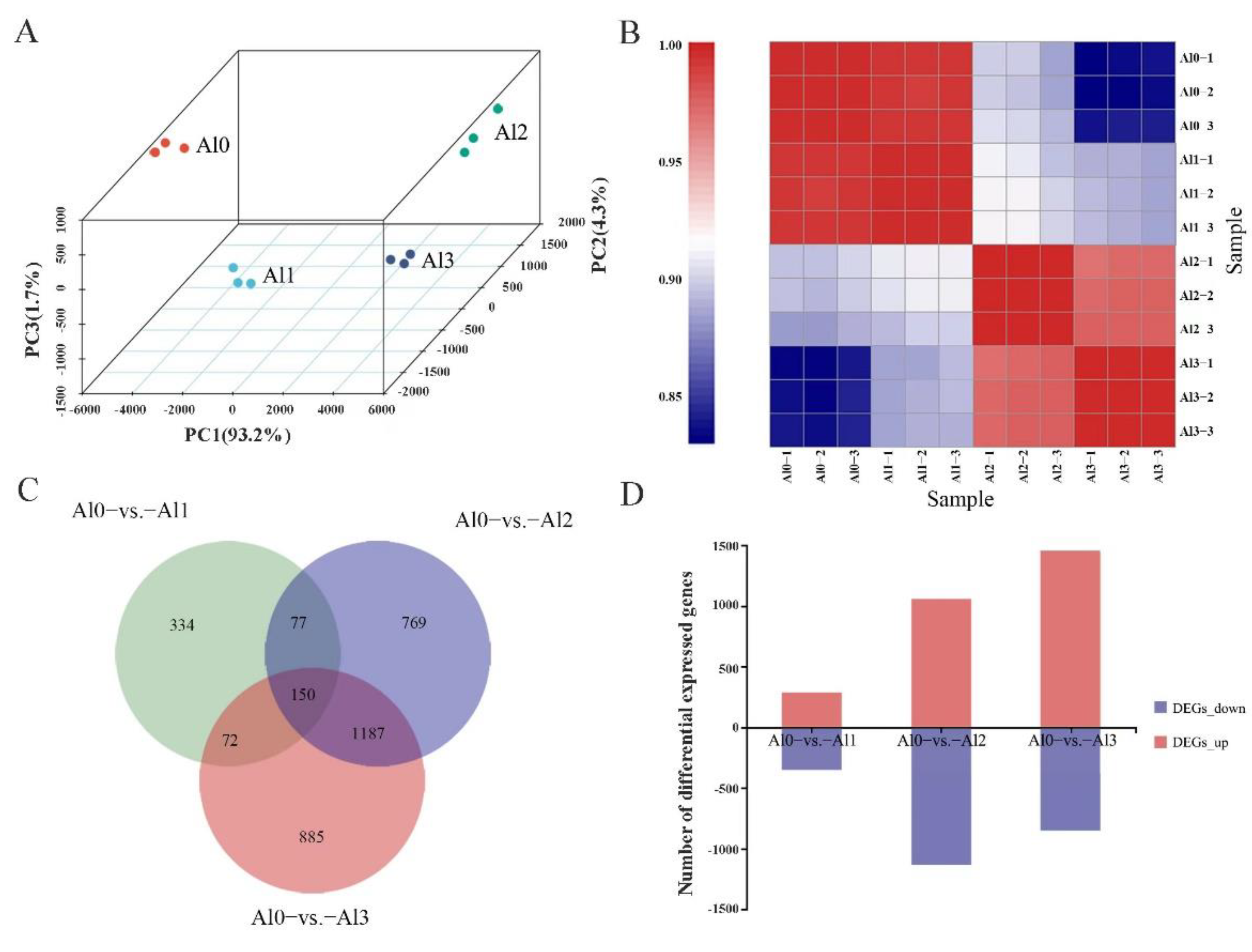
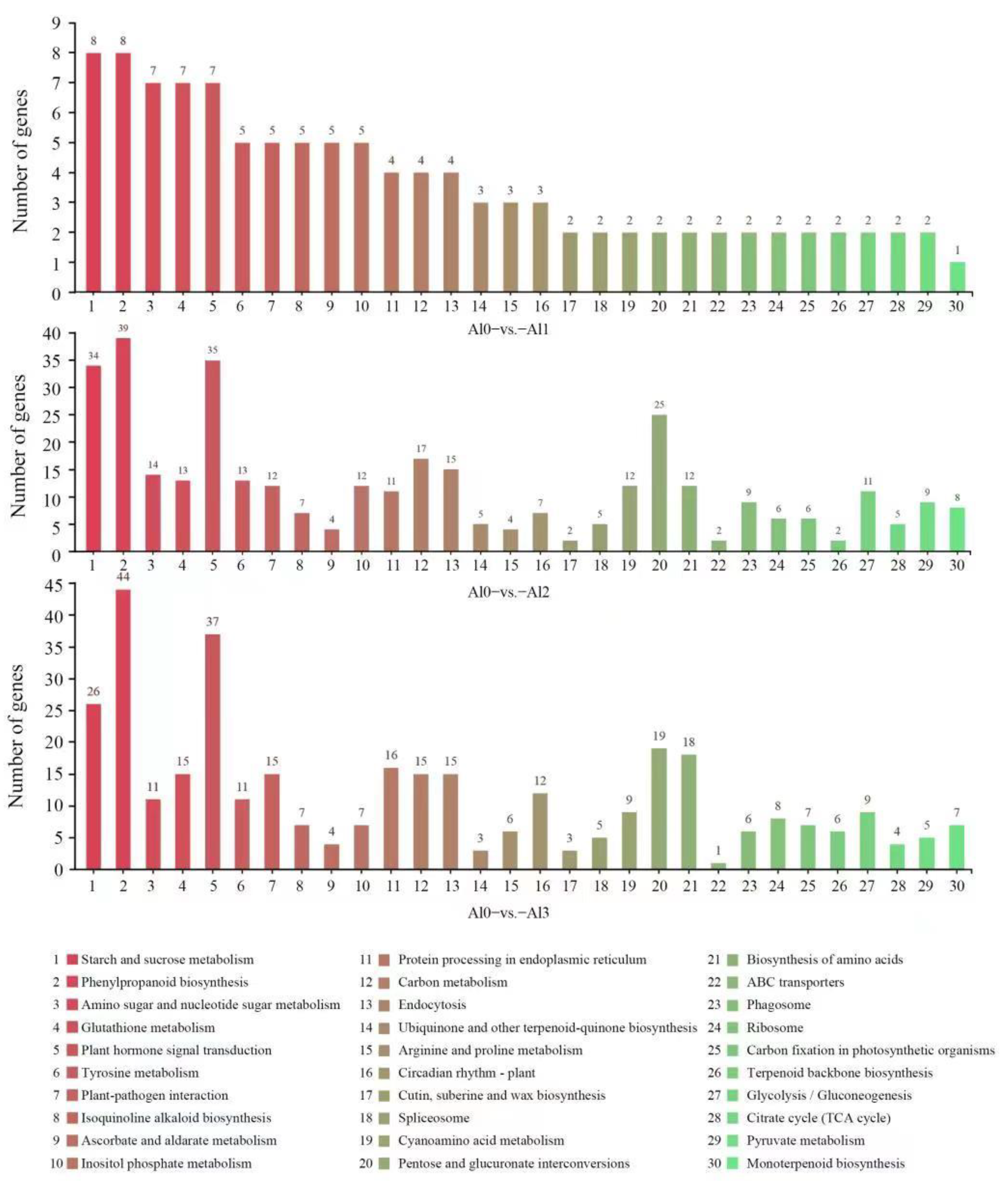
3.4. DEGs Identification and KEGG Pathway Analysis
3.5. Effect of Different Al3+ Concentrations on the Soluble Sugar Content and AhERD6 Transcript Levels
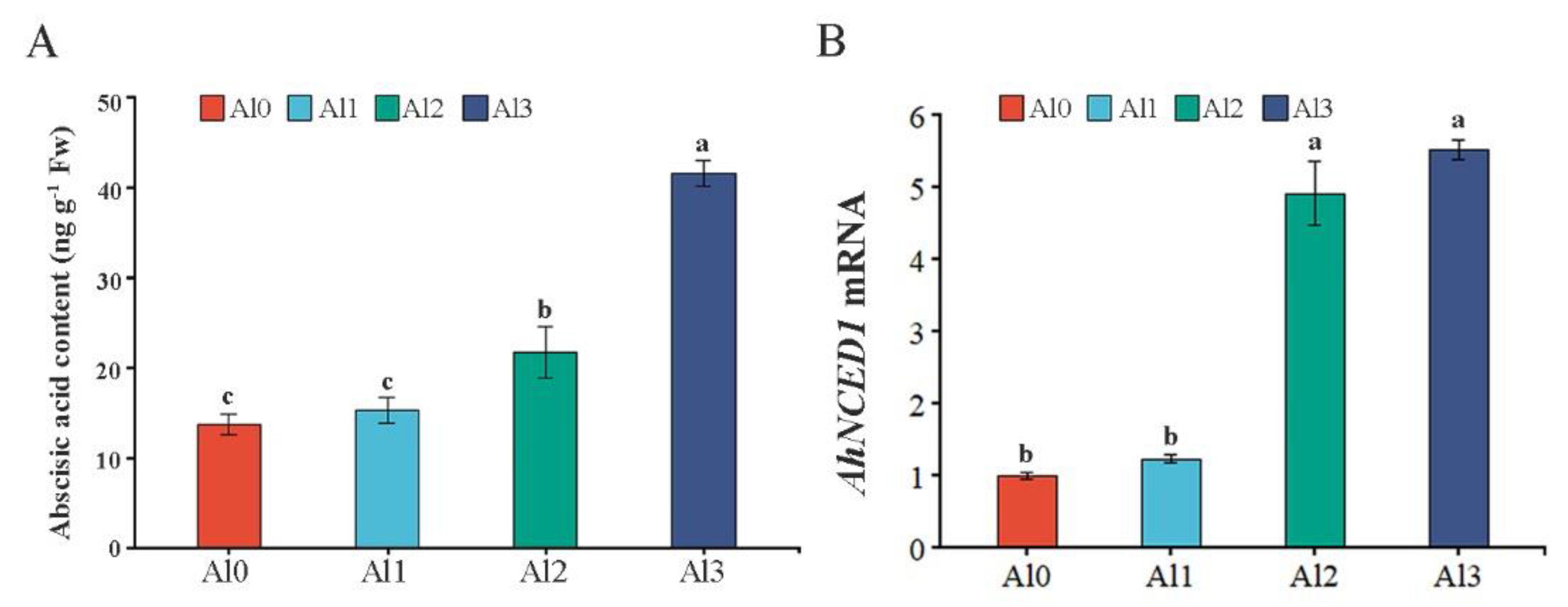
3.6. Effect of Different Al3+ Concentrations on the ABA Content and AhNCED1 Transcript Levels
3.7. Effect of Different Al3+ Concentrations on the Regulation of TFs Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zheng, A.Z.; Li, C.X. Influences and improvement of Al toxin on plants in acid red soil. Hubei Agr. Sci. 2004, 6, 41–43. [Google Scholar] [CrossRef]
- Chauhan, D.K.; Yadav, V.; Vaculík, M.; Gassmann, W.; Pike, S.; Arif, N.; Singh, V.P.; Deshmukh, R.; Sahi, S.; Tripathi, D.K. Aluminum toxicity and aluminum stress-induced physiological tolerance responses in higher plants. Crit. Rev. Biotechnol. 2021, 41, 715–730. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.C.; He, Q.; Wang, W.J. Aluminum morphology in red soil. Acta Pedol. Sin. 1998, 35, 38–48. [Google Scholar] [CrossRef]
- Dahlgren, R.A.; Ugolini, F.C. Aluminum fractionation of soil solutions from unperturbed and tephra-treated spodosols, Cascade Range, Washington, USA. Soil Sci. Soc. Am. J. 1989, 53, 559–566. [Google Scholar] [CrossRef]
- Wang, J.W.; Luo, S.M.; Feng, Y.Q. Aluminum forms in Acid Sulfate Soils. Chin. J. Appl. Ecol. 2000, 11, 735–740. [Google Scholar] [CrossRef]
- Ma, J.F. Plant root responses to three abundant soil minerals: Silicon, aluminum and iron. Crit. Rev. Plant Sci. 2005, 24, 267–281. [Google Scholar] [CrossRef]
- Ma, J.F. Syndrome of aluminum toxicity and diversity of aluminum resistance in higher plants. Int. Rev. Cytol. 2007, 264, 225–252. [Google Scholar] [CrossRef]
- Rahman, M.A.; Lee, S.H.; Ji, H.C.; Kabir, A.H.; Jones, C.S.; Lee, K.W. Molecular sciences importance of mineral nutrition for mitigating aluminum toxicity in plants on acidic soils: Current status and opportunities. Int. J. Mol. Sci. 2018, 19, 3073. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.L.; Tian, J.; Zhang, H.Y.; Liao, H. Phytohormone regulation of root growth triggered by P deficiency or Al toxicity. J. Exp. Bot. 2016, 67, 3655–3664. [Google Scholar] [CrossRef] [Green Version]
