Regulation of DNA Methylation by Cannabidiol and Its Implications for Psychiatry: New Insights from In Vivo and In Silico Models
Abstract
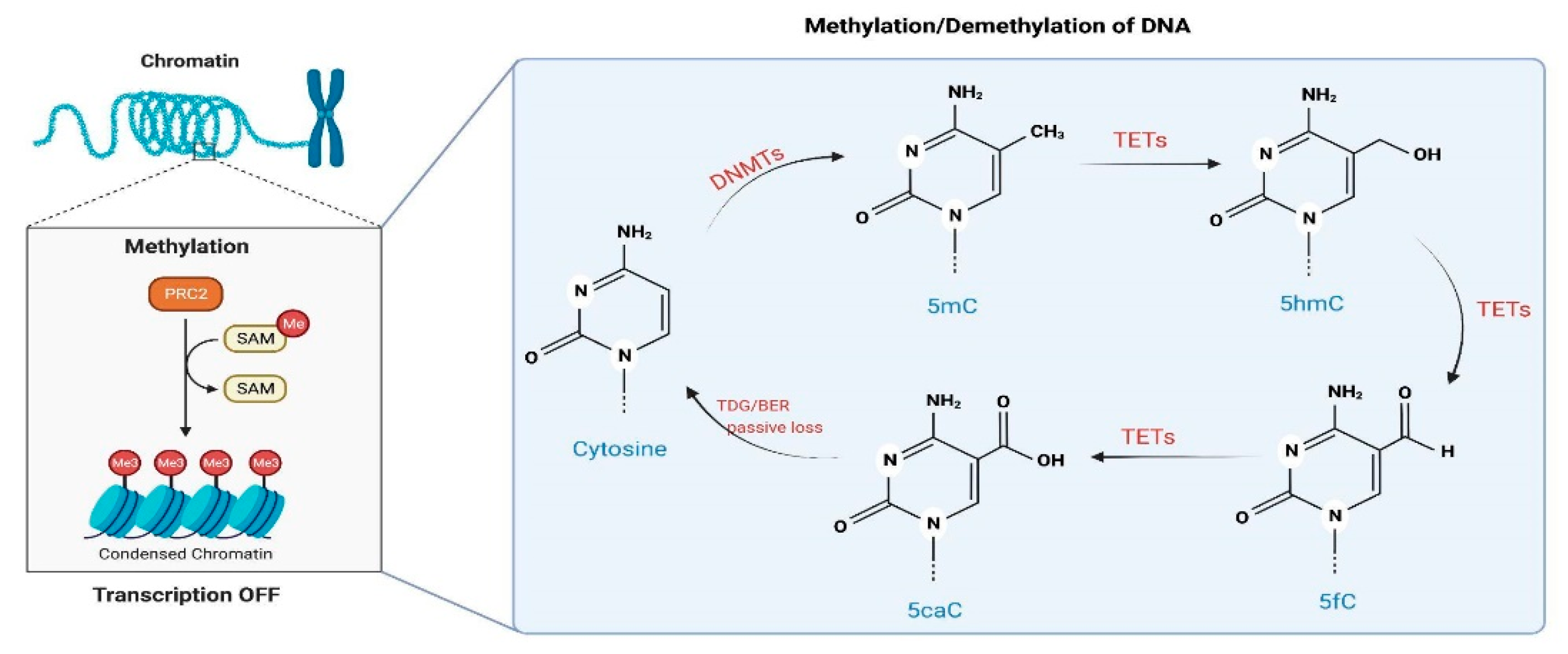
:1. Introduction
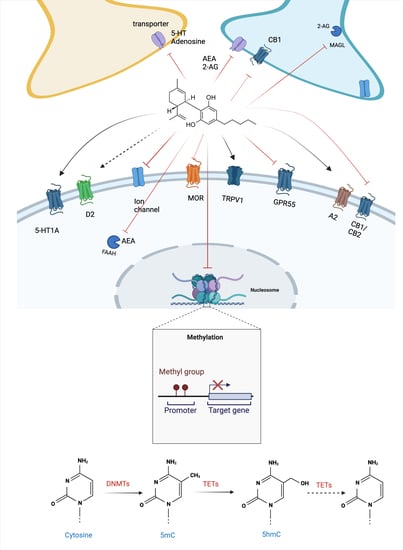
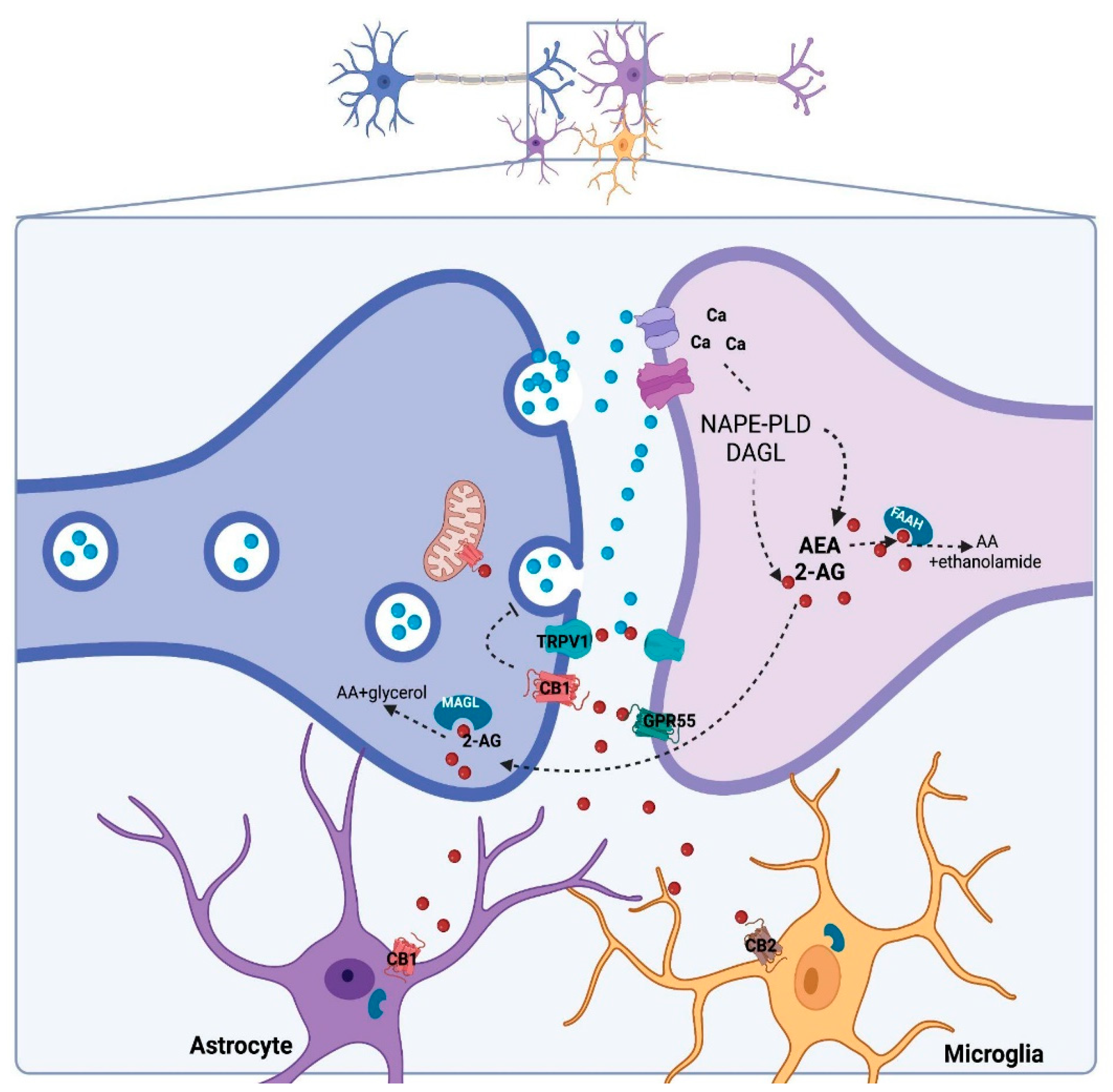
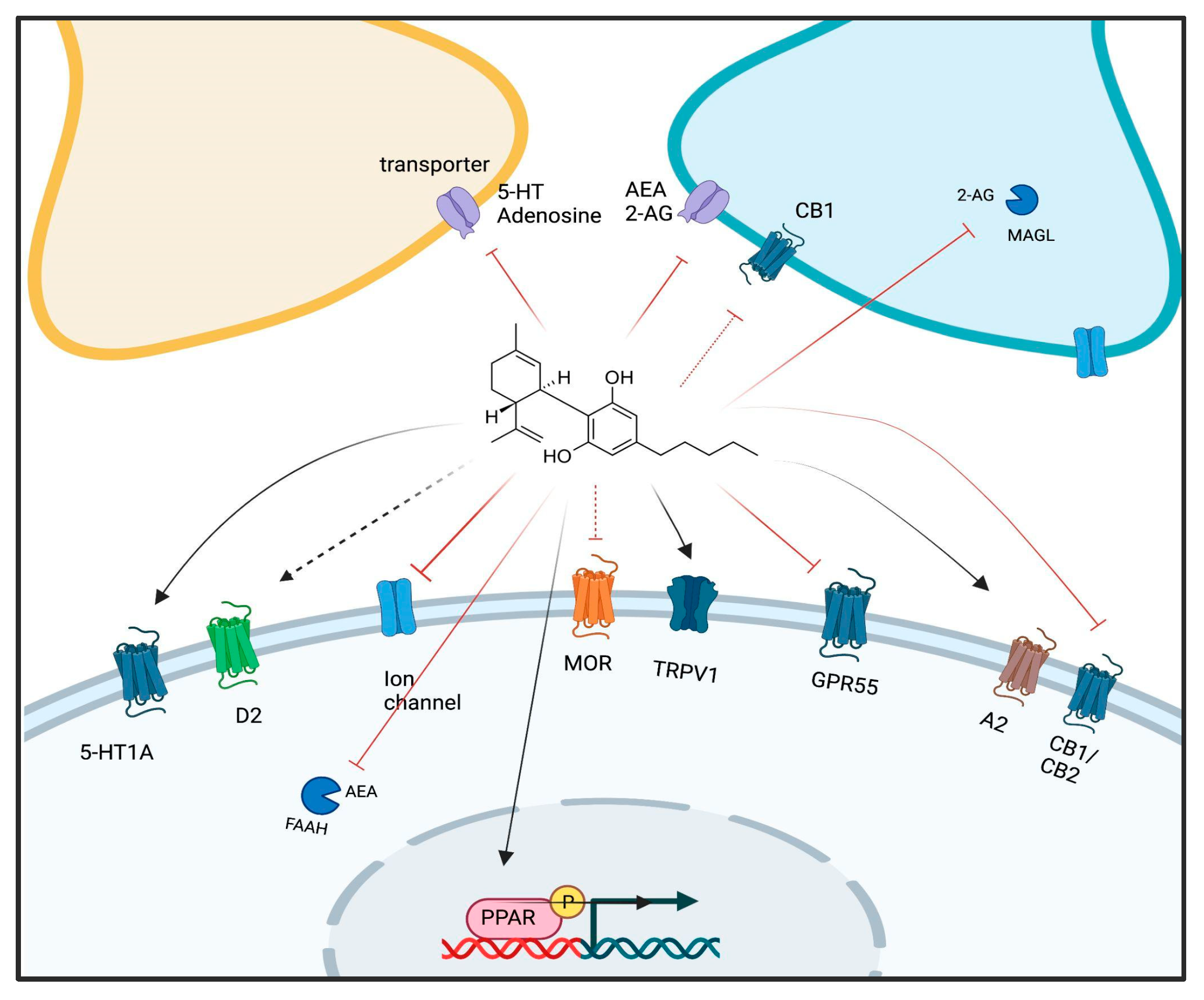
2. Cannabidiol and Its Molecular Targets
3. Regulation of DNAm by CBD
3.1. DNAm and Psychiatric Disorders: A Brief Overview
3.2. CBD Effects on DNAm: In Vivo Evidence
- (a)
- CBD could regulate DNAm by indirectly changing the availability of neurotransmitters, such as eCB and glutamate. As previously mentioned herein, CBD can increase AEA availability [14], and AEA can induce DNMT activity in a CB1-dependent manner involving p38 MAPK signaling in differentiated keratinocytes [120]. CBD can also regulate glutamate levels by blocking its reuptake or indirectly by increasing eCB levels with subsequent CB1 activation, thereby inhibiting neurotransmitter release [121]. Activation of NMDA receptors can regulate DNMT3 activity/expression levels through a CREB-dependent mechanism [122].
- (b)
- CBD could directly target enzymes involved in methylation and demethylation of the DNA, such as DNMTs and TETs, respectively. Currently, there is no evidence that CBD could bind and/or regulate the activity of DNMTs. However, a recent publication indicates that CBD and other related cannabinoids exhibit potent inhibitory activities towards the TET1 protein in vitro, most likely due to interaction with amino acid residues in the active center of the enzyme, according to an in silico molecular docking approach [108].
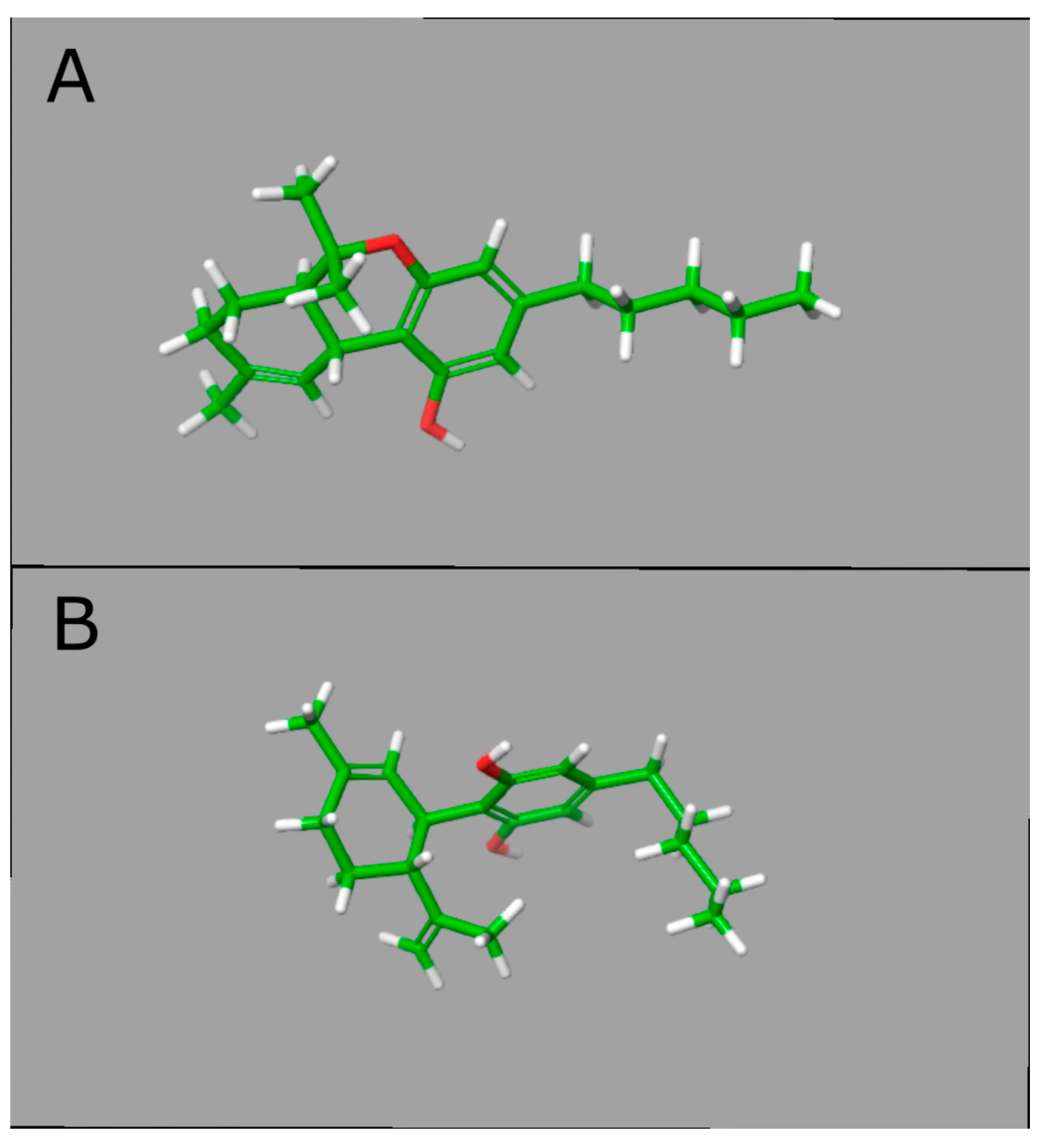
3.3. CBD Effects on DNAm: In Silico Evidence
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zuardi, A.W. History of Cannabis as a Medicine: A Review. Braz. J. Psychiatry 2006, 28, 153–157. [Google Scholar] [CrossRef] [Green Version]
- Li, H.-L. An Archaeological and Historical Account of Cannabis in China. Econ. Bot. 1974, 28, 437–448. [Google Scholar] [CrossRef]
- Ren, M.; Tang, Z.; Wu, X.; Spengler, R.; Jiang, H.; Yang, Y.; Boivin, N. The Origins of Cannabis Smoking: Chemical Residue Evidence from the First Millennium BCE in the Pamirs. Sci. Adv. 2019, 5, eaaw1391. [Google Scholar] [CrossRef] [Green Version]
- Warf, B. High Points: An Historical Geography of Cannabis. Geogr. Rev. 2014, 104, 414–438. [Google Scholar] [CrossRef]
- Mechoulam, R.; Shani, A.; Edery, H.; Grunfeld, Y. Chemical Basis of Hashish Activity. Science 1970, 169, 611–612. [Google Scholar] [CrossRef]
- Gaoni, Y.; Mechoulam, R. Isolation, Structure, and Partial Synthesis of an Active Constituent of Hashish. J. Am. Chem. Soc. 1964, 86, 1646–1647. [Google Scholar] [CrossRef]
- Turner, S.E.; Williams, C.M.; Iversen, L.; Whalley, B.J. Molecular Pharmacology of Phytocannabinoids. Prog. Chem. Org. Nat. Prod. 2017, 103, 61–101. [Google Scholar] [CrossRef]
- Ladha, K.S.; Ajrawat, P.; Yang, Y.; Clarke, H. Understanding the Medical Chemistry of the Cannabis Plant Is Critical to Guiding Real World Clinical Evidence. Molecules 2020, 25, 4042. [Google Scholar] [CrossRef]
- Britch, S.C.; Babalonis, S.; Walsh, S.L. Cannabidiol: Pharmacology and Therapeutic Targets. Psychopharmacology 2021, 238, 9–28. [Google Scholar] [CrossRef]
- Bonaccorso, S.; Ricciardi, A.; Zangani, C.; Chiappini, S.; Schifano, F. Cannabidiol (CBD) Use in Psychiatric Disorders: A Systematic Review. Neurotoxicology 2019, 74, 282–298. [Google Scholar] [CrossRef]
- Schoedel, K.A.; Szeto, I.; Setnik, B.; Sellers, E.M.; Levy-Cooperman, N.; Mills, C.; Etges, T.; Sommerville, K. Abuse Potential Assessment of Cannabidiol (CBD) in Recreational Polydrug Users: A Randomized, Double-Blind, Controlled Trial. Epilepsy Behav. 2018, 88, 162–171. [Google Scholar] [CrossRef] [Green Version]
- National Academies of Sciences, Engineering, and Medicine; Health and Medicine Division; Board on Population Health and Public Health Practice; Committee on the Health Effects of Marijuana: An Evidence Review and Research Agenda. The Health Effects of Cannabis and Cannabinoids: The Current State of Evidence and Recommendations for Research; National Academies Press: Washington, DC, USA, 2017. [Google Scholar]
- Ożarowski, M.; Karpiński, T.M.; Zielińska, A.; Souto, E.B.; Wielgus, K. Cannabidiol in Neurological and Neoplastic Diseases: Latest Developments on the Molecular Mechanism of Action. Int. J. Mol. Sci. 2021, 22, 4294. [Google Scholar] [CrossRef]
- Peng, J.; Fan, M.; An, C.; Ni, F.; Huang, W.; Luo, J. A Narrative Review of Molecular Mechanism and Therapeutic Effect of Cannabidiol (CBD). Basic Clin. Pharm. Toxicol. 2022, 130, 439–456. [Google Scholar] [CrossRef]
