Integrative Meta-Analysis of Huntington’s Disease Transcriptome Landscape
Abstract
1. Introduction
2. Methods
2.1. Dataset and Pre-Processing
2.2. Alignment: Reference Genome
2.3. SNP Calling
2.4. Effect of Noncoding Variants
2.5. Effect of Variants on miRNA Binding
2.6. Effect of Variants on Regulatory and Epigenetic Modifications
2.7. Differential Gene/Transcript/Transcript Proportion Expression Analysis
2.8. Functional Interaction Network & Module Enrichment
3. Results
3.1. Elucidating Interlinks between Variant Effect and Transcriptome Expression
3.1.1. Role of Variants in Epigenetic Modifications
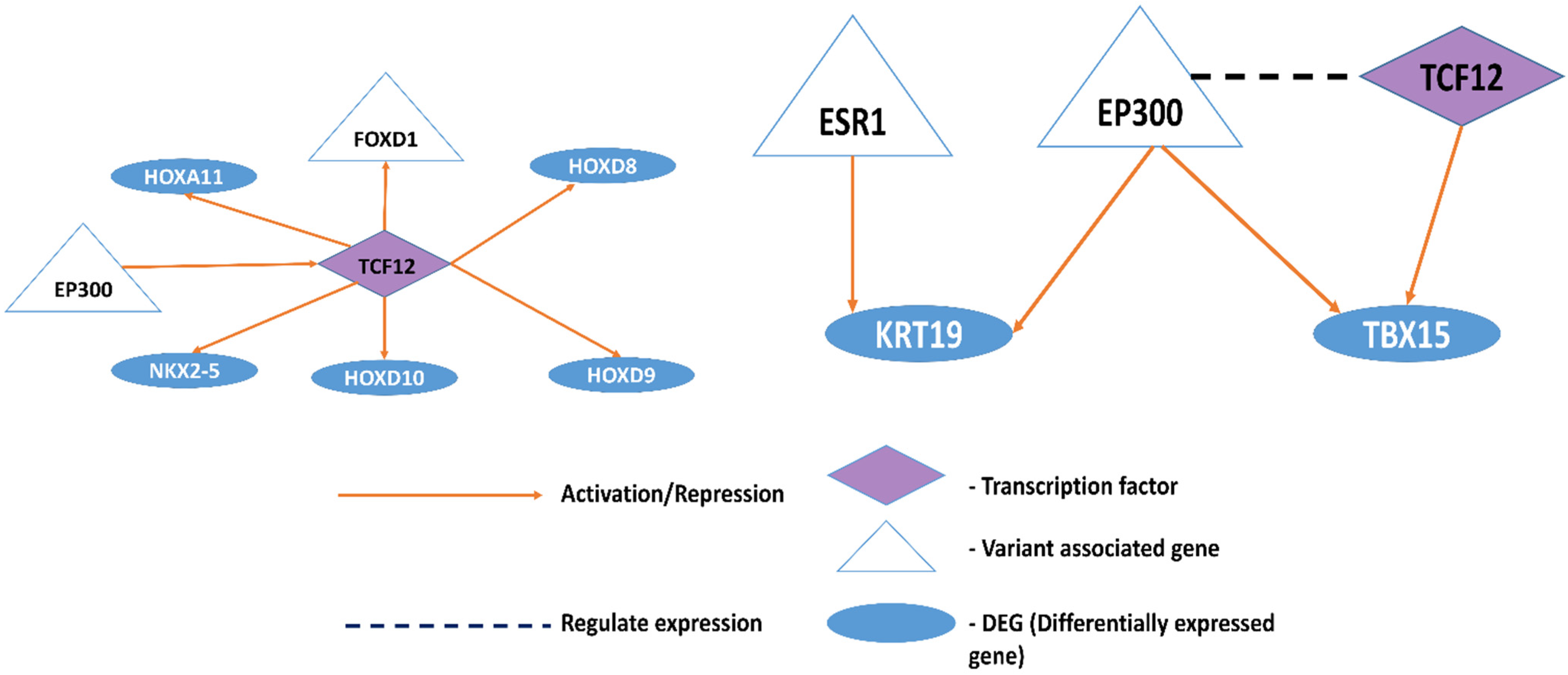
3.1.2. Effect of Variants on Transcription Factor Binding and Histone Methylation/Acetylation Modification
3.1.3. Effect of Variants on miRNA Binding and Its Expression
3.2. Differential Gene Expression/Transcript Expression
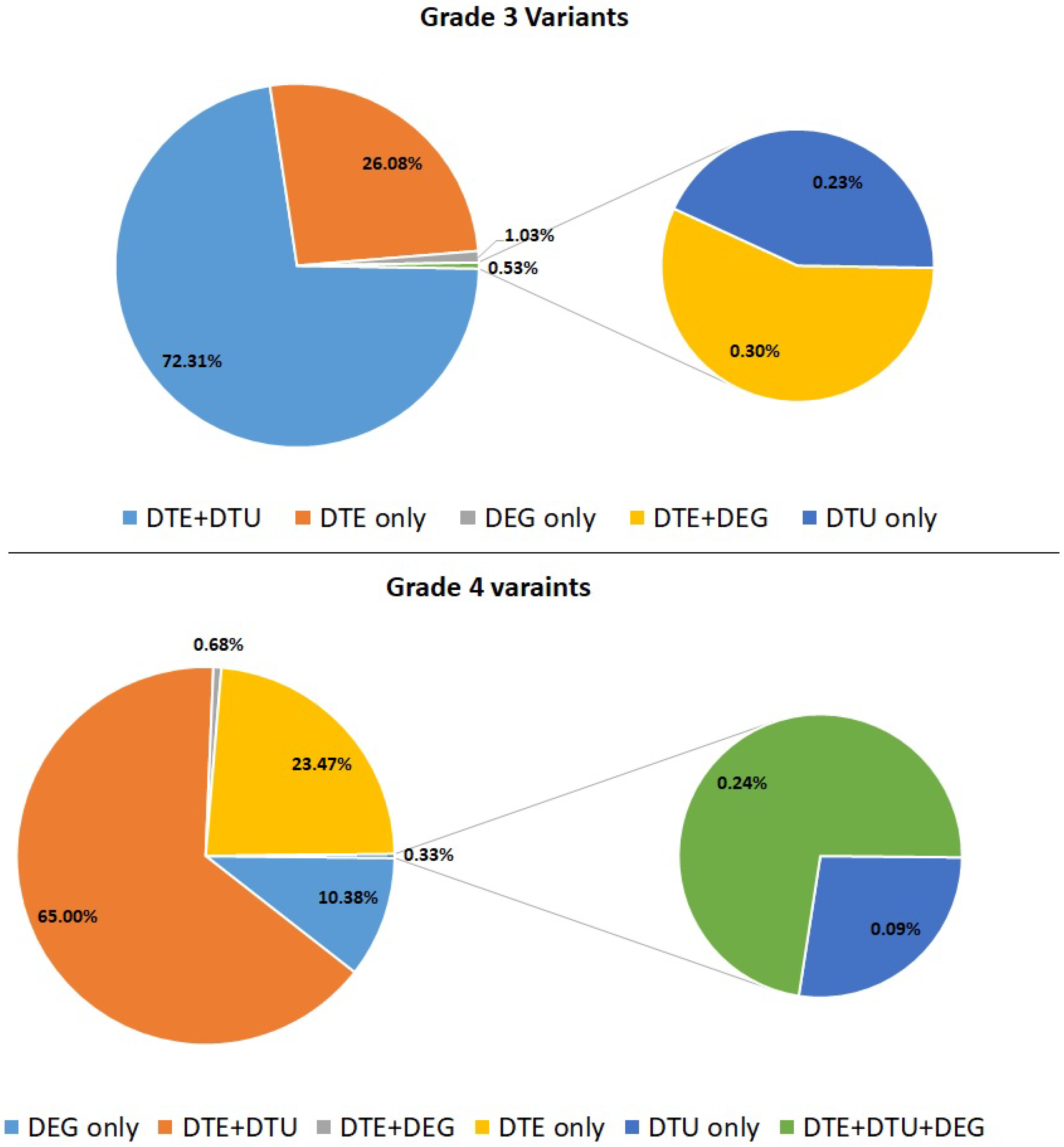
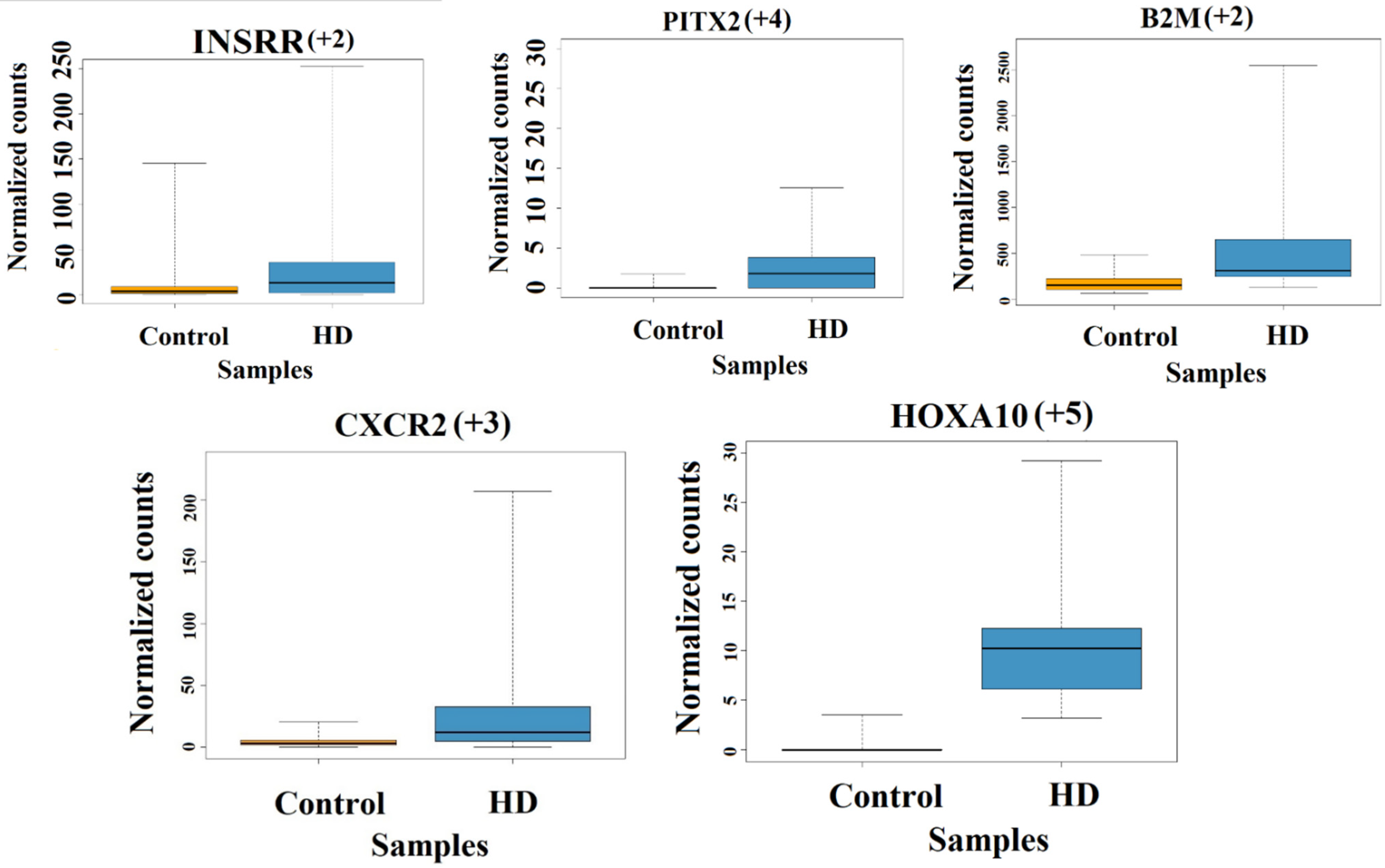
3.2.1. Grade-3 Specific Gene Expression
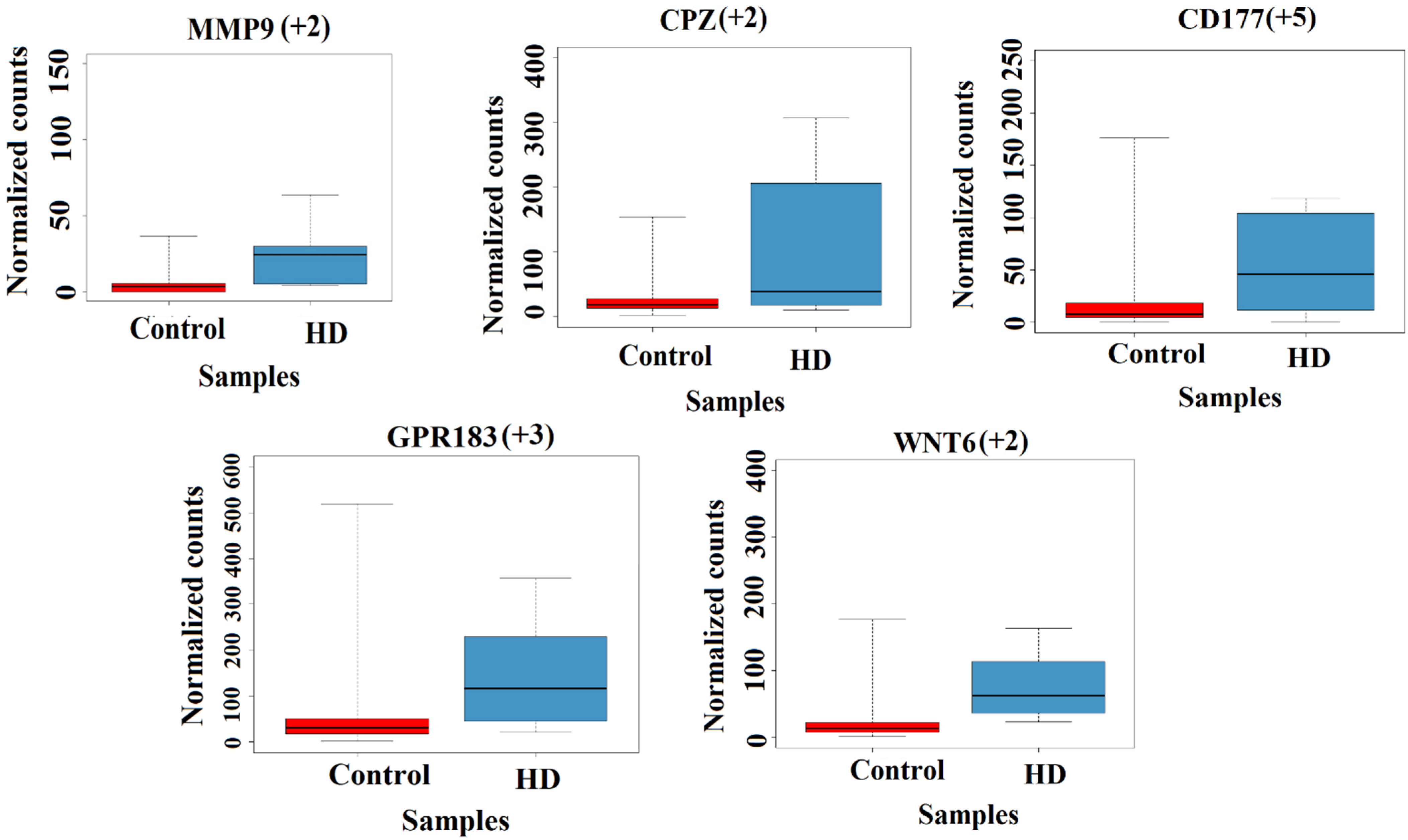
3.2.2. Grade-4 Specific Gene Expression
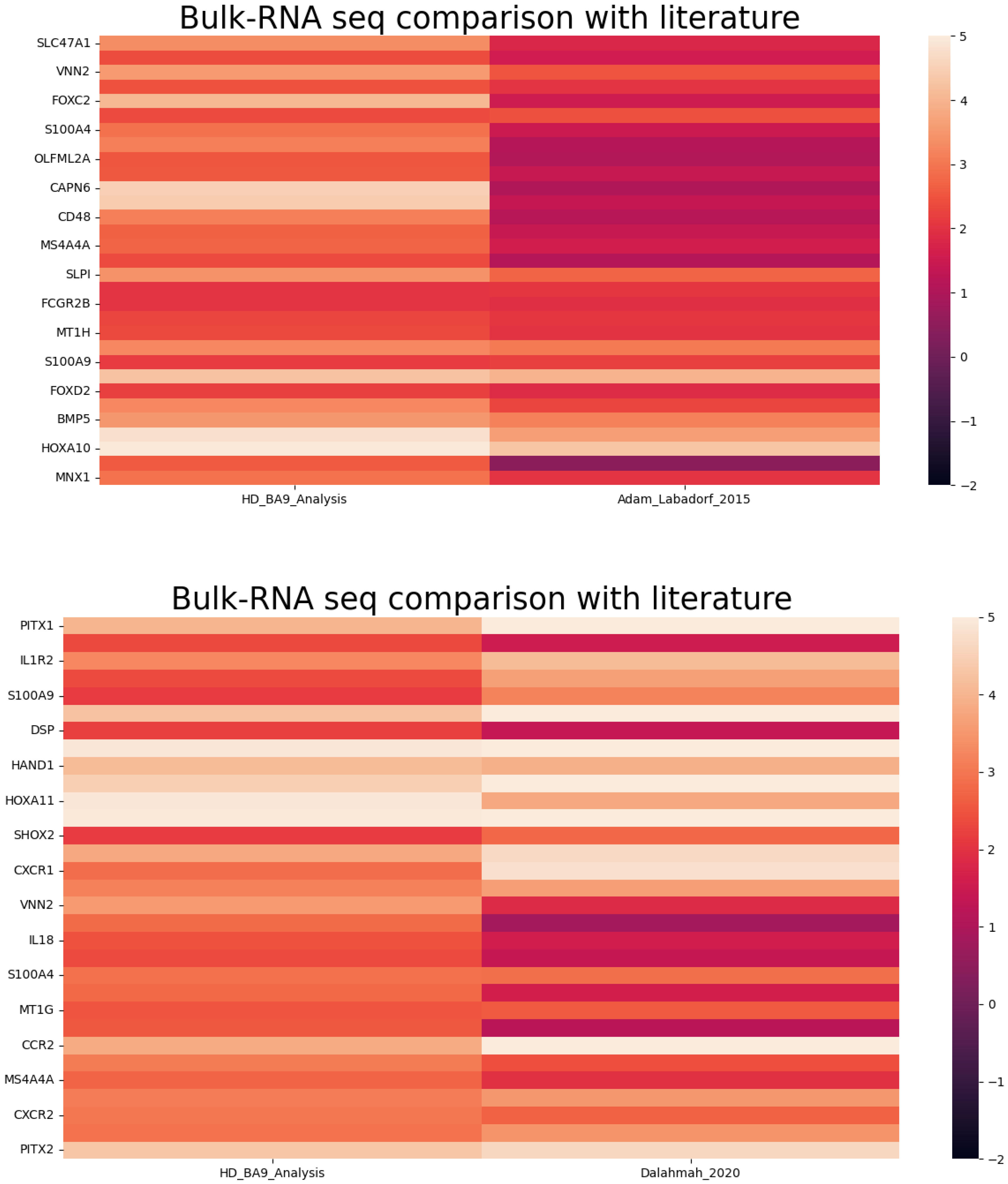
3.2.3. Comparison of Gene Expression (BA9/HD) with Literature Bulk RNA-Seq
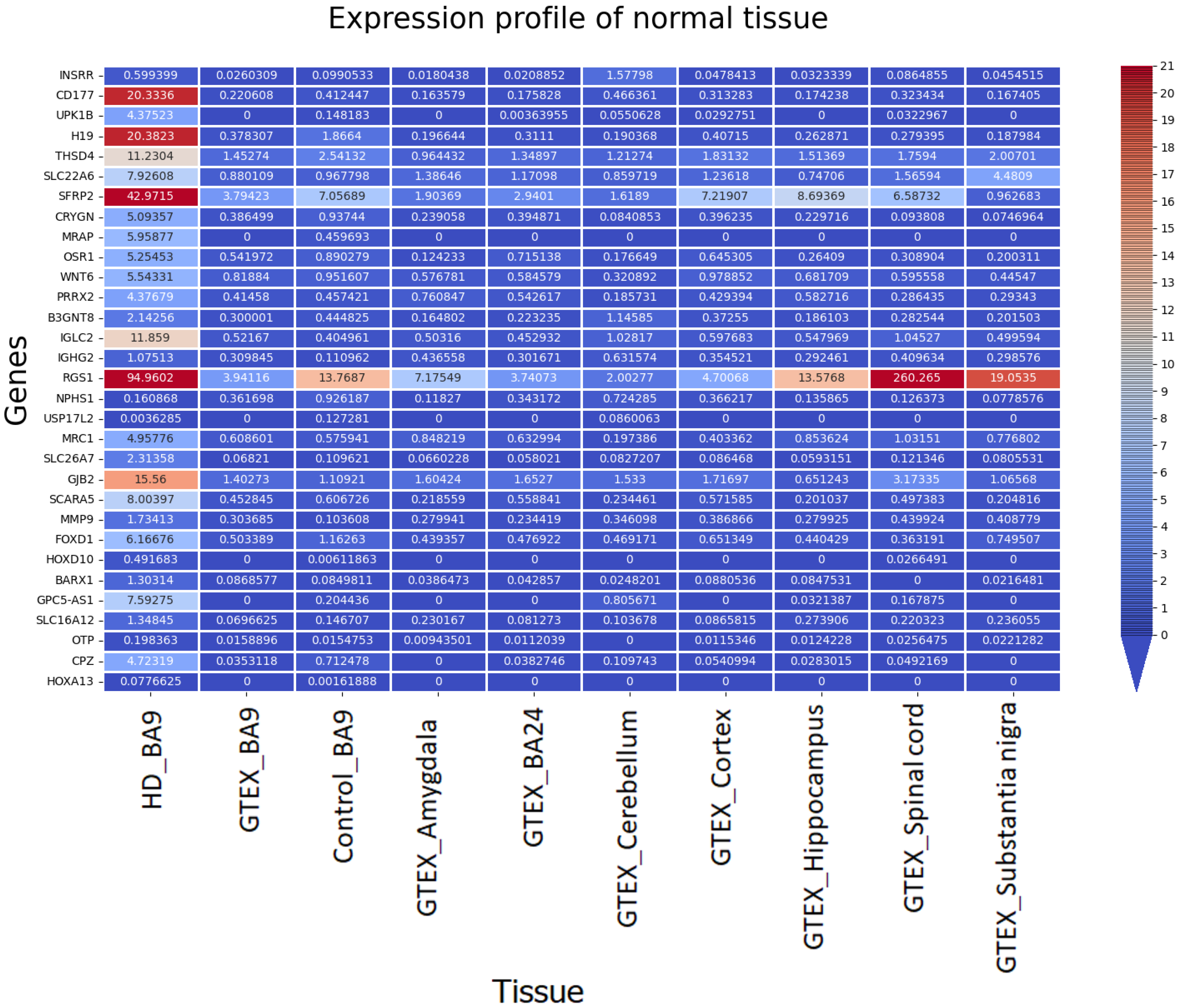
3.2.4. Comparison of Gene Expression (BA9/HD) with GTEX (BA9) and Other Tissue
3.3. Network and Tissue-Specific Functional Enrichment
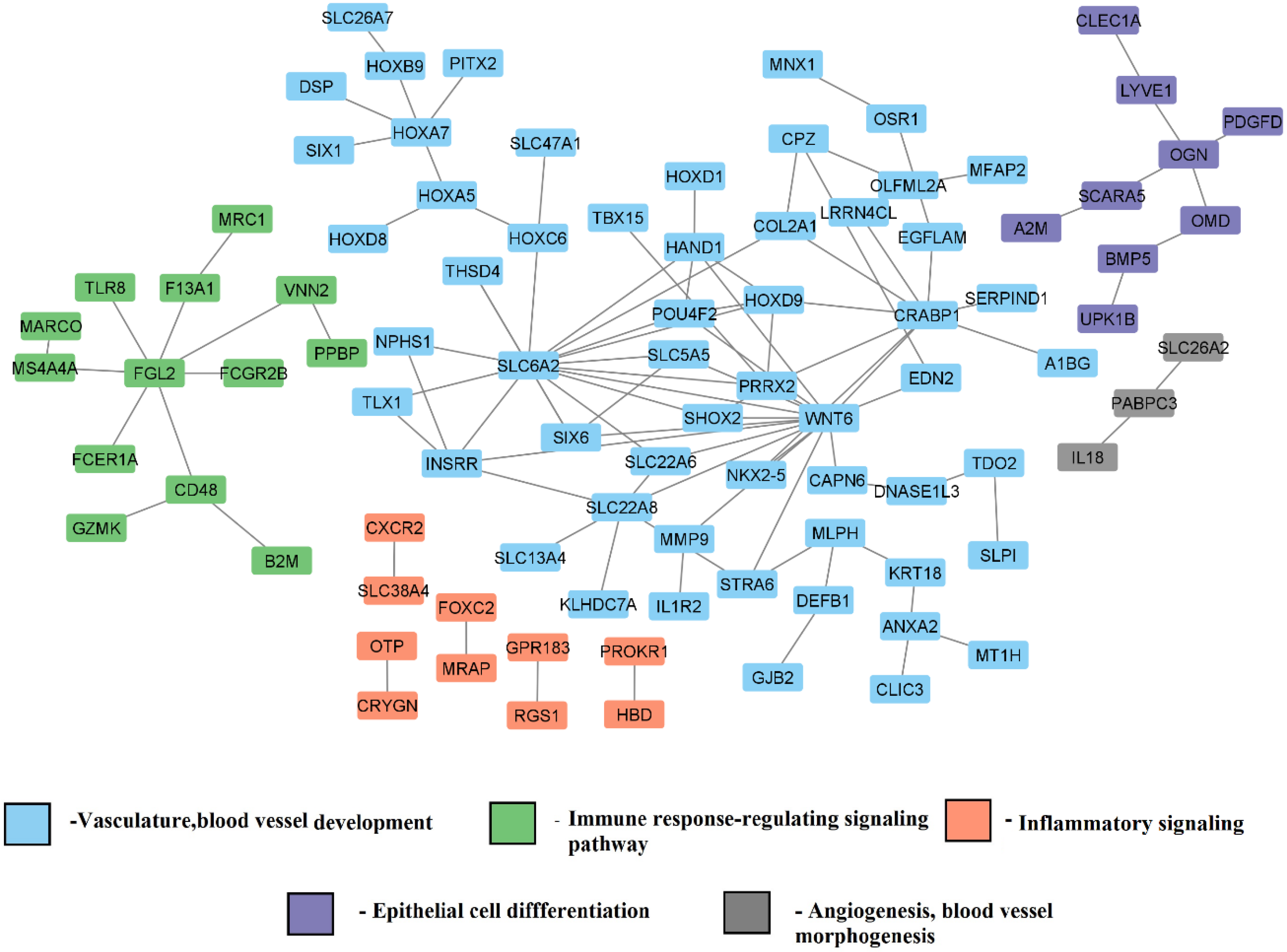
3.3.1. Gene Co-Expression Network and Function Module Network for Differentially Expressed Genes
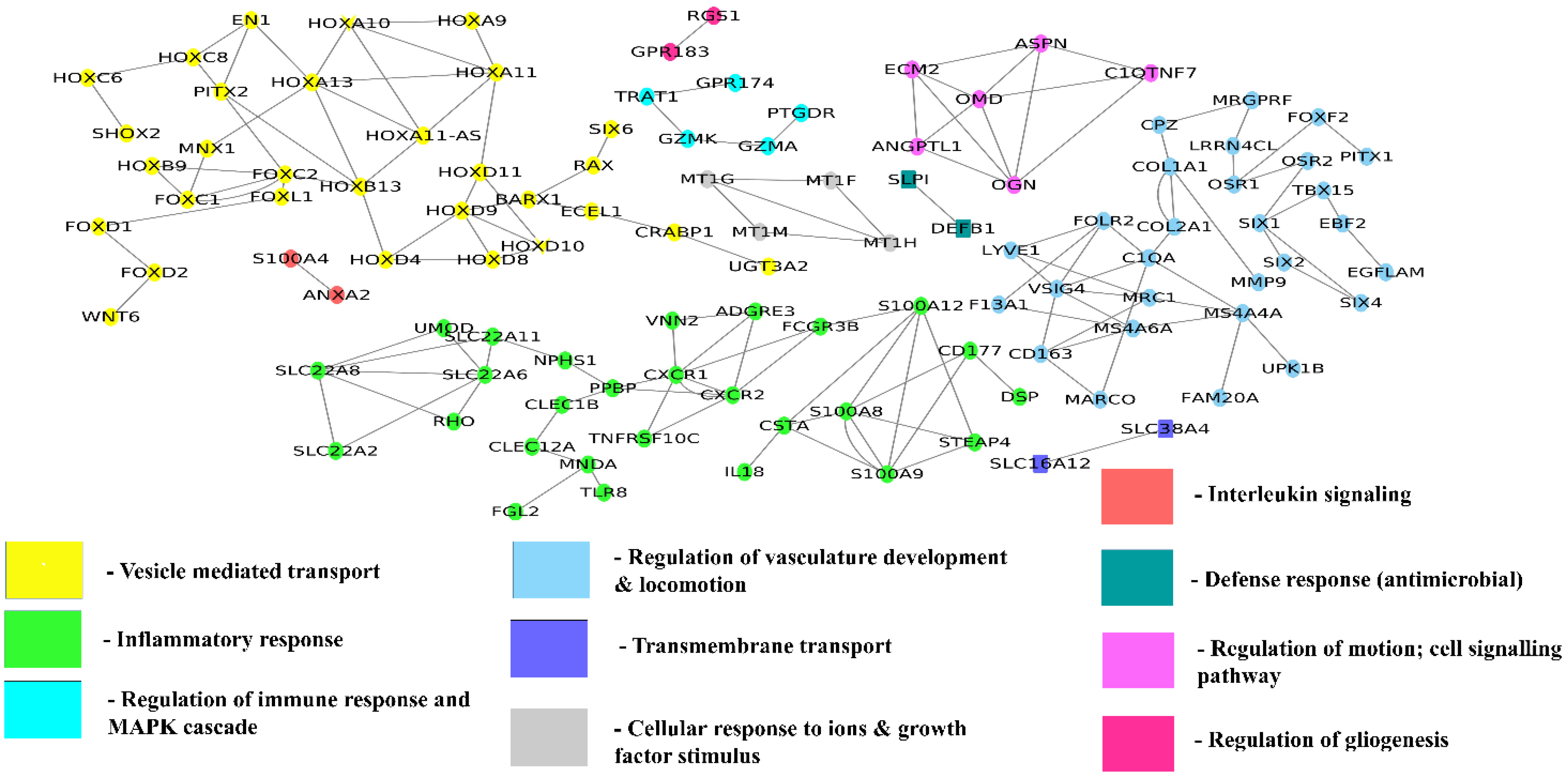
3.3.2. Tissue-Specific Function-Interaction Network
3.3.3. Tissue-Specific Functional Module Network
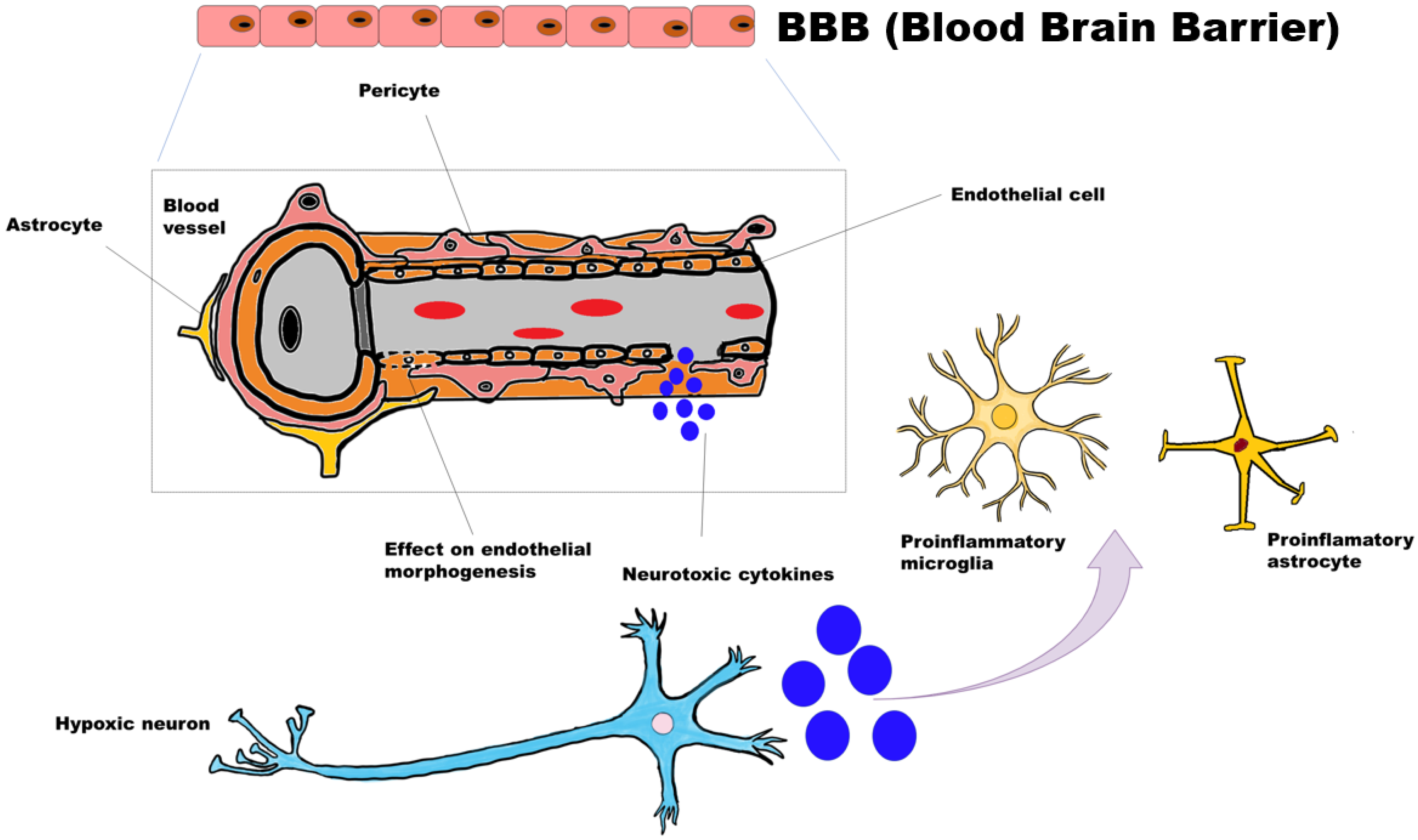
3.4. Proposed Mechanism for Neurodegeneration
3.5. Limitations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cattaneo, E.; Zuccato, C.; Tartari, M. Normal huntingtin function: An alternative approach to Huntington’s disease. Nat. Rev. Neurosci. 2005, 6, 919–930. [Google Scholar] [CrossRef] [PubMed]
- Bhide, P.; Day, M.; Sapp, E.; Schwarz, C.; Sheth, A.; Kim, J.; Young, A.B.; Penney, J.; Golden, J.; Aronin, N.; et al. Expression of Normal and Mutant Huntingtin in the Developing Brain. J. Neurosci. 1996, 16, 5523–5535. [Google Scholar] [CrossRef] [PubMed]