- Qiu, W.; Wang, N.Q.; Dai, J.; Wang, T.Q.; Kochian, L.V.; Liu, J.P.; Zuo, Y.M. AhFRDL1-mediated citrate secretion contributes to adaptation to iron deficiency and aluminum stress in peanuts. J. Exp. Bot. 2019, 70, 2783–2886. [Google Scholar] [CrossRef]
- Moreno-Alvarado, M.; García-Morales, S.; Trejo-Téllez, L.I.; Hidalgo-Contreras, J.V.; Gómez-Merino, F.C. Aluminum enhances growth and sugar concentration, alters macronutrient status and regulates the expression of NAC transcription factors in rice. Front. Plant Sci. 2017, 8, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, T.; Jansen, S.; Osaki, M. Al-Fe interactions and growth enhancement in Melastoma malabathricum and Miscanthus sinensis dominating acid sulphate soils. Plant Cell Environ. 2006, 29, 2124–2132. [Google Scholar] [CrossRef] [PubMed]
- Osaki, M.; Watanabe, T.; Tadano, T. Beneficial effect of aluminum on growth of plants adapted to low pH soils. Soil Sci. Plant Nutr. 1997, 43, 551–563. [Google Scholar] [CrossRef]
- Yamaji, N.; Huang, C.F.; Nagao, S.; Yano, M.; Sato, Y.; Nagamura, Y.; Ma, J.F. A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell 2009, 21, 3339–3349. [Google Scholar] [CrossRef] [Green Version]
- Bao, G.G.; Zhou, Q.; Li, S.Y.; Ashraf, U.; Huang, S.; Miao, A.M.; Cheng, Z.S.; Wan, X.R.; Zheng, Y.X. Transcriptome analysis revealed the mechanisms involved in ultrasonic seed treatment-induced aluminum tolerance in peanut. Front. Plant Sci. 2022, 12, 807021. [Google Scholar] [CrossRef]
- Chen, J.G.; Lai, Q.; Zeng, B.Q.; Guo, L.B.; Ye, G.Y. Progress on Molecular Mechanism of Aluminum Resistance in Rice. Rice Sci. 2020, 27, 454–467. [Google Scholar] [CrossRef]
- Ma, J.F.; Ryan, P.R.; Delhaize, E. Aluminium tolerance in plants and the complexing role of organic acids. Trends Plant Sci. 2001, 6, 273–278. [Google Scholar] [CrossRef]
- Huang, K.; Zhang, H.Y.; Zhang, H.Q.; Li, Y.; Zu, Y.Q.; Chen, J.J. Research Progress on the Molecular Mechanism of Plants Response to Aluminum Toxicity. Biotechnol. Bull. 2021, 37, 125–135. [Google Scholar] [CrossRef]
- Jia, Y.J.; Pei, Y.K.; Zhu, Y.T.; Hu, X.Q.; Hou, Y.X. Research progress on pectin methesterase response to disease resistance and aluminum tolerance. Mol. Plant Breed. 2020, 19, 4127–4132. [Google Scholar] [CrossRef]
- Ranjan, A.; Sinha, R.; Lal, S.K.; Bishi, S.K.; Singh, A.K. Phytohormone signalling and cross-talk to alleviate aluminium toxicity in plants. Plant Cell Rep. 2021, 40, 1331–1343. [Google Scholar] [CrossRef]