- Mechoulam, R. The Pharmacohistory of Cannabis Sativa. In Cannabinoids as Therapeutic Agents; Chapman and Hall/CRC: Boca Raton, FL, USA, 1986; ISBN 978-0-429-26066-7. [Google Scholar]
- Pacher, P.; Kogan, N.M.; Mechoulam, R. Beyond THC and Endocannabinoids. Annu. Rev. Pharm. Toxicol. 2020, 60, 637–659. [Google Scholar] [CrossRef] [Green Version]
- Zanelati, T.V.; Biojone, C.; Moreira, F.A.; Guimarães, F.S.; Joca, S.R.L. Antidepressant-like Effects of Cannabidiol in Mice: Possible Involvement of 5-HT1A Receptors. Br. J. Pharm. 2010, 159, 122–128. [Google Scholar] [CrossRef] [Green Version]
- Joca, S.; Silote, G.P.; Sartim, A.; Sales, A.; Guimarães, F.; Wegener, G. Chapter 45—Putative Effects of Cannabidiol in Depression and Synaptic Plasticity. In The Neuroscience of Depression; Martin, C.R., Hunter, L.-A., Patel, V.B., Preedy, V.R., Rajendram, R., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 459–467. ISBN 978-0-12-817933-8. [Google Scholar]
- Paulus, V.; Billieux, J.; Benyamina, A.; Karila, L. Cannabidiol in the Context of Substance Use Disorder Treatment: A Systematic Review. Addict. Behav. 2022, 132, 107360. [Google Scholar] [CrossRef]
- Filippini, G.; Minozzi, S.; Borrelli, F.; Cinquini, M.; Dwan, K. Cannabis and Cannabinoids for Symptomatic Treatment for People with Multiple Sclerosis. Cochrane Database Syst. Rev. 2022, 5, CD013444. [Google Scholar] [CrossRef]
- Sainsbury, B.; Bloxham, J.; Pour, M.H.; Padilla, M.; Enciso, R. Efficacy of Cannabis-Based Medications Compared to Placebo for the Treatment of Chronic Neuropathic Pain: A Systematic Review with Meta-Analysis. J. Dent. Anesth. Pain. Med. 2021, 21, 479–506. [Google Scholar] [CrossRef]
- Khan, R.; Naveed, S.; Mian, N.; Fida, A.; Raafey, M.A.; Aedma, K.K. The Therapeutic Role of Cannabidiol in Mental Health: A Systematic Review. J. Cannabis Res. 2020, 2, 2. [Google Scholar] [CrossRef] [Green Version]
- Pavel, A.N.; Paun, R.; Matei, V.P. The Use of Cannabidiol in Treating Psychiatric Disorders: A Systematic Review. Psychiatry Clin. Psychopharmacol. 2021, 31, 226–232. [Google Scholar] [CrossRef]
- Ibeas Bih, C.; Chen, T.; Nunn, A.V.W.; Bazelot, M.; Dallas, M.; Whalley, B.J. Molecular Targets of Cannabidiol in Neurological Disorders. Neurotherapeutics 2015, 12, 699–730. [Google Scholar] [CrossRef] [Green Version]
- Elsaid, S.; Le Foll, B. The Complexity of Pharmacology of Cannabidiol (CBD) and Its Implications in the Treatment of Brain Disorders. Neuropsychopharmacology 2020, 45, 229–230. [Google Scholar] [CrossRef]
- Keverne, J.; Binder, E.B. A Review of Epigenetics in Psychiatry: Focus on Environmental Risk Factors. Med. Genet. 2020, 32, 57–64. [Google Scholar] [CrossRef]
- Liu, C.; Jiao, C.; Wang, K.; Yuan, N. DNA Methylation and Psychiatric Disorders. Prog. Mol. Biol. Transl. Sci. 2018, 157, 175–232. [Google Scholar] [CrossRef]
- Starnawska, A.; Demontis, D. Role of DNA Methylation in Mediating Genetic Risk of Psychiatric Disorders. Front. Psychiatry 2021, 12, 596821. [Google Scholar] [CrossRef]
- Reggio, P.H.; Bramblett, R.D.; Yuknavich, H.; Seltzman, H.H.; Fleming, D.N.; Fernando, S.R.; Stevenson, L.A.; Pertwee, R.G. The Design, Synthesis and Testing of Desoxy-CBD: Further Evidence for a Region of Steric Interference at the Cannabinoid Receptor. Life. Sci. 1995, 56, 2025–2032. [Google Scholar] [CrossRef]
- Pertwee, R.G. The Diverse CB1 and CB2 Receptor Pharmacology of Three Plant Cannabinoids: Delta9-Tetrahydrocannabinol, Cannabidiol and Delta9-Tetrahydrocannabivarin. Br. J. Pharm. 2008, 153, 199–215. [Google Scholar] [CrossRef] [Green Version]
- Silveira, K.M.; Wegener, G.; Joca, S.R.L. Targeting 2-Arachidonoylglycerol Signalling in the Neurobiology and Treatment of Depression. Basic Clin. Pharm. Toxicol. 2021, 129, 3–14. [Google Scholar] [CrossRef]
- Freeman, A.M.; Petrilli, K.; Lees, R.; Hindocha, C.; Mokrysz, C.; Curran, H.V.; Saunders, R.; Freeman, T.P. How Does Cannabidiol (CBD) Influence the Acute Effects of Delta-9-Tetrahydrocannabinol (THC) in Humans? A Systematic Review. Neurosci. Biobehav. Rev. 2019, 107, 696–712. [Google Scholar] [CrossRef]
- McPartland, J.M.; Duncan, M.; Di Marzo, V.; Pertwee, R.G. Are Cannabidiol and Δ(9) -Tetrahydrocannabivarin Negative Modulators of the Endocannabinoid System? A Systematic Review. Br. J. Pharm. 2015, 172, 737–753. [Google Scholar] [CrossRef]
- Thomas, A.; Baillie, G.L.; Phillips, A.M.; Razdan, R.K.; Ross, R.A.; Pertwee, R.G. Cannabidiol Displays Unexpectedly High Potency as an Antagonist of CB1 and CB2 Receptor Agonists in Vitro. Br. J. Pharm. 2007, 150, 613–623. [Google Scholar] [CrossRef] [Green Version]
- Thiele, E.A.; Bebin, E.M.; Bhathal, H.; Jansen, F.E.; Kotulska, K.; Lawson, J.A.; O’Callaghan, F.J.; Wong, M.; Sahebkar, F.; Checketts, D.; et al. Add-on Cannabidiol Treatment for Drug-Resistant Seizures in Tuberous Sclerosis Complex: A Placebo-Controlled Randomized Clinical Trial. JAMA Neurol. 2021, 78, 285–292. [Google Scholar] [CrossRef]