- Cattaneo, E.; Rigamonti, D.; Goffredo, D.; Zuccato, C.; Squitieri, F.; Sipione, S. Loss of normal huntingtin function: New developments in Huntington’s disease research. Trends Neurosci. 2001, 24, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Arrasate, M.; Finkbeiner, S. Protein aggregates in Huntington’s disease. Exp. Neurol. 2012, 238, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Diana, P. Tetrabenazine in the treatment of Huntington’s disease. Expert Opin. 2007, 3, 545–551. [Google Scholar]
- Li, H.; Li, S.-H.; Johnston, H.; Shelbourne, P.F.; Li, X.-J. Amino-terminal fragments of mutant huntingtin show selective accumulation in striatal neurons and synaptic toxicity. Nat. Genet. 2000, 25, 385–389. [Google Scholar] [CrossRef]
- Mitchell, I.; Cooper, A.; Griffiths, M. The selective vulnerability of striatopallidal neurons. Prog. Neurobiol. 1999, 59, 691–719. [Google Scholar] [CrossRef]
- Hodges, A.; Strand, A.D.; Aragaki, A.K.; Kuhn, A.; Sengstag, T.; Hughes, G.; Luthi-Carter, R. Regional and cellular gene expression changes in human Huntington’s disease brain. Hum. Mol. Genet. 2006, 15, 965–977. [Google Scholar] [CrossRef]
- Miller, J.R.C.; Lo, K.K.; Andre, R.; Moss, D.J.H.; Träger, U.; Stone, T.C.; Jones, L.; Holmans, P.; Plagnol, V.; Tabrizi, S.J. RNA-Seq of Huntington’s disease patient myeloid cells reveals innate transcriptional dysregulation associated with proinflammatory pathway activation. Hum. Mol. Genet. 2016, 25, 2893–2904. [Google Scholar] [CrossRef]
- Consortium, H.D.I. Developmental Alterations in Huntington’s Disease Neural Cells and Pharmacological rescue in Cells and Mice. Nat. Neurosci. 2017, 20, 648–660. [Google Scholar] [CrossRef]
- Ooi, J.; Langley, S.R.; Xu, X.; Utami, K.H.; Sim, B.; Huang, Y.; Harmston, N.P.; Tay, Y.L.; Ziaei, A.; Zeng, R.; et al. Unbiased Profiling of Isogenic Huntington Disease hPSC-Derived CNS and Peripheral Cells Reveals Strong Cell-type Specificity of CAG Length Effects. Cell Rep. 2019, 26, 2494–2508. [Google Scholar] [CrossRef]
- Lim, R.G.; Quan, C.; Reyes-Ortiz, A.M.; Lutz, S.E.; Kedaigle, A.J.; Gipson, T.A.; Wu, J.; Vatine, G.D.; Stocksdale, J.; Casale, M.S.; et al. Huntington’s Disease iPSC-Derived Brain Microvascular Endothelial Cells Reveal WNT-Mediated Angiogenic and Blood-Brain Barrier Deficits. Cell Rep. 2017, 19, 1365–1377. [Google Scholar] [CrossRef]
- Al-Dalahmah, O.; Sosunov, A.A.; Shaik, A.; Ofori, K.; Liu, Y.; Vonsattel, J.P.; Adorjan, I.; Menon, V.; Goldman, J.E. Single-nucleus RNA-seq identifies Huntington disease astrocyte states. Acta Neuropathol. Commun. 2020, 8, 19. [Google Scholar] [CrossRef]