- Vishwakarma, K.; Upadhyay, N.; Kumar, N.; Yadav, G.; Singh, J.; Mishra, R.K.; Kumar, V.; Verma, R.; Upadhyay, R.G.; Pandey, M.; et al. Abscisic acid signaling and abiotic stress tolerance in plants: A review on current knowledge and future prospects. Front. Plant Sci. 2017, 8, 161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.X.; Yan, J.Y.; Ren, J.Y.; Sun, L.; Xu, C.; Li, G.X.; Ding, Z.J.; Zheng, S.J. A WRKY transcription factor confers aluminum tolerance via regulation of cell wall modifying genes. J. Integr. Plant Biol. 2020, 8, 1176–1192. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, J.; Guo, J.; Zhou, F.; Singh, S.; Xu, X.; Xie, Q.; Yang, Z.; Huang, C.F. F-box protein RAE1 regulates the stability of the aluminum-resistance transcription factor STOP1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 319–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.C.; Lin, Z.; Lin, H.M. Research on the land degradation status andprevention countermeasures in south China. J. Anhui Agric. Sci. 2013, 41, 418. [Google Scholar] [CrossRef]
- Bao, G.G.; Zhuo, C.; Qian, C.; Xiao, T.; Guo, Z.; Lu, S. Co-expression of NCED and ALO improves vitamin C level and tolerance to drought and chilling in transgenic tobacco and Stylothanthes plants. Plant Biotechnol. J. 2016, 14, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Ashrafab, U.; Mahmoodc, M.H.; Hussaind, S.; Abbase, F.; Anjumd, S.A.; Tang, X.R. Lead (Pb) distribution and accumulation in different plant parts and its associations with grain Pb contents in fragrant rice. Chemosphere 2020, 248, 126003. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, U.; Mahmood, S.; Anjum, S.A.; Abbas, R.N.; Rasul, F.; Iqbal, J.; Mo, Z.W.; Tang, X.R. Exogenous Gamma-Aminobutyric Acid Application Induced Modulations in the Performance of Aromatic Rice Under Lead Toxicity. Front Plant Sci. 2022, 22, 933694–933709. [Google Scholar] [CrossRef]
- Anjum, S.A.; Ashrafab, U.; Khan, I.; Tanveer, M.; Shahid, M.; Shakoor, A.; Wang, L.C. Phyto-toxicity of chromium in maize: Oxidative damage, osmolyte accumulation, anti-oxidative defense and chromium uptake. Pedosphere 2017, 27, 262–273. [Google Scholar] [CrossRef]
- Anjum, S.A.; Tanveer, M.; Hussain, S.; Ashraf, U.; Khan, I.; Wang, L.C. Alteration in growth, leaf gas exchange, and photosynthetic pigments of maize plants under combined cadmium and arsenic stress. Water Air Soil Pollut. 2017, 228, 1–12. [Google Scholar] [CrossRef]