- Zuardi, A.W.; Shirakawa, I.; Finkelfarb, E.; Karniol, I.G. Action of Cannabidiol on the Anxiety and Other Effects Produced by Delta 9-THC in Normal Subjects. Psychopharmacology 1982, 76, 245–250. [Google Scholar] [CrossRef]
- Fowler, C.J. Anandamide Uptake Explained? Trends Pharm. Sci. 2012, 33, 181–185. [Google Scholar] [CrossRef]
- Elmes, M.W.; Kaczocha, M.; Berger, W.T.; Leung, K.; Ralph, B.P.; Wang, L.; Sweeney, J.M.; Miyauchi, J.T.; Tsirka, S.E.; Ojima, I.; et al. Fatty Acid-Binding Proteins (FABPs) Are Intracellular Carriers for Δ9-Tetrahydrocannabinol (THC) and Cannabidiol (CBD). J. Biol. Chem. 2015, 290, 8711–8721. [Google Scholar] [CrossRef] [Green Version]
- Sartim, A.G.; Guimarães, F.S.; Joca, S.R.L. Antidepressant-like Effect of Cannabidiol Injection into the Ventral Medial Prefrontal Cortex-Possible Involvement of 5-HT1A and CB1 Receptors. Behav. Brain. Res. 2016, 303, 218–227. [Google Scholar] [CrossRef]
- Casarotto, P.C.; Gomes, F.V.; Resstel, L.B.M.; Guimarães, F.S. Cannabidiol Inhibitory Effect on Marble-Burying Behaviour: Involvement of CB1 Receptors. Behav. Pharm. 2010, 21, 353–358. [Google Scholar] [CrossRef]
- Campos, A.C.; Ortega, Z.; Palazuelos, J.; Fogaça, M.V.; Aguiar, D.C.; Díaz-Alonso, J.; Ortega-Gutiérrez, S.; Vázquez-Villa, H.; Moreira, F.A.; Guzmán, M.; et al. The Anxiolytic Effect of Cannabidiol on Chronically Stressed Mice Depends on Hippocampal Neurogenesis: Involvement of the Endocannabinoid System. Int. J. Neuropsychopharmacol. 2013, 16, 1407–1419. [Google Scholar] [CrossRef] [Green Version]
- de Almeida, D.L.; Devi, L.A. Diversity of Molecular Targets and Signaling Pathways for CBD. Pharm. Res. Perspect. 2020, 8, e00682. [Google Scholar] [CrossRef]
- Abyadeh, M.; Gupta, V.; Paulo, J.A.; Gupta, V.; Chitranshi, N.; Godinez, A.; Saks, D.; Hasan, M.; Amirkhani, A.; McKay, M.; et al. A Proteomic View of Cellular and Molecular Effects of Cannabis. Biomolecules 2021, 11, 1411. [Google Scholar] [CrossRef]
- Maguire, R.F.; Wilkinson, D.J.; England, T.J.; O’Sullivan, S.E. The Pharmacological Effects of Plant-Derived versus Synthetic Cannabidiol in Human Cell Lines. MCA 2021, 4, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Zuardi, A.W.; Rodrigues, N.P.; Silva, A.L.; Bernardo, S.A.; Hallak, J.E.C.; Guimarães, F.S.; Crippa, J.A.S. Inverted U-Shaped Dose-Response Curve of the Anxiolytic Effect of Cannabidiol during Public Speaking in Real Life. Front. Pharm. 2017, 8, 259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fogaça, M.V.; Campos, A.C.; Coelho, L.D.; Duman, R.S.; Guimarães, F.S. The Anxiolytic Effects of Cannabidiol in Chronically Stressed Mice Are Mediated by the Endocannabinoid System: Role of Neurogenesis and Dendritic Remodeling. Neuropharmacology 2018, 135, 22–33. [Google Scholar] [CrossRef] [PubMed]
- da Silva, N.R.; Gomes, F.V.; Sonego, A.B.; da Silva, N.R.; Guimarães, F.S. Cannabidiol Attenuates Behavioral Changes in a Rodent Model of Schizophrenia through 5-HT1A, but Not CB1 and CB2 Receptors. Pharm. Res. 2020, 156, 104749. [Google Scholar] [CrossRef] [PubMed]
- Batalla, A.; Janssen, H.; Gangadin, S.S.; Bossong, M.G. The Potential of Cannabidiol as a Treatment for Psychosis and Addiction: Who Benefits Most? A Systematic Review. J. Clin. Med. 2019, 8, 1058. [Google Scholar] [CrossRef] [Green Version]
- Silote, G.P.; Gatto, M.C.; Eskelund, A.; Guimarães, F.S.; Wegener, G.; Joca, S.R.L. Strain-, Sex-, and Time-Dependent Antidepressant-like Effects of Cannabidiol. Pharmaceuticals 2021, 14, 1269. [Google Scholar] [CrossRef]
- Silote, G.P.; Sartim, A.; Sales, A.; Eskelund, A.; Guimarães, F.S.; Wegener, G.; Joca, S. Emerging Evidence for the Antidepressant Effect of Cannabidiol and the Underlying Molecular Mechanisms. J. Chem. Neuroanat. 2019, 98, 104–116. [Google Scholar] [CrossRef]
- Bitencourt, R.M.; Takahashi, R.N. Cannabidiol as a Therapeutic Alternative for Post-Traumatic Stress Disorder: From Bench Research to Confirmation in Human Trials. Front. Neurosci. 2018, 12, 502. [Google Scholar] [CrossRef] [Green Version]
- Campos, A.C.; Fogaça, M.V.; Sonego, A.B.; Guimarães, F.S. Cannabidiol, Neuroprotection and Neuropsychiatric Disorders. Pharm. Res. 2016, 112, 119–127. [Google Scholar] [CrossRef]
- Luján, M.Á.; Valverde, O. The Pro-Neurogenic Effects of Cannabidiol and Its Potential Therapeutic Implications in Psychiatric Disorders. Front. Behav. Neurosci. 2020, 14, 109. [Google Scholar] [CrossRef]
- Philpott, H.T.; O’Brien, M.; McDougall, J.J. Attenuation of Early Phase Inflammation by Cannabidiol Prevents Pain and Nerve Damage in Rat Osteoarthritis. Pain 2017, 158, 2442–2451. [Google Scholar] [CrossRef] [PubMed]