- Lee, H.; Fenster, R.J.; Pineda, S.S.; Gibbs, W.S.; Mohammadi, S.; Davila-Velderrain, J.; Garcia, F.J.; Therrien, M.; Novis, H.S.; Gao, F.; et al. Cell Type-specific Tran-scriptomics Reveals that Mutant Huntingtin Leads to Mitochondrial RNA Release and Neuronal Innate Immune Activation. Neuron 2020, 107, 891–908. [Google Scholar] [CrossRef]
- Seredenina, T.; Luthi-Carter, R. What have we learned from gene expression profiles in Huntington’s disease? Neurobiol. Dis. 2012, 45, 83–98. [Google Scholar] [CrossRef]
- Vashishtha, M.; Ng, C.W.; Yildirim, F.; Gipson, T.A.; Kratter, I.H.; Bodai, L.; Song, W.; Lau, A.; Labadorf, A.; Vogel-Ciernia, A.; et al. Targeting H3K4 trimethylation in Huntington disease. Proc. Natl. Acad. Sci. USA 2013, 110, E3027–E3036. [Google Scholar] [CrossRef]
- Ng, C.W.; Yildirim, F.; Yap, Y.S.; Dalin, S.; Matthews, B.J.; Velez, P.J.; Labadorf, A.; Housman, D.E.; Fraenkel, E. Extensive changes in DNA methylation are associated with expression of mutant huntingtin. Proc. Natl. Acad. Sci. USA 2013, 110, 2354–2359. [Google Scholar] [CrossRef] [PubMed]
- Yildirim, F.; Ng, C.W.; Kappes, V.; Ehrenberger, T.; Rigby, S.K.; Stivanello, V.; Gipson, T.A.; Soltis, A.R.; Vanhoutte, P.; Caboche, J.; et al. Early Epigenomic and Tran-scriptional Changes Reveal Elk-1 Transcription Factor as a Therapeutic Target in Huntington’s Disease. Proc. Natl. Acad. Sci. USA 2019, 116, 24840–24851. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.L.; Marcora, E.; Pimenova, A.A.; Di Narzo, A.F.; Kapoor, M.; Jin, S.C.; Harari, O.; Bertelsen, S.; Fairfax, B.P.; Czajkowski, J.; et al. A common haplotype lowers PU.1 expression in myeloid cells and delays onset of Alzheimer’s disease. Nat. Neurosci. 2017, 20, 1052–1061. [Google Scholar] [CrossRef] [PubMed]
- Deming, Y.; Li, Z.; Kapoor, M.; Harari, O.; Del Aguila, J.L.; Black, K.; Carrell, D.; Cai, Y.; Fernandez, M.V.; Budde, J.; et al. Genome-wide association study identifies four novel loci associated with Alzheimer’s endophenotypes and disease modifiers. Acta Neuropathol. 2017, 133, 839–856. [Google Scholar] [CrossRef]
- Fritsche, L.G.; Igl, W.; Bailey, J.N.C.; Grassmann, F.; Sengupta, S.; Bragg-Gresham, J.L.; Zhang, K. A large ge-nome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet. 2016, 48, 134–143. [Google Scholar] [CrossRef]
- Nicolas, A.; Kenna, K.P.; Renton, A.E.; Ticozzi, N.; Faghri, F.; Chia, R.; Dominov, J.A.; Kenna, B.J.; Nalls, M.A.; Keagle, P.; et al. Genome-wide analyses identify KIF5A as a novel ALS gene. Neuron 2018, 97, 1268–1283.e6. [Google Scholar] [CrossRef]
