Analysis of Genomic Alternative Splicing Patterns in Rat under Heat Stress Based on RNA-Seq Data
Abstract
:1. Introduction
2. Materials and Methods
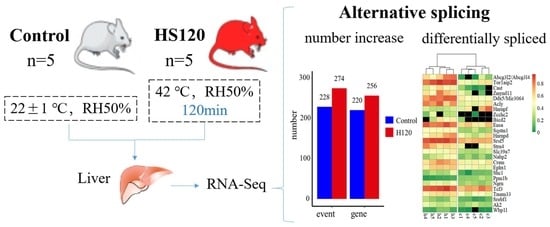
2.1. Animals and Treatments
2.2. RNA Extraction, cDNA Library Construction and Illumina Deep Sequencing
2.3. Data Filtering and Transcriptome Alignment
2.4. Identification of Differential Expression Genes
2.5. Identification of Differential Alternative Splicing Events
2.6. Integrated Gene Expression and Alternative Splicing Results
2.7. Functional Analysis of Differentially Expressed Genes and Differentially Alternatively Spliced Genes
2.8. Validation of the Expression Level of the DAS Genes by Real-Time Quantitative PCR
3. Results
3.1. Summary of the Basic Information
3.2. Identification of Differentially Expressed Genes
3.3. Functional Enrichment Analysis for Differentially Expressed Genes
3.4. Summary of Alternative Splicing Events
3.5. Identification of Differential Alternative Splicing Events and Genes
3.6. Functional Enrichment Analysis for Differentially Alternatively Spliced Genes
3.7. The Differentially Alternatively Spliced Genes with Different Expression Levels
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Collier, R.J.; Baumgard, L.H.; Zimbelman, R.B.; Xiao, Y. Heat stress: Physiology of acclimation and adaptation. Anim. Front. 2019, 9, 12–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baumgard, L.H.; Rhoads, R.J. Effects of heat stress on postabsorptive metabolism and energetics. Annu. Rev. Anim. Biosci. 2013, 1, 311–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Summer, A.; Lora, I.; Formaggioni, P.; Gottardo, F. Impact of heat stress on milk and meat production. Anim. Front. 2019, 9, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Sohail, M.U.; Ijaz, A.; Yousaf, M.S.; Ashraf, K.; Zaneb, H.; Aleem, M.; Rehman, H. Alleviation of cyclic heat stress in broilers by dietary supplementation of mannan-oligosaccharide and Lactobacillus-based probiotic: Dynamics of cortisol, thyroid hormones, cholesterol, C-reactive protein, and humoral immunity. Poultry Sci. 2010, 89, 1934–1938. [Google Scholar] [CrossRef]
- Mayorga, E.J.; Renaudeau, D.; Ramirez, B.C.; Ross, J.W.; Baumgard, L.H. Heat stress adaptations in pigs. Anim. Front. 2019, 9, 54–61. [Google Scholar] [CrossRef]
- Padua, J.T.; Dasilva, R.G. Effect of high environmental temperatures on weight gain and food intake of Suffolk lambs reared in a tropical environment. In Proceedings of the 5th International Symposium, Bloomington, MI, USA, 29–31 May 1997; pp. 809–815. [Google Scholar]
- Hansen, P.J. Effects of heat stress on mammalian reproduction. Philos. Trans. R. Soc. B 2009, 364, 3341–3350. [Google Scholar] [CrossRef] [PubMed]
- Lacetera, N. Impact of climate change on animal health and welfare. Anim. Front. 2019, 9, 26–31. [Google Scholar] [CrossRef] [Green Version]
- St-Pierre, N.R.; Cobanov, B.; Schnitkey, G. Economic Losses from Heat Stress by US Livestock Industries1. J. Dairy Sci. 2003, 86, E52–E77. [Google Scholar] [CrossRef] [Green Version]
- Herbut, P.; Angrecka, S. Forming of temperature-humidity index (THI) and milk production of cows in the free-stall barn during the period of summer heat. Anim. Sci. Pap. Rep. 2012, 30, 363–372. [Google Scholar]
- Lallo, C.H.O.; Cohen, J.; Rankine, D.; Taylor, M.; Cambell, J.; Stephenson, T. Characterizing heat stress on livestock using the temperature humidity index (THI)—prospects for a warmer Caribbean. Reg. Environ. Chang. 2018, 18, 2329–2340. [Google Scholar] [CrossRef] [Green Version]
- Dou, J.; Cánovas, A.; Brito, L.F.; Yu, Y.; Schenkel, F.S.; Wang, Y. Comprehensive RNA-Seq Profiling Reveals Temporal and Tissue-Specific Changes in Gene Expression in Sprague–Dawley Rats as Response to Heat Stress Challenges. Front. Genet. 2021, 12, 651979. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, X.; Sun, F.; Zhang, J.; Feng, J.; Liu, H.; Rajendran, K.V.; Sun, L.; Zhang, Y.; Jiang, Y.; et al. RNA-Seq reveals expression signatures of genes involved in oxygen transport, protein synthesis, folding, and degradation in response to heat stress in catfish. Physiol. Genom. 2013, 45, 462–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dou, J.; Khan, A.; Khan, M.Z.; Mi, S.; Wang, Y.; Yu, Y.; Wang, Y. Heat Stress Impairs the Physiological Responses and Regulates Genes Coding for Extracellular Exosomal Proteins in Rat. Genes 2020, 11, 306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dado-Senn, B.; Skibiel, A.L.; Fabris, T.F.; Zhang, Y.; Dahl, G.E.; Penagaricano, F.; Laporta, J. RNA-Seq reveals novel genes and pathways involved in bovine mammary involution during the dry period and under environmental heat stress. Sci. Rep. 2018, 8, 11096. [Google Scholar] [CrossRef]