- Burstein, S. Cannabidiol (CBD) and Its Analogs: A Review of Their Effects on Inflammation. Bioorg. Med. Chem. 2015, 23, 1377–1385. [Google Scholar] [CrossRef] [PubMed]
- Sales, A.J.; Crestani, C.C.; Guimarães, F.S.; Joca, S.R.L. Antidepressant-like Effect Induced by Cannabidiol Is Dependent on Brain Serotonin Levels. Prog. Neuropsychopharmacol. Biol. Psychiatry 2018, 86, 255–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linge, R.; Jiménez-Sánchez, L.; Campa, L.; Pilar-Cuéllar, F.; Vidal, R.; Pazos, A.; Adell, A.; Díaz, Á. Cannabidiol Induces Rapid-Acting Antidepressant-like Effects and Enhances Cortical 5-HT/Glutamate Neurotransmission: Role of 5-HT1A Receptors. Neuropharmacology 2016, 103, 16–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakas, T.; van Nieuwenhuijzen, P.S.; Devenish, S.O.; McGregor, I.S.; Arnold, J.C.; Chebib, M. The Direct Actions of Cannabidiol and 2-Arachidonoyl Glycerol at GABAA Receptors. Pharm. Res. 2017, 119, 358–370. [Google Scholar] [CrossRef] [PubMed]
- Maggio, N.; Shavit Stein, E.; Segal, M. Cannabidiol Regulates Long Term Potentiation Following Status Epilepticus: Mediation by Calcium Stores and Serotonin. Front. Mol. Neurosci. 2018, 11, 32. [Google Scholar] [CrossRef]
- Sweatt, J.D. The Emerging Field of Neuroepigenetics. Neuron 2013, 80, 624–632. [Google Scholar] [CrossRef] [Green Version]
- Coda, D.M.; Gräff, J. Neurogenetic and Neuroepigenetic Mechanisms in Cognitive Health and Disease. Front. Mol. Neurosci. 2020, 13, 205. [Google Scholar] [CrossRef]
- Stylianou, E. Epigenetics of Chronic Inflammatory Diseases. J. Inflamm. Res. 2018, 12, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Karpova, N.N.; Sales, A.J.; Joca, S.R. Epigenetic Basis of Neuronal and Synaptic Plasticity. Curr. Top. Med. Chem. 2017, 17, 771–793. [Google Scholar] [CrossRef]
- Sales, A.J.; Maciel, I.S.; Suavinha, A.C.D.R.; Joca, S.R.L. Modulation of DNA Methylation and Gene Expression in Rodent Cortical Neuroplasticity Pathways Exerts Rapid Antidepressant-Like Effects. Mol. Neurobiol. 2021, 58, 777–794. [Google Scholar] [CrossRef] [PubMed]
- Gujar, H.; Weisenberger, D.J.; Liang, G. The Roles of Human DNA Methyltransferases and Their Isoforms in Shaping the Epigenome. Genes 2019, 10, 172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christopher, M.A.; Kyle, S.M.; Katz, D.J. Neuroepigenetic Mechanisms in Disease. Epigenetics Chromatin. 2017, 10, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bannister, A.J.; Kouzarides, T. Regulation of Chromatin by Histone Modifications. Cell. Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Ravichandran, M.; Jurkowska, R.Z.; Jurkowski, T.P. Target Specificity of Mammalian DNA Methylation and Demethylation Machinery. Org. Biomol. Chem. 2018, 16, 1419–1435. [Google Scholar] [CrossRef]
- Ito, S.; Shen, L.; Dai, Q.; Wu, S.C.; Collins, L.B.; Swenberg, J.A.; He, C.; Zhang, Y. Tet Proteins Can Convert 5-Methylcytosine to 5-Formylcytosine and 5-Carboxylcytosine. Science 2011, 333, 1300–1303. [Google Scholar] [CrossRef] [Green Version]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.U.; Su, Y.; Zhong, C.; Ming, G.; Song, H. Hydroxylation of 5-Methylcytosine by TET1 Promotes Active DNA Demethylation in the Adult Brain. Cell 2011, 145, 423–434. [Google Scholar] [CrossRef] [Green Version]
- Kaas, G.A.; Zhong, C.; Eason, D.E.; Ross, D.L.; Vachhani, R.V.; Ming, G.-L.; King, J.R.; Song, H.; Sweatt, J.D. TET1 Controls CNS 5-Methylcytosine Hydroxylation, Active DNA Demethylation, Gene Transcription, and Memory Formation. Neuron 2013, 79, 1086–1093. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.; Su, Y.; Shin, J.; Zhong, C.; Guo, J.U.; Weng, Y.-L.; Gao, F.; Geschwind, D.H.; Coppola, G.; Ming, G.; et al. Tet3 Regulates Synaptic Transmission and Homeostatic Plasticity via DNA Oxidation and Repair. Nat. Neurosci. 2015, 18, 836–843. [Google Scholar] [CrossRef]
- Shi, D.-Q.; Ali, I.; Tang, J.; Yang, W.-C. New Insights into 5hmC DNA Modification: Generation, Distribution and Function. Front. Genet. 2017, 8, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goud Alladi, C.; Etain, B.; Bellivier, F.; Marie-Claire, C. DNA Methylation as a Biomarker of Treatment Response Variability in Serious Mental Illnesses: A Systematic Review Focused on Bipolar Disorder, Schizophrenia, and Major Depressive Disorder. Int. J. Mol. Sci. 2018, 19, 3026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gatta, E.; Saudagar, V.; Auta, J.; Grayson, D.R.; Guidotti, A. Epigenetic Landscape of Stress Surfeit Disorders: Key Role for DNA Methylation Dynamics. Int. Rev. Neurobiol. 2021, 156, 127–183. [Google Scholar] [CrossRef]
- Cao-Lei, L.; Veru, F.; Elgbeili, G.; Szyf, M.; Laplante, D.P.; King, S. DNA Methylation Mediates the Effect of Exposure to Prenatal Maternal Stress on Cytokine Production in Children at Age 13½ Years: Project Ice Storm. Clin. Epigenetics 2016, 8, 54. [Google Scholar] [CrossRef] [Green Version]