- Labadorf, A.; Hoss, A.G.; Lagomarsino, V.; Latourelle, J.C.; Hadzi, T.C.; Bregu, J.; MacDonald, M.E.; Gusella, J.F.; Chen, J.-F.; Akbarian, S.; et al. Correction: RNA Sequence Analysis of Human Huntington Disease Brain Reveals an Extensive Increase in Inflammatory and Developmental Gene Expression. PLoS ONE 2016, 11, e0160295. [Google Scholar] [CrossRef]
- Vonsattel, J.-P.; Myers, R.H.; Stevens, T.J.; Ferrante, R.J.; Bird, E.D.; Richardson, E.P., Jr. Neuropathological Classification of Huntington’s Disease. J. Neuropathol. Exp. Neurol. 1985, 44, 559–577. [Google Scholar] [CrossRef]
- Patel, R.K.; Jain, M. NGS QC Toolkit: A Toolkit for Quality Control of Next Generation Sequencing Data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef]
- Blankenberg, D.; Gordon, A.; Von Kuster, G.; Coraor, N.; Taylor, J.; Nekrutenko, A.; The Galaxy Team. Manipulation of FASTQ data with Galaxy. Bioinformatics 2010, 26, 1783–1785. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef]
- Koboldt, D.C.; Chen, K.; Wylie, T.; Larson, D.E.; McLellan, M.D.; Mardis, E.R.; Weinstock, G.M.; Wilson, R.K.; Ding, L. VarScan: Variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 2009, 25, 2283–2285. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Ullah, A.D.; Lemoine, N.; Chelala, C. SNPnexus: A web server for functional annotation of novel and publicly known genetic variants (2012 update). Nucleic Acids Res. 2012, 40, W65–W70. [Google Scholar] [CrossRef] [PubMed]
- Rentzsch, P.; Witten, D.; Cooper, G.M.; Shendure, J.; Kircher, M. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019, 47, D886–D894. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, G.R.S.; Dunham, I.; Zeggini, E.; Flicek, P. Functional annotation of noncoding sequence variants. Nat. Methods 2014, 11, 294–296. [Google Scholar] [CrossRef] [PubMed]
- Gulko, B.; Hubisz, M.J.; Gronau, I.; Siepel, A. A method for calculating probabilities of fitness consequences for point mutations across the human genome. Nat. Genet. 2015, 47, 276–283. [Google Scholar] [CrossRef]
- Ionita-Laza, I.; McCallum, K.J.; Xu, B.; Buxbaum, J. A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat. Genet. 2016, 48, 214–220. [Google Scholar] [CrossRef]
- Shihab, H.A.; Gough, J.; Cooper, D.N.; Stenson, P.D.; Barker, G.L.A.; Edwards, K.J.; Day, I.N.M.; Gaunt, T.R. Predicting the Functional, Molecular, and Phenotypic Consequences of Amino Acid Substitutions using Hidden Markov Models. Hum. Mutat. 2013, 34, 57–65. [Google Scholar] [CrossRef]