- Ma, L.; Kong, L.L.; Gui, R.F.; Yang, X.J.; Zhang, J.W.; Gong, Q.; Qin, D.J.; Zhuang, M.S.; Umair, A.; Mo, Z.W. Application of hydrogen-rich water modulates physio-biochemical functions and early growth of fragrant rice under Cd and Pb stress. Environ. Sci. Pollut. Res. 2021, 28, 58558–58569. [Google Scholar] [CrossRef]
- Huang, S.H.; Rao, G.S.; Ashraf, U.; He, L.X.; Zhang, Z.Z.; Zhang, H.L.; Mo, Z.W.; Pan, S.G.; Tang, X.R. Application of inorganic passivators reduced Cd contents in brown rice in oilseed rape-rice rotation under Cd contaminated soil. Chemosphere 2020, 259, 127404. [Google Scholar] [CrossRef] [PubMed]
- Fang, Q.; Zhou, F.; Zhang, Y.; Singh, S.; Huang, C.F. Degradation of STOP1 mediated by the F-box proteins RAH1 and RAE1 balances aluminum resistance and plant growth in Arabidopsis thaliana. Plant J. 2021, 106, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Fang, Q.; Zhang, J.; Zhang, Y.; Fan, N.; van den Burg, H.A.; Huang, C.F. Regulation of aluminum resistance in Arabidopsis involves the SUMOylation of the zinc finger transcription Factor STOP1. Plant Cell 2020, 32, 3921–3938. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, U.; Hussain, S.; Anjum, S.A.; Abbas, F.; Tanveer, M.; Noor, M.A.; Tang, X.R. Alterations in growth, oxidative damage, and metal uptake of five aromatic rice cultivars under lead toxicity. Plant Physiol. Biochem. 2017, 115, 461–471. [Google Scholar] [CrossRef]
- Ashraf, U.; Tang, X.R. Yield and quality responses, plant metabolism and metal distribution pattern in aromatic rice under lead (Pb) toxicity. Chemosphere 2017, 176, 141–155. [Google Scholar] [CrossRef]
- Kong, X.; Pan, J.; Zhang, M.; Xing, X.; Zhou, Y.; Liu, Y.; Li, D.; Li, D. ZmMKK4, a novel group C mitogen-activated protein kinase kinase in maize (Zea mays), confers salt and cold tolerance in transgenic Arabidopsis. Plant Cell Environ. 2011, 34, 1291–1303. [Google Scholar] [CrossRef]
- Yan, X.X.; Yang, G.Z.; Yin, W.; Xiong, L.L.; Ma, R.R.; Li, C.D. Effects of lead and aluminum stress on germination and physiological and biochemical characteristics of two kinds of leguminous forage. J. Qinghai University. 2018, 36, 1–7. [Google Scholar] [CrossRef]