- Choudhury, A.; Sahu, T.; Ramanujam, P.L.; Banerjee, A.K.; Chakraborty, I.; Ramachandravarapu, A.K.; Arora, N. Neurochemicals, Behaviours and Psychiatric Perspectives of Neurological Diseases. Neuropsychiatry 2018, 8, 395–424. [Google Scholar] [CrossRef]
- Zannas, A.S.; West, A.E. Epigenetics and the Regulation of Stress Vulnerability and Resilience. Neuroscience 2014, 264, 157–170. [Google Scholar] [CrossRef] [Green Version]
- Heyn, H.; Moran, S.; Hernando-Herraez, I.; Sayols, S.; Gomez, A.; Sandoval, J.; Monk, D.; Hata, K.; Marques-Bonet, T.; Wang, L.; et al. DNA Methylation Contributes to Natural Human Variation. Genome Res. 2013, 23, 1363–1372. [Google Scholar] [CrossRef] [Green Version]
- Cross-Disorder Group of the Psychiatric Genomics Consortium; Lee, S.H.; Ripke, S.; Neale, B.M.; Faraone, S.V.; Purcell, S.M.; Perlis, R.H.; Mowry, B.J.; Thapar, A.; Goddard, M.E.; et al. Genetic Relationship between Five Psychiatric Disorders Estimated from Genome-Wide SNPs. Nat. Genet. 2013, 45, 984–994. [Google Scholar] [CrossRef] [Green Version]
- Kochunov, P.; Ma, Y.; Hatch, K.S.; Jahanshad, N.; Thompson, P.M.; Adhikari, B.M.; Bruce, H.; Van der Vaart, A.; Goldwaser, E.L.; Sotiras, A.; et al. Brain-Wide versus Genome-Wide Vulnerability Biomarkers for Severe Mental Illnesses. Hum. Brain Mapp. 2022, 43, 4970–4983. [Google Scholar] [CrossRef]
- Wu, Y.; Cao, H.; Baranova, A.; Huang, H.; Li, S.; Cai, L.; Rao, S.; Dai, M.; Xie, M.; Dou, Y.; et al. Multi-Trait Analysis for Genome-Wide Association Study of Five Psychiatric Disorders. Transl. Psychiatry 2020, 10, 209. [Google Scholar] [CrossRef]
- Cross-Disorder Group of the Psychiatric Genomics Consortium. Genomic Relationships, Novel Loci, and Pleiotropic Mechanisms across Eight Psychiatric Disorders. Cell 2019, 179, 1469–1482.e11. [Google Scholar] [CrossRef] [Green Version]
- Kular, L.; Kular, S. Epigenetics Applied to Psychiatry: Clinical Opportunities and Future Challenges. Psychiatry Clin. Neurosci. 2018, 72, 195–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Li, M.; Wang, X.; He, Y.; Xia, Y.; Sweeney, J.A.; Kopp, R.F.; Liu, C.; Chen, C. Drug Response-Related DNA Methylation Changes in Schizophrenia, Bipolar Disorder, and Major Depressive Disorder. Front. Neurosci. 2021, 15, 536. [Google Scholar] [CrossRef] [PubMed]
- Sales, A.J.; Guimarães, F.S.; Joca, S.R.L. DNA Methylation in Stress and Depression: From Biomarker to Therapeutics. Acta Neuropsychiatr. 2021, 33, 217–241. [Google Scholar] [CrossRef] [PubMed]
- Sales, A.J.; Joca, S.R.L. Effects of DNA Methylation Inhibitors and Conventional Antidepressants on Mice Behaviour and Brain DNA Methylation Levels. Acta Neuropsychiatr. 2016, 28, 11–22. [Google Scholar] [CrossRef] [Green Version]
- Sales, A.J.; Biojone, C.; Terceti, M.S.; Guimarães, F.S.; Gomes, M.V.; Joca, S.R. Antidepressant-like Effect Induced by Systemic and Intra-Hippocampal Administration of DNA Methylation Inhibitors. Br. J. Pharm. 2011, 164, 1711–1721. [Google Scholar] [CrossRef] [Green Version]
- La Plant, Q.; Vialou, V.; Covington, H.E.; Dumitriu, D.; Feng, J.; Warren, B.L.; Maze, I.; Dietz, D.M.; Watts, E.L.; Iñiguez, S.D.; et al. Dnmt3a Regulates Emotional Behavior and Spine Plasticity in the Nucleus Accumbens. Nat. Neurosci. 2010, 13, 1137–1143. [Google Scholar] [CrossRef] [Green Version]
- Uchida, S.; Hara, K.; Kobayashi, A.; Otsuki, K.; Yamagata, H.; Hobara, T.; Suzuki, T.; Miyata, N.; Watanabe, Y. Epigenetic Status of Gdnf in the Ventral Striatum Determines Susceptibility and Adaptation to Daily Stressful Events. Neuron 2011, 69, 359–372. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Zhang, Y. Reversing DNA Methylation: Mechanisms, Genomics, and Biological Functions. Cell 2014, 156, 45–68. [Google Scholar] [CrossRef] [Green Version]
- Darigh, F.; Iranbakhsh, A.; Oraghi Ardebili, Z.; Ebadi, M.; Hassanpour, H. Simulated Microgravity Contributed to Modification of Callogenesis Performance and Secondary Metabolite Production in CannabisIndica. Plant. Physiol. Biochem. 2022, 186, 157–168. [Google Scholar] [CrossRef]
- Darigh, F.; Iranbakhsh, A.; Oraghi Ardebili, Z.; Ebadi, M. Non-Thermal Plasma Improved Callogenesis Performance and Elicited the Production of Cannabinoids by Modifying DNA Methylome, Expression of WRKY1 and ERF1B Transcription Factors, and Expression of Genes That Contributed to the Biosynthesis of Cannabinoids. Protoplasma 2022, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Massimini, M.; Dalle Vedove, E.; Bachetti, B.; Di Pierro, F.; Ribecco, C.; D’Addario, C.; Pucci, M. Polyphenols and Cannabidiol Modulate Transcriptional Regulation of Th1/Th2 Inflammatory Genes Related to Canine Atopic Dermatitis. Front. Vet. Sci. 2021, 8, 606197. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.; Sominsky, L.; Walder, K.R.; Berk, M.; Marx, W.; Carvalho, A.F.; Bortolasci, C.C.; Maes, M.; Puri, B.K. Inflammation and Nitro-Oxidative Stress as Drivers of Endocannabinoid System Aberrations in Mood Disorders and Schizophrenia. Mol. Neurobiol. 2022, 59, 3485–3503. [Google Scholar] [CrossRef] [PubMed]