- Zhou, J.; Troyanskaya, O.G. Predicting effects of noncoding variants with deep learning–based sequence model. Nat. Methods 2015, 12, 931–934. [Google Scholar] [CrossRef]
- Fu, Y.; Liu, Z.; Lou, S.; Bedford, J.; Mu, X.J.; Yip, K.Y.; Khurana, E.; Gerstein, M. FunSeq2: A framework for prioritizing noncoding regulatory variants in cancer. Genome Biol. 2014, 15, 480. [Google Scholar] [CrossRef]
- Kumar, S.; Ambrosini, G.; Bucher, P. SNP2TFBS—a database of regulatory SNPs affecting predicted transcription factor binding site affinity. Nucleic Acids Res. 2017, 45, D139–D144. [Google Scholar] [CrossRef]
- Ward, L.D.; Kellis, M. HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012, 40, D930–D934. [Google Scholar] [CrossRef]
- Thomas, L.F.; Saito, T.; Sætrom, P. Inferring causative variants in microRNA target sites. Nucleic Acids Res. 2011, 39, e109. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Ziebarth, J.D.; Cui, Y. PolymiRTS Database 3.0: Linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res. 2014, 42, D86–D91. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef]
- Zheng, Z.; Huang, D.; Wang, J.; Zhao, K.; Zhou, Y.; Guo, Z.; Zhai, S.; Xu, H.; Cui, H.; Yao, H.; et al. QTLbase: An integrative resource for quantitative trait loci across multiple human molecular phenotypes. Nucleic Acids Res. 2020, 48, D983–D991. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Smith, R.N.; Aleksic, J.; Butano, D.; Carr, A.; Contrino, S.; Hu, F.; Lyne, M.; Lyne, R.; Kalderimis, A.; Rutherford, K.; et al. InterMine: A flexible data warehouse system for the integration and analysis of heterogeneous biological data. Bioinformatics 2012, 28, 3163–3165. [Google Scholar] [CrossRef]
- Robinson, M.D.; Nowicka, M. DRIMSeq: A Dirichlet-multinomial framework for multivariate count outcomes in ge-nomics. F1000Research 2016, 5, 1356. [Google Scholar] [CrossRef]
- Berge, K.V.D.; Soneson, C.; Robinson, M.D.; Clement, L. stageR: A general stage-wise method for controlling the gene-level false discovery rate in differential expression and differential transcript usage. Genome Biol. 2017, 18, 151. [Google Scholar] [CrossRef]
- Wu, G.; Dawson, E.; Duong, A.; Haw, R.; Stein, L. ReactomeFIViz: A cytoscape app for pathway and network-based data analysis. F1000Research 2014, 3, 146. [Google Scholar] [CrossRef]
- Zhou, J.; Theesfeld, C.L.; Yao, K.; Chen, K.M.; Wong, A.K.; Troyanskaya, O.G. Deep learning sequence-based ab initio prediction of variant effects on expression and disease risk. Nat. Genet. 2018, 50, 1171–1179. [Google Scholar] [CrossRef] [PubMed]