- Hajiboland, R.; Rad, S.B.; Barceló, J.; Poschenrieder, C. Mechanisms of aluminum-induced growth stimulation in tea (Camellia sinensis). J. Inorg. Biochem. 2013, 176, 616–625. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Kobayashi, Y.; Sugimoto, M.; Lakshmanan, V.; Iuchi, S.; Kobayashi, M.; Bais, H.P.; Koyama, H. Characterization of the complex regulation of AtALMT1 expression in response to phytohormones and other inducers. Plant Physiol. 2013, 162, 732–740. [Google Scholar] [CrossRef] [Green Version]
- Gavassi, M.A.; Silva, G.S.; Silva, C.; Thompson, A.J.; Habermann, G. NCED expression is related to increased ABA biosynthesis and stomatal closure under aluminum stress. Environ. Exp. Bot. 2021, 185, 104404. [Google Scholar] [CrossRef]
- Fan, W.; Xu, J.M.; Wu, P.; Yang, Z.X.; Lou, H.Q.; Chen, W.W.; Jin, J.F.; Zheng, S.J.; Yang, J.L. Alleviation by abscisic acid of Al toxicity in rice bean is not associated with citrate efflux but depends on ABI5-mediated signal transduction pathways. J. Integr. Plant Biol. 2019, 61, 140–154. [Google Scholar] [CrossRef] [PubMed]
- Li, G.Z.; Wang, Z.Q.; Yokosho, K.; Ding, B.; Fan, W.; Gong, Q.Q.; Li, G.X.; Wu, Y.R.; Yang, J.L.; Ma, J.F.; et al. Transcription factor WRKY22 promotes aluminum tolerance via activation of OsFRDL4 expression and enhancement of citrate secretion in rice (Oryza sativa). New Phytol. 2018, 219, 149–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arenhart, R.A.; Schunemann, M.; Neto, L.B.; Margis, R.; Wang, Z.Y.; Margis-Pinheiro, M. Rice ASR1 and ASR5 are complementary transcription factors regulating aluminium responsive genes. Plant Cell Environ. 2016, 39, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.F.; Yamaji, N.; Mitani, N.; Yano, M.; Nagamura, Y.; Ma, J.F. A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell 2009, 21, 655–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Li, L.; Yang, C.; Cheng, Y.; Han, Z.; Cai, Z.; Nian, H.; Ma, Q. GsMAS1 encoding a MADS-box transcription factor enhances the tolerance to aluminum stress in Arabidopsis thaliana. Int. J. Mol. Sci. 2020, 21, 2004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Escobar-Sepúlveda, H.F.; Trejo-Téllez, L.I.; García-Morales, S.; Gómez-Merino, F.C. Expression patterns and promoter analyses of aluminum-responsive NAC genes suggest a possible growth regulation of rice mediated by aluminum, hormones and NAC transcription factors. PLoS ONE 2017, 12, e0186084. [Google Scholar] [CrossRef]