- Melas, P.A.; Scherma, M.; Fratta, W.; Cifani, C.; Fadda, P. Cannabidiol as a Potential Treatment for Anxiety and Mood Disorders: Molecular Targets and Epigenetic Insights from Preclinical Research. Int. J. Mol. Sci. 2021, 22, 1863. [Google Scholar] [CrossRef] [PubMed]
- Reece, A.S.; Hulse, G.K. Impacts of Cannabinoid Epigenetics on Human Development: Reflections on Murphy et. al. “Cannabinoid Exposure and Altered DNA Methylation in Rat and Human Sperm” Epigenetics 2018, 13, 1208–1221. Epigenetics 2019, 14, 1041–1056. [Google Scholar] [CrossRef] [Green Version]
- Protic, D.; Salcedo-Arellano, M.J.; Dy, J.B.; Potter, L.A.; Hagerman, R.J. New Targeted Treatments for Fragile X Syndrome. Curr. Pediatr. Rev. 2019, 15, 251–258. [Google Scholar] [CrossRef]
- Pucci, M.; Rapino, C.; Di Francesco, A.; Dainese, E.; D’Addario, C.; Maccarrone, M. Epigenetic Control of Skin Differentiation Genes by Phytocannabinoids. Br. J. Pharm. 2013, 170, 581–591. [Google Scholar] [CrossRef] [Green Version]
- da Silva, V.K.; de Freitas, B.S.; Dornelles, V.C.; Kist, L.W.; Bogo, M.R.; Silva, M.C.; Streck, E.L.; Hallak, J.E.; Zuardi, A.W.; Crippa, J.A.S.; et al. Novel Insights into Mitochondrial Molecular Targets of Iron-Induced Neurodegeneration: Reversal by Cannabidiol. Brain Res. Bull. 2018, 139, 1–8. [Google Scholar] [CrossRef]
- Stark, T.; Ruda-Kucerova, J.; Iannotti, F.A.; D’Addario, C.; Di Marco, R.; Pekarik, V.; Drazanova, E.; Piscitelli, F.; Bari, M.; Babinska, Z.; et al. Peripubertal Cannabidiol Treatment Rescues Behavioral and Neurochemical Abnormalities in the MAM Model of Schizophrenia. Neuropharmacology 2019, 146, 212–221. [Google Scholar] [CrossRef]
- Sales, A.J.; Guimarães, F.S.; Joca, S.R.L. CBD Modulates DNA Methylation in the Prefrontal Cortex and Hippocampus of Mice Exposed to Forced Swim. Behav. Brain Res. 2020, 388, 112627. [Google Scholar] [CrossRef]
- Wanner, N.M.; Colwell, M.; Drown, C.; Faulk, C. Subacute Cannabidiol Alters Genome-Wide DNA Methylation in Adult Mouse Hippocampus. Env. Mol. Mutagen. 2020, 61, 890–900. [Google Scholar] [CrossRef] [PubMed]
- Di Bartolomeo, M.; Stark, T.; Maurel, O.M.; Iannotti, F.A.; Kuchar, M.; Ruda-Kucerova, J.; Piscitelli, F.; Laudani, S.; Pekarik, V.; Salomone, S.; et al. Crosstalk between the Transcriptional Regulation of Dopamine D2 and Cannabinoid CB1 Receptors in Schizophrenia: Analyses in Patients and in Perinatal Δ9-Tetrahydrocannabinol-Exposed Rats. Pharm. Res. 2021, 164, 105357. [Google Scholar] [CrossRef] [PubMed]
- Wanner, N.M.; Colwell, M.; Drown, C.; Faulk, C. Developmental Cannabidiol Exposure Increases Anxiety and Modifies Genome-Wide Brain DNA Methylation in Adult Female Mice. Clin. Epigenetics 2021, 13, 4. [Google Scholar] [CrossRef] [PubMed]
- Pedrazzi, J.F.C.; Sales, A.J.; Guimarães, F.S.; Joca, S.R.L.; Crippa, J.A.S.; Del Bel, E. Cannabidiol Prevents Disruptions in Sensorimotor Gating Induced by Psychotomimetic Drugs That Last for 24-h with Probable Involvement of Epigenetic Changes in the Ventral Striatum. Prog. Neuropsychopharmacol. Biol. Psychiatry 2021, 111, 110352. [Google Scholar] [CrossRef]
- Antonyová, V.; Kejík, Z.; Brogyanyi, T.; Kaplánek, R.; Veselá, K.; Abramenko, N.; Ocelka, T.; Masařík, M.; Matkowski, A.; Gburek, J.; et al. Non-Psychotropic Cannabinoids as Inhibitors of TET1 Protein. Bioorg. Chem. 2022, 124, 105793. [Google Scholar] [CrossRef]
- Mohammad, G.S.; Joca, S.; Starnawska, A. The Cannabis-Induced Epigenetic Regulation of Genes Associated with Major Depressive Disorder. Genes 2022, 13, 1435. [Google Scholar] [CrossRef]
- Kasahara, T.; Kato, T. What Can Mitochondrial DNA Analysis Tell Us About Mood Disorders? Biol. Psychiatry 2018, 83, 731–738. [Google Scholar] [CrossRef] [Green Version]
- Shao, L.; Martin, M.V.; Watson, S.J.; Schatzberg, A.; Akil, H.; Myers, R.M.; Jones, E.G.; Bunney, W.E.; Vawter, M.P. Mitochondrial Involvement in Psychiatric Disorders. Ann. Med. 2008, 40, 281–295. [Google Scholar] [CrossRef] [Green Version]
- Weger, M.; Alpern, D.; Cherix, A.; Ghosal, S.; Grosse, J.; Russeil, J.; Gruetter, R.; de Kloet, E.R.; Deplancke, B.; Sandi, C. Mitochondrial Gene Signature in the Prefrontal Cortex for Differential Susceptibility to Chronic Stress. Sci. Rep. 2020, 10, 18308. [Google Scholar] [CrossRef]