- Obayashi, T.; Kagaya, Y.; Aoki, Y.; Tadaka, S.; Kinoshita, K. COXPRESdb v7: A gene coexpression database for 11 animal species supported by 23 coexpression platforms for technical evaluation and evolutionary inference. Nucleic Acids Res. 2019, 47, D55–D62. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Yu, G. clusterProfiler 4.0: A universal enrichment tool for inter-preting omics data. Innovation 2021, 2, 100141. [Google Scholar]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Hunter, S.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Bork, P.; Das, U.; Daugherty, L.; Duquenne, L.; et al. InterPro: The integrative protein signature database. Nucleic Acids Res. 2008, 37, D211–D215. [Google Scholar] [CrossRef]
- Cunningham, F.; Allen, J.E.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Flicek, P. Ensembl 2022. Nucleic Acids Res. 2022, 50, D988–D995. [Google Scholar] [CrossRef]
- Chen, K.-P.; Hua, K.-F.; Tsai, F.-T.; Lin, T.-Y.; Cheng, C.-Y.; Yang, D.-I.; Hsu, H.-T.; Ju, T.-C. A selective inhibitor of the NLRP3 inflammasome as a potential therapeutic approach for neuroprotection in a transgenic mouse model of Huntington’s disease. J. Neuroinflamm. 2022, 19, 56. [Google Scholar] [CrossRef]
- Lin, A.W.; Gill, K.K.; Castañeda, M.S.; Matucci, I.; Eder, N.; Claxton, S.; Flynn, H.; Snijders, A.P.; George, R.; Ultanir, S.K. Chemical genetic identification of GAK substrates reveals its role in regulating Na+/K+-ATPase. Life Sci. Alliance 2018, 1. [Google Scholar] [CrossRef]
- Norris, R.A.; Scott, K.K.; Moore, C.S.; Stetten, G.; Brown, C.R.; Jabs, E.W.; Wulfsberg, E.A.; Yu, J.; Kern, M.J. Human PRRX1 and PRRX2 genes: Cloning, expression, genomic localization, and exclusion as disease genes for Nager syndrome. Mamm. Genome 2000, 11, 1000–1005. [Google Scholar] [CrossRef]
- Bodai, L.; Marsh, J.L. A novel target for Huntington’s disease: ERK at the crossroads of signaling: The ERK signaling pathway is implicated in Huntington’s disease and its upregulation ameliorates pathology. Bioessays 2012, 34, 142–148. [Google Scholar] [CrossRef]
- Bhat, R.A.; Stauffer, B.; Komm, B.S.; Bodine, P.V. Structure–Function analysis of secreted frizzled-related protein-1 for its Wnt antagonist function. J. Cell. Biochem. 2007, 102, 1519–1528. [Google Scholar] [CrossRef]
- Goodwin, A.; D’Amore, P. Wnt signaling in the vasculature. Angiogenesis 2002, 5, 1–9. [Google Scholar] [CrossRef]
- Palpagama, T.H.; Waldvogel, H.J.; Faull, R.L.M.; Kwakowsky, A. The Role of Microglia and Astrocytes in Huntington’s Disease. Front. Mol. Neurosci. 2019, 12, 258. [Google Scholar] [CrossRef]