- Iuchi, S.; Koyama, H.; Iuchi, A.; Kobayashi, Y.; Kitabayashi, S.; Kobayashi, Y.; Ikka, T.; Hirayama, T.; Shinozaki, K.; Kobayashi, M. Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc. Natl. Acad. Sci. USA 2007, 104, 9900–9905. [Google Scholar] [CrossRef]



| Gene Name | Gene ID | Primer Sequences |
|---|---|---|
| AhFRDL1 | LOC112769177 | F 5′-TGATGCTGAAACCAAAGAGTTCCTACC-3′ |
| R 5′-CAGATGAAGCCGAAGGGATATGCC-3′ | ||
| AhALMT9 | LOC112802199 | F 5′-ACCACACTCTCTCCCTCCAAATCC-3′ |
| R 5′-ACAGCAGCCTCTCCTTCTTCTCC-3′ | ||
| AhWRKY1 | LOC112766106 | F 5′-TTGCTTGGATACTTGGATTGGACTCTC-3′ |
| R 5′-TGGTTGGTTGGTTGGTTGAATTGAATG-3′ | ||
| AhWRKY23 | LOC112710566 | F 5′-AAGCGTGTGGAGCGGTCATTTAC-3′ |
| R 5′-AATGGCTGAACGAGGCATAACTGG-3′ | ||
| AhWRKY44 | LOC112759075 | F 5′-ACAGTCACAGCATTACACCACCTTC-3′ |
| R 5′-AGCCTTCTGATCCTCCTCTATGTTCTG-3′ | ||
| AhOFP1 | LOC112728150 | F 5′-ACCCCAAAGATTCACCACCACAAAG-3′ |
| R 5′-AGAAGAGACAGAGGAGGAGGAAGTAAC-3′ | ||
| AhOFP13 | LOC112780116 | F 5′-GGAGATGGTTGAGTGTCATGGAGTG-3′ |
| R 5′-GAAGCAGAAGCAGCAATGGTGATAATC-3′ | ||
| AhbZIP60 | LOC112727401 | F 5′-AACCATGCTTTGCGTCTTTGCTTAC-3′ |
| R 5′-AGTCAATGTGTTACTGTAGGGCTTACC-3′ | ||
| AhAP2-AIL5 | LOC112723564 | F 5′-TGGCAATGGCATGATGGACTTCTC-3′ |
| R 5′-ACCACCACTCTCTTGTTGATGATGATG-3′ | ||
| AhAP2-ANT | LOC112756010 | F 5′-CTTCAAGCACTGCCTCAACAACAAG-3′ |
| R 5′-GCCAGAGAAGGAGAAGAATGAAAGGG-3′ | ||
| AhNAC29 | LOC112759194 | F 5′-TGGTCATGCCGTTGCGTTCTAC-3′ |
| R 5′-ACCACCCTCACTCACATCGTCTC-3′ | ||
| AhNAC56 | LOC112707370 | F 5′-TTCTGGAAGGTATGATGCTAGGAGGAG-3′ |
| R 5′-TGTTAATGGTGTTTGAGGCTGATCCC-3′ | ||
| AhbHLH13 | LOC112758465 | F 5′-TGCTGTTAGGAAGATGGAGGAGAGG-3′ |
| R 5′-GATGAAGACCATTGCGAGGAGGAC-3′ | ||
| AhbHLH18 | LOC112720570 | F 5′-GCCACCACCAGAAACCCAAC-3′ |
| R 5′-AGTCCAGGAACAATGGCGGA-3′ | ||
| AhbHLH30 | LOC112802694 | F 5′-TCACGGCGGACGAAGAAGATTATTC-3′ |
| R 5′-AGCAGATTCATCACCACCACCTTTC-3′ | ||
| AhbHLH96 | LOC112765920 | F 5′-GGACATAGAAGTGACAATGGTGGACAG-3′ |
| R 5′-AGGTGGAGAATGGTGAGGCTTAGAG-3′ | ||
| AhbHLH115 | LOC112772203 | F 5′-GGACATAGATTCTTCAGGAGGCACATC-3′ |
| R 5′-CAGCCAGGTGAAAGCAGCAGAG-3′ | ||
| AhbHLH122 | LOC112706635 | F 5′-ATGTTGTTGATATGCGAGGGATGGG-3′ |
| R 5′-GAGGCGGAGGAGGAGTAGTTCTG-3′ | ||
| AhMYB14 | LOC112722417 | F 5′-AACCTGCACCGTGTCTGATGTTG-3′ |
| R 5′-AAGCCGAATGTCTTGATGGAGTCTG-3′ | ||