- Hannon, E.; Dempster, E.L.; Mansell, G.; Burrage, J.; Bass, N.; Bohlken, M.M.; Corvin, A.; Curtis, C.J.; Dempster, D.; Di Forti, M.; et al. DNA Methylation Meta-Analysis Reveals Cellular Alterations in Psychosis and Markers of Treatment-Resistant Schizophrenia. eLife 2021, 10, e58430. [Google Scholar] [CrossRef]
- Ovenden, E.S.; McGregor, N.W.; Emsley, R.A.; Warnich, L. DNA Methylation and Antipsychotic Treatment Mechanisms in Schizophrenia: Progress and Future Directions. Prog. Neuropsychopharmacol. Biol. Psychiatry 2018, 81, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Gururajan, A.; Reif, A.; Cryan, J.F.; Slattery, D.A. The Future of Rodent Models in Depression Research. Nat. Rev. Neurosci. 2019, 20, 686–701. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-C.; Wang, K.-Y.; Shen, C.-K.J. DNA 5-Methylcytosine Demethylation Activities of the Mammalian DNA Methyltransferases. J. Biol. Chem. 2013, 288, 9084–9091. [Google Scholar] [CrossRef] [Green Version]
- Sales, A.J.; Joca, S.R.L. Antidepressant Administration Modulates Stress-Induced DNA Methylation and DNA Methyltransferase Expression in Rat Prefrontal Cortex and Hippocampus. Behav. Brain. Res. 2018, 343, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Sales, A.J.; Fogaça, M.V.; Sartim, A.G.; Pereira, V.S.; Wegener, G.; Guimarães, F.S.; Joca, S.R.L. Cannabidiol Induces Rapid and Sustained Antidepressant-Like Effects Through Increased BDNF Signaling and Synaptogenesis in the Prefrontal Cortex. Mol. Neurobiol. 2019, 56, 1070–1081. [Google Scholar] [CrossRef] [PubMed]
- Zheleznyakova, G.Y.; Cao, H.; Schiöth, H.B. BDNF DNA Methylation Changes as a Biomarker of Psychiatric Disorders: Literature Review and Open Access Database Analysis. Behav. Brain Funct. 2016, 12, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paradisi, A.; Pasquariello, N.; Barcaroli, D.; Maccarrone, M. Anandamide Regulates Keratinocyte Differentiation by Inducing DNA Methylation in a CB1 Receptor-Dependent Manner. J. Biol. Chem. 2008, 283, 6005–6012. [Google Scholar] [CrossRef] [Green Version]
- Sugaya, Y.; Kano, M. Endocannabinoid-Mediated Control of Neural Circuit Excitability and Epileptic Seizures. Front. Neural. Circuits 2022, 15, 781113. [Google Scholar] [CrossRef]
- Lv, J.; Xin, Y.; Zhou, W.; Qiu, Z. The Epigenetic Switches for Neural Development and Psychiatric Disorders. J. Genet. Genom. 2013, 40, 339–346. [Google Scholar] [CrossRef]
- Feng, J.; Pena, C.J.; Purushothaman, I.; Engmann, O.; Walker, D.; Brown, A.N.; Issler, O.; Doyle, M.; Harrigan, E.; Mouzon, E.; et al. Tet1 in Nucleus Accumbens Opposes Depression- and Anxiety-Like Behaviors. Neuropsychopharmacology 2017, 42, 1657–1669. [Google Scholar] [CrossRef]
- Pappalardi, M.B.; Keenan, K.; Cockerill, M.; Kellner, W.A.; Stowell, A.; Sherk, C.; Wong, K.; Pathuri, S.; Briand, J.; Steidel, M.; et al. Discovery of a First-in-Class Reversible DNMT1-Selective Inhibitor with Improved Tolerability and Efficacy in Acute Myeloid Leukemia. Nat. Cancer 2021, 2, 1002–1017. [Google Scholar] [CrossRef]
- Zhang, Z.-M.; Lu, R.; Wang, P.; Yu, Y.; Chen, D.; Gao, L.; Liu, S.; Ji, D.; Rothbart, S.B.; Wang, Y.; et al. Structural Basis for DNMT3A-Mediated de Novo DNA Methylation. Nature 2018, 554, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Emperle, M.; Guo, Y.; Grimm, S.A.; Ren, W.; Adam, S.; Uryu, H.; Zhang, Z.-M.; Chen, D.; Yin, J.; et al. Comprehensive Structure-Function Characterization of DNMT3B and DNMT3A Reveals Distinctive de Novo DNA Methylation Mechanisms. Nat. Commun. 2020, 11, 3355. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein-Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [PubMed]

| Article Categories | Cell or Tissue Type | Type of Study | CBD Effects Related Psychiatric Disorder | Gene | DNAm Measure | Main Findings | Assessment | Reference |
|---|---|---|---|---|---|---|---|---|
| Comparative study | Keratinocytes human HaCaT cells | In vitro | N.A. | Keratin 10 and global DNAm | DNAm-specific primed PCR Methyl-accepting assay with CpG methylase SssI | CBD (0.5 µM) increased DNAm of keratin 10 gene CBD (0.5 µM) increased global DNAm by selectively enhancing DNMT expression | First evidence that CBD could target DNAm | [100] |
| Research study | Hippocampal mitochondria | Animal model (male Wistar rats) | Neurodegenerative | N.A. | Methylated DNA quantification ELISA kit | CBD (10 mg/kg/day, i.p.; 14 consecutive days) attenuated iron-induced decrease in global DNAm | First evidence that CBD restores hippocampal DNAm of mitochondrial mtDNA | [101] |
| Research study | Prefrontal cortex | Animal model (male Sprague-Dawley rats) | Schizophrenia | CNR1 | Pyrosequencing of bisulfite converted DNA | CBD (30 mg/kg/day, i.p.; 20 days) reduced DNAm in the CNR1 promoter | First evidence regarding the involvement of DNAm in the antipsychotic properties of CBD | [102] |