- Bowles, K.R.; Jones, L. Kinase signalling in Huntington’s disease. J. Huntingt. Dis. 2014, 3, 89–123. [Google Scholar] [CrossRef]
- Kang, J.; Shi, Y.; Xiang, B.; Qu, B.; Su, W.; Zhu, M.; Zhang, M.; Bao, G.; Wang, F.; Zhang, X.; et al. A Nuclear Function of β-Arrestin1 in GPCR Signaling: Regulation of Histone Acetylation and Gene Transcription. Cell 2005, 123, 833–847. [Google Scholar] [CrossRef]
- Di Leva, G.; Gasparini, P.; Piovan, C.; Ngankeu, A.; Garofalo, M.; Taccioli, C.; Iorio, M.; Li, M.; Volinia, S.; Alder, H.; et al. MicroRNA Cluster 221-222 and Estrogen Receptor α Interactions in Breast Cancer. Gynecol. Oncol. 2010, 102, 706–721. [Google Scholar] [CrossRef]
- Angeles-López, D.Q.; García-Lara, L.; Aguirre-Pineda, N.; Castañeda-Arellano, R.; Eli-zondo-Azuela, G.; Pérez-Severiano, F.; Segovia, J. The absence of the aryl hydrocarbon receptor in the R6/1 transgenic mouse model of Huntington’s disease improves the neurological phenotype. Behav. Brain Res. 2021, 408, 113230. [Google Scholar] [CrossRef]
| Chr | REF/ALT | Gene | log2FC | Variant | Transcripts | Novel/Reported | QTL Category | Expression |
|---|---|---|---|---|---|---|---|---|
| Chr11 | A/G | IL18 (Grades 4 & 3) | 2.484262691 | rs360720 | ENST00000280357.11, ENST00000524595.5, ENST00000525547.5, ENST00000534225.1 | PD, AD | hQTL, eQTL, mQTL, sQTL | UP |
| chr11 | G/A | USH1C (Grades 4 & 3) | 11.8 | rs2041027 | ENST00000529563.5 | PD, AD | hQTL | UP |
| Chr16 | G/C | SLC9A5 (Grades 4 & 3) | 11.1 | rs61740454 | ENST00000564704.5 | Novel | hQTL | UP |
| Chr17 | C/A | MPRIP (Grades 4 & 3) | 5.1 | rs3826304 | ENST00000579361.1 | Novel | hQTL | UP |
| Chr22 | G/A | MPPED1 (Grades 4 & 3) | 4.9 | rs739018 | ENST00000334209.9, ENST00000417669.6 | PD | hQTL, Eqtl | UP |
| Chr4 | G/A | GAK (Grades 4 & 3) | 3.2 | rs17165089 | ENST00000504668.5 | Novel | hQTL | UP |
| Chr7 | C/T | SLC13A4 (Grades 4 & 3) | 2.588 | rs3112355 | ENST00000471405.5, ENST00000480376.1 | PD, AD | eQTL | UP |
| Chr3 | C/T | GOLGB1 (Grades 4 & 3) | −5.4 | rs77570895 | ENST00000491690.1 | Novel | eQTL, mQTL | DOWN |
| Chr18 | G/A | CYB5A (Grades 4 & 3) | −7.6 | rs2032263 | ENST00000299438.13, ENST00000397914.4 | PD | eQTL, sQTL | DOWN |
| Chr15 | T/C | ADAMTSL3 (Grades 4) | NA | rs1480822 | ENST00000569510.5 | PD | hQTL, eQTL | Novel |