- Wu, Z.L.; Yang, X.; Chen, S.Y.; Deng, F.L.; Jia, X.B.; Hu, S.Q.; Wang, J.; Lai, S.J. Liver Transcriptome Changes of Hyla Rabbit in Response to Chronic Heat Stress. Animals 2019, 9, 1141. [Google Scholar] [CrossRef] [Green Version]
- Moreira, C.; Stillman, J.H.; Lima, F.P.; Xavier, R.; Seabra, R.; Gomes, F.; Verissimo, A.; Silva, S.M. Transcriptomic response of the intertidal limpet Patella vulgata to temperature extremes. J. Therm. Biol. 2021, 101, 103096. [Google Scholar] [CrossRef]
- Gilbert, W. Why Genes in Pieces? Nature 1978, 271, 501. [Google Scholar] [CrossRef]
- Chen, M.; Manley, J.L. Mechanisms of alternative splicing regulation: Insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 2009, 10, 741–754. [Google Scholar] [CrossRef]
- Black, D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003, 72, 291–336. [Google Scholar] [CrossRef] [Green Version]
- Laloum, T.; Martin, G.; Duque, P. Alternative Splicing Control of Abiotic Stress Responses. Trends Plant Sci. 2018, 23, 140–150. [Google Scholar] [CrossRef] [Green Version]
- Staiger, D.; Brown, J.W. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 2013, 25, 3640–3656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mastrangelo, A.M.; Marone, D.; Laido, G.; De Leonardis, A.M.; De Vita, P. Alternative splicing: Enhancing ability to cope with stress via transcriptome plasticity. Plant Sci. 2012, 185–186, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Wang, Y.; Qiu, C.; Qian, W.; Xie, H.; Ding, Z. Alternative splicing in tea plants was extensively triggered by drought, heat and their combined stresses. PeerJ 2020, 8, e8258. [Google Scholar] [CrossRef]
- Zhang, X.; Yuan, J.; Zhang, X.; Liu, C.; Xiang, J.; Li, F. Genome-Wide Analysis of Alternative Splicing Provides Insights Into Stress Response of the Pacific White Shrimp Litopenaeus vanname. Front. Genet. 2019, 10, 845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takechi, H.; Hosokawa, N.; Hirayoshi, K.; Nagata, K. Alternative 5′ splice site selection induced by heat shock. Mol. Cell Biol. 1994, 14, 567–575. [Google Scholar] [CrossRef] [Green Version]
- Nevo, Y.; Kamhi, E.; Jacob-Hirsch, J.; Amariglio, N.; Rechavi, G.; Sperling, J.; Sperling, R. Genome-wide activation of latent donor splice sites in stress and disease. Nucleic Acids Res. 2012, 40, 10980–10994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nevo, Y.; Sperling, J.; Sperling, R. Heat shock activates splicing at latent alternative 5′ splice sites in nematodes. Nucleus 2015, 6, 225–235. [Google Scholar] [CrossRef] [Green Version]
- Ju, X.; Xu, H.; Yong, Y.; An, L.; Xu, Y.; Jiao, P.; Liao, M. Heat Stress Upregulates the Expression of TLR4 and Its Alternative Splicing Variant in Bama Miniature Pigs. J. Integr. Agr. 2014, 13, 2479–2487. [Google Scholar] [CrossRef] [Green Version]
- Fujikake, N.; Nagai, Y.; Popiel, H.A.; Kano, H.; Yamaguchi, M.; Toda, T. Alternative splicing regulates the transcriptional activity of Drosophila heat shock transcription factor in response to heat/cold stress. FEBS Lett. 2005, 579, 3842–3848. [Google Scholar] [CrossRef] [Green Version]
- Kaitsuka, T.; Tomizawa, K.; Matsushita, M. Transformation of eEF1Bdelta into heat-shock response transcription factor by alternative splicing. EMBO Rep. 2011, 12, 673–681. [Google Scholar] [CrossRef] [Green Version]