| AhMYB44 | LOC112766377 | F 5′-GACCCGTTGACTGCGTTGACTC-3′ |
| R 5′-CTCACTTCCCTGGCAATCACATCC-3′ | ||
| AhERD6 | LOC112741105 | F 5′-ACAGCATTGGGAGCAATACTGATGG-3′ |
| R 5′-CAGCCTACGAAAGTGCCACTAGC-3′ | ||
| AhNCED1 | LOC112703460 | F 5′-TGGAAGGAAGACACAGTTCGCATAC-3′ |
| R 5′-CTTCGCCGCTGGACAGATCAAC-3′ | ||
| Ahactin | LOC112787680 | F 5′-AAGCTGGCTTACATTGCCCT-3′ |
| R 5′-TGACCTGTCCATCAGGCAAC-3′ |
| Sample | Clean Reads | Clean Bases | GC Content | % ≥ Q30 | SRX |
|---|---|---|---|---|---|
| Al0-1 | 20,553,687 | 6,146,509,322 | 46.01% | 93.00% | 11,763,364 |
| Al0-2 | 23,425,837 | 7,009,817,148 | 45.85% | 92.97% | 11,763,363 |
| Al0-3 | 24,343,990 | 7,275,220,472 | 45.96% | 93.16% | 11,763,362 |
| Al1-1 | 23,330,520 | 6,975,644,336 | 45.84% | 93.14% | 11,763,361 |
| Al1-2 | 22,315,847 | 6,672,118,448 | 45.85% | 92.97% | 11,763,360 |
| Al1-3 | 20,976,609 | 6,272,951,552 | 45.98% | 92.96% | 11,763,359 |
| Al2-1 | 19,314,435 | 5,777,435,056 | 45.50% | 92.89% | 11,763,358 |
| Al2-2 | 21,413,255 | 6,403,612,920 | 45.53% | 93.07% | 11,763,357 |
| Al2-3 | 21,233,487 | 6,346,252,156 | 45.43% | 92.81% | 11,763,356 |
| Al3-1 | 21,037,988 | 6,290,647,134 | 45.44% | 92.81% | 11,763,355 |
| Al3-2 | 22,477,591 | 6,720,640,752 | 45.43% | 92.77% | 11,763,354 |
| Al3-3 | 22,844,859 | 6,834,289,998 | 45.49% | 92.88% | 11,763,353 |
| Sample | Total Reads | Mapped Reads | Unique Mapped Reads | Multiple Map Reads |
|---|---|---|---|---|
| Al0-1 | 41,107,374 | 38,674,767 (94.08%) | 35,195,591 (85.62%) | 3,479,176 (8.46%) |
| Al0-2 | 46,851,674 | 44,198,905 (94.34%) | 40,263,962 (85.94%) | 3,934,943 (8.40%) |
| Al0-3 | 48,687,980 | 45,441,812 (93.33%) | 41,288,361 (84.80%) | 4,153,451 (8.53%) |
| Al1-1 | 46,661,040 | 43,883,594 (94.05%) | 39,896,921 (85.50%) | 3,986,673 (8.54%) |
| Al1-2 | 44,631,694 | 42,028,809 (94.17%) | 38,235,448 (85.67%) | 3,793,361 (8.50%) |
| Al1-3 | 41,953,218 | 39,454,492 (94.04%) | 35,906,710 (85.59%) | 3,547,782 (8.46%) |
| Al2-1 | 38,628,870 | 36,691,674 (94.99%) | 33,371,783 (86.39%) | 3,319,891 (8.59%) |
| Al2-2 | 42,826,510 | 40,650,589 (94.92%) | 36,919,727 (86.21%) | 3,730,862 (8.71%) |
| Al2-3 | 42,466,974 | 40,121,437 (94.48%) | 36,392,658 (85.70%) | 3,728,779 (8.78%) |
| Al3-1 | 42,075,976 | 39,782,778 (94.55%) | 36,169,774 (85.96%) | 3,613,004 (8.59%) |
| Al3-2 | 44,955,182 | 42,699,985 (94.98%) | 38,830,480 (86.38%) | 3,869,505 (8.61%) |