| Review | N.A. | N.A. | Fragile X Syndrome | N.A. | N.A. | N.A. | None about CBD and DNAm | [99] |
| Review | N.A. | N.A. | N.A. | N.A. | N.A. | N.A. | Brief review of published articles | [98] |
| Research study | Prefrontal cortex and hippocampus | Animal model (male Swiss mice) | Depression | Global DNAm | DNAm ELISA kit | CBD (10 mg/kg, i.p.) attenuated the DNAm changes induced by stress (increasing DNAm in the prefrontal cortex and the decreasing DNAm in the hippocampus) | First evidence regarding the involvement of DNAm in the antidepressant-like action of CBD | [103] |
| Research study | Hippocampus | Animal model (male Agouti viable yellow- Avy mice) | Autism, schizophrenia | N.A. | Genome-wide DNAm (reduced-representation bisulfite sequencing) | 3323 genes’ differentially methylated loci were found in CBD-exposed animals (20 mg/kg/day, p.o.; 14 days) | CBD modifies DNAm in genes relevant for psychiatric diseases | [104] |
| Research study | Prefrontal cortex and PBMCs | Animal model (male Sprague-Dawley) | Schizophrenia | DRD2 dopamine D2 receptor | Pyrosequencing | DRD2 DNAm (CpG site 1) was reduced in the PBMCs of schizophrenic subjects CBD (30 mg/kg/day, i.p.; 21 consecutive days) attenuated the DRD2 DNAm reduction in the prefrontal cortex of rats exposed to the THC | Peripubertal CBD treatment reverted DNAm modulation of DRD2 in rats | [105] |
| Comparative study | Cortex and hippocampus | Animal model (female Avy mice) | Neurodevelopmental disorders, epilepsy and others | N.A. | Genome-wide DNAm (reduced-representation bisulfite sequencing) | CBD (20 mg/kg/day, i.p.; 14 consecutive days) | First evidence that developmental CBD exposure modified DNAm | [106] |
| Review | N.A. | N.A. | Mood disorders and anxiety | N.A. | N.A. | N.A. | Showed evidence that therapeutic effects of CBD could involve DNAm | [97] |
| Research study | Canine monocyte-macrophage (DH82) and epidermal keratinocytes cells | In vitro | Canine atopic dermatitis | Ccl2, ccl17 and tslp, il31ra | Bisulfite-treated DNAm pyrosequencing | The nutraceutical mixture induced a significant downregulation of many genes in immune cells, along with increased DNAm | CBD effects were not investigated isolated, but only as part of the nutraceutical treatment (mixture containing polyphenols and cannabinoids), making it difficult to assess CBD effects | [95] |
| Research study | Ventral striatum and prefrontal cortex | Animal model (male Swiss mice) | Schizophrenia | Global DNAm | DNAm ELISA kit | CBD (30 and 60 mg/kg, i.p.) prevented the amphetamine-induced DNAm increase in the ventral striatum | First evidence that CBD has sustained antipsychotic-like action, suggesting the involvement of DNAm in these effects | [107] |
| Review | N.A. | N.A. | Mood disorders and schizophrenia | N.A. | N.A. | N.A. | None about CBD and DNAm | [96] |
| Research study | N.A. | Silico molecular docking | N.A. | N.A. | N.A. | Cannabinoids, including CBD, inhibited the activity of TET1 protein | First in silico evidence that CBD can regulate DNAm through direct interaction with TET1 Did not investigate CBD effects on DNAm | [108] |
| Research study | Leaf-originated explant | In vitro | N.A. | N.A. | Methylation-sensitive amplification polymorphism | The plasma treatment induced differential DNA methylome | Did not investigate CBD effects on DNAm | [94] |
| Research study | Callus cells of Cannabis indica | In vitro | N.A. | N.A. | Methylation-sensitive amplification polymorphism | Simulated microgravity-triggered changes in the DNAm profile | Did not investigate CBD effects on DNAm | [93] |
| Review | N.A. | N.A. | Depression | N.A. | N.A. | N.A. | Showed that multiple genes related with depression are differentially methylated upon exposure to the cannabis or cannabis-derived compounds, including CBD | [109] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Domingos, L.B.; Silva, N.R.; Chaves Filho, A.J.M.; Sales, A.J.; Starnawska, A.; Joca, S. Regulation of DNA Methylation by Cannabidiol and Its Implications for Psychiatry: New Insights from In Vivo and In Silico Models. Genes 2022, 13, 2165. https://doi.org/10.3390/genes13112165
Domingos LB, Silva NR, Chaves Filho AJM, Sales AJ, Starnawska A, Joca S. Regulation of DNA Methylation by Cannabidiol and Its Implications for Psychiatry: New Insights from In Vivo and In Silico Models. Genes. 2022; 13(11):2165. https://doi.org/10.3390/genes13112165
Chicago/Turabian StyleDomingos, Luana B., Nicole R. Silva, Adriano J. M. Chaves Filho, Amanda J. Sales, Anna Starnawska, and Sâmia Joca. 2022. "Regulation of DNA Methylation by Cannabidiol and Its Implications for Psychiatry: New Insights from In Vivo and In Silico Models" Genes 13, no. 11: 2165. https://doi.org/10.3390/genes13112165