| CHR | REF/ALT | GENE | Variant Gene Expression | Variant Gene Expression Pattern | SNP | Category (Methylation, Acetylation, Gene Expression, Splicing) | Transcription Factor Affected | Transcription Factor Expression | GWAS Study |
|---|---|---|---|---|---|---|---|---|---|
| chr11 | G/A | LRRN4CL | Novel | UP | rs2512561 | eqtl, sqtl, mqtl | NFKB1 | DOWN | PD, AD |
| chr19 | T/C | B3GNT8 | Novel | UP | rs284662 | hqtl, eqtl, sqtl, mqtl | CTCF | DOWN | PD |
| chr4 | G/T | CPZ | Novel | UP | rs13121547 | Hqtl | CTCF | DOWN | Novel |
| chr7 | C/T | SLC13A4 | Reported | UP | rs3112355 | Eqtl | Prrx2 | UP | PD, AD |
| chr11 | C/G | PDGFD | Reported | UP | rs11226059 | Eqtl | MAFF | UP | AD |
| chr12 | C/T | COL2A1 | Reported | UP | rs2070739 | hqtl, sqtl | Tcf3 | DOWN | PD, AD |
| chr14 | A/G | PTGDR | Reported | UP | rs4898758 | hqtl, eqtl | RUNX1 | UP | PD |
| chr14 | C/G | PDGFD | Reported | DOWN | rs813921 | Hqtl | Pdx1 | DOWN | Novel |
| chr6 | G/A | DSP | Reported | UP | rs6929069 | Eqtl | SOX10 | UP | PD |
| Chr | Alt/Ref | Gene | SNP | GWAS/Novel SNP | Affected miRNA | miRNA Expression | miRNA Target Gene/s | Expression of Gene |
|---|---|---|---|---|---|---|---|---|
| Chr12 | C/T | DRAM1 (DOWN) | rs17032062 | PD, AD | miR-302e, miR-101 | Up | SLC16A12, SCARA5, HOXD8, PTGDR, FGL2, FCGR2B, SLC26A2, DSC3, SLC26A7, OGN | DOWN |
| Chr13 | A/G | FARP1 (DOWN) | rs2282048 | PD, AD | miR-100 | Up | SIX1 | DOWN |
| Chr5 | T/A | CSNK1A1 (DOWN) | rs11167469 | PD | miR-374a | Up | CD177, CAPN6, OGN, HOXA10, SLC26A2, RGS1, MS4A4A, FOXD2, | DOWN |
| Chr3 | G/T | TMEM43 (DOWN) | rs3796308 | PD, AD | miR-30a | Up | HOXA13, CLEC12A, MRC1, SIX1, DSP, FOXD1, SCARA5, SPTLC3, RGS1 | DOWN |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sneha, N.P.; Dharshini, S.A.P.; Taguchi, Y.-H.; Gromiha, M.M. Integrative Meta-Analysis of Huntington’s Disease Transcriptome Landscape. Genes 2022, 13, 2385. https://doi.org/10.3390/genes13122385
Sneha NP, Dharshini SAP, Taguchi Y-H, Gromiha MM. Integrative Meta-Analysis of Huntington’s Disease Transcriptome Landscape. Genes. 2022; 13(12):2385. https://doi.org/10.3390/genes13122385
Chicago/Turabian StyleSneha, Nela Pragathi, S. Akila Parvathy Dharshini, Y.-H. Taguchi, and M. Michael Gromiha. 2022. "Integrative Meta-Analysis of Huntington’s Disease Transcriptome Landscape" Genes 13, no. 12: 2385. https://doi.org/10.3390/genes13122385
APA StyleSneha, N. P., Dharshini, S. A. P., Taguchi, Y.-H., & Gromiha, M. M. (2022). Integrative Meta-Analysis of Huntington’s Disease Transcriptome Landscape. Genes, 13(12), 2385. https://doi.org/10.3390/genes13122385