| Al3-3 | 45,689,718 | 43,437,766 (95.07%) | 39,532,547 (86.52%) | 3,905,219 (8.55%) |
| Gene | Sample | FC | Gene | Sample | FC |
|---|---|---|---|---|---|
| AhOFP1 | Al0_RNA-seq | AhOFP1 | Al0_qRT-PCR | ||
| AhOFP1 | Al1_RNA-seq | 4.2249179 | AhOFP1 | Al1_qRT-PCR | 4.4242215 |
| AhbZIP60 | Al0_RNA-seq | AhbZIP60 | Al0_qRT-PCR | ||
| AhbZIP60 | Al1_RNA-seq | 1.9782843 | AhbZIP60 | Al1_qRT-PCR | 6.1841118 |
| AhbHlH122 | Al0_RNA-seq | AhbHlH122 | Al0_qRT-PCR | ||
| AhbHlH122 | Al1_RNA-seq | 0.7069601 | AhbHlH122 | Al1_qRT-PCR | 2.0390607 |
| AhbHlH18 | Al0_RNA-seq | AhbHlH18 | Al0_qRT-PCR | ||
| AhbHlH18 | Al1_RNA-seq | 1.4979671 | AhbHlH18 | Al1_qRT-PCR | 3.4426249 |
| AhMYB44 | Al0_RNA-seq | AhMYB44 | Al0_qRT-PCR | ||
| AhMYB44 | Al1_RNA-seq | 2.9794002 | AhMYB44 | Al1_qRT-PCR | 2.4741158 |
| AhAP2-AIl5 | Al0_RNA-seq | AhAP2-AIl5 | Al0_qRT-PCR | ||
| AhAP2-AIl5 | Al1_RNA-seq | 1.1578537 | AhAP2-AIl5 | Al1_qRT-PCR | 2.1525224 |
| AhAP2-ANT | Al0_RNA-seq | AhAP2-ANT | Al0_qRT-PCR | ||
| AhAP2-ANT | Al1_RNA-seq | 0.8790023 | AhAP2-ANT | Al1_qRT-PCR | 0.3706083 |
| AhbHlH30 | Al0_RNA-seq | AhbHlH30 | Al0_qRT-PCR | ||
| AhbHlH30 | Al1_RNA-seq | 1.1577513 | AhbHlH30 | Al1_qRT-PCR | 1.1665719 |
| AhNAC56 | Al0_RNA-seq | AhNAC56 | Al0_qRT-PCR | ||
| AhNAC56 | Al1_RNA-seq | 6.5449812 | AhNAC56 | Al1_qRT-PCR | 20.860103 |
| AhNAC29 | Al0_RNA-seq | AhNAC29 | Al0_qRT-PCR | ||
| AhNAC29 | Al1_RNA-seq | 1.0325376 | AhNAC29 | Al1_qRT-PCR | 15.497673 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bao, G.; Li, S.; Zhou, Q.; Ashraf, U.; Qiao, J.; Li, X.; Wan, X.; Zheng, Y. Transcriptomic Analysis Provides Insights into the Differential Effects of Aluminum on Peanut (Arachis hypogaea L.). Genes 2022, 13, 1830. https://doi.org/10.3390/genes13101830
Bao G, Li S, Zhou Q, Ashraf U, Qiao J, Li X, Wan X, Zheng Y. Transcriptomic Analysis Provides Insights into the Differential Effects of Aluminum on Peanut (Arachis hypogaea L.). Genes. 2022; 13(10):1830. https://doi.org/10.3390/genes13101830
Chicago/Turabian StyleBao, Gegen, Shengyu Li, Qi Zhou, Umair Ashraf, Jingxuan Qiao, Xiaolin Li, Xiaorong Wan, and Yixiong Zheng. 2022. "Transcriptomic Analysis Provides Insights into the Differential Effects of Aluminum on Peanut (Arachis hypogaea L.)" Genes 13, no. 10: 1830. https://doi.org/10.3390/genes13101830