- Tan, S.; Wang, W.; Tian, C.; Niu, D.; Zhou, T.; Jin, Y.; Yang, Y.; Gao, D.; Dunham, R.; Liu, Z. Heat stress induced alternative splicing in catfish as determined by transcriptome analysis. Comp. Biochem. Physiol. Part D Genom. Proteom. 2019, 29, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.W.; Madkour, M.; Kuenzel, W.J. Tissue-Specific Expression of DNA Methyltransferases Involved in Early-Life Nutritional Stress of Chicken, Gallus gallus. Front. Genet. 2017, 8, 204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madkour, M.; Aboelenin, M.M.; Aboelazab, O.; Elolimy, A.A.; El-Azeem, N.A.; El-Kholy, M.S.; Alagawany, M.; Shourrap, M. Hepatic expression responses of DNA methyltransferases, heat shock proteins, antioxidant enzymes, and NADPH 4 to early life thermal conditioning in broiler chickens. Ital. J. Anim. Sci. 2021, 20, 433–446. [Google Scholar] [CrossRef]
- Hadfield, J.; Eldridge, M.D. Multi-genome alignment for quality control and contamination screening of next-generation sequencing data. Front. Genet. 2014, 5, 31. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Goldstein, L.D.; Cao, Y.; Pau, G.; Lawrence, M.; Wu, T.D.; Seshagiri, S.; Gentleman, R. Prediction and Quantification of Splice Events from RNA-Seq Data. PLoS ONE 2016, 11, e156132. [Google Scholar] [CrossRef]
- Anders, S.; Reyes, A.; Huber, W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012, 22, 2008–2017. [Google Scholar] [CrossRef]
- Rodriguez, J.M.; Maietta, P.; Ezkurdia, I.; Pietrelli, A.; Wesselink, J.J.; Lopez, G.; Valencia, A.; Tress, M.L. APPRIS: Annotation of principal and alternative splice isoforms. Nucleic Acids Res. 2013, 41, D110–D117. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- UP Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.R.; Hayman, G.T.; Wang, S.J.; Laulederkind, S.; Hoffman, M.J.; Kaldunski, M.L.; Tutaj, M.; Thota, J.; Nalabolu, H.S.; Ellanki, S.; et al. The Year of the Rat: The Rat Genome Database at 20: A multi-species knowledgebase and analysis platform. Nucleic Acids Res. 2020, 48, D731–D742. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dou, J.; Montanholi, Y.R.; Wang, Z.; Li, Z.; Yu, Y.; Martell, J.E.; Wang, Y.J.; Wang, Y. Corticosterone tissue-specific response in Sprague Dawley rats under acute heat stress. J. Therm. Biol. 2019, 81, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Qu, Q.; Li, H.; Bai, L.; Zhang, S.; Sun, J.; Lv, W.; Ye, C.; Liu, C.; Shi, D. Effects of Heat Stress on Gut Microbiome in Rats. Indian J. Microbiol. 2021, 61, 338–347. [Google Scholar] [CrossRef]
- Yu, J.; Liu, F.; Yin, P.; Zhu, X.; Cheng, G.; Wang, N.; Lu, A.; Luan, W.; Zhang, N.; Li, J.; et al. Integrating miRNA and mRNA expression profiles in response to heat stress-induced injury in rat small intestine. Funct. Integr. Genom. 2011, 11, 203–213. [Google Scholar] [CrossRef]
- Sinha, R.K. Study of changes in some pathophysiological stress markers in different age groups of an animal model of acute and chronic heat stress. Iran. Biomed. J. 2007, 11, 101–111. [Google Scholar]
- Sammad, A.; Wang, Y.J.; Umer, S.; Lirong, H.; Khan, I.; Khan, A.; Ahmad, B.; Wang, Y. Nutritional Physiology and Biochemistry of Dairy Cattle under the Influence of Heat Stress: Consequences and Opportunities. Animals 2020, 10, 793. [Google Scholar] [CrossRef]
- Barrett, N.W.; Rowland, K.; Schmidt, C.J.; Lamont, S.J.; Rothschild, M.F.; Ashwell, C.M.; Persia, M.E. Effects of acute and chronic heat stress on the performance, egg quality, body temperature, and blood gas parameters of laying hens. Poult. Sci. 2019, 98, 6684–6692. [Google Scholar] [CrossRef]
- Xie, J.; Tang, L.; Lu, L.; Zhang, L.; Xi, L.; Liu, H.C.; Odle, J.; Luo, X. Differential expression of heat shock transcription factors and heat shock proteins after acute and chronic heat stress in laying chickens (Gallus gallus). PLoS ONE 2014, 9, e102204. [Google Scholar] [CrossRef] [Green Version]
- Vijayan, M.M. Gene expression pattern in the liver during recovery from an acute stressor in rainbow trout. Comp. Biochem. Physiol. Part D Genom. Proteom. 2007, 2, 244. [Google Scholar]
- Wang, Y.; Li, C.; Pan, C.; Liu, E.; Zhao, X.; Ling, Q. Alterations to transcriptomic profile, histopathology, and oxidative stress in liver of pikeperch (Sander lucioperca) under heat stress. Fish Shellfish. Immunol. 2019, 95, 659–669. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Bao, E. Effect of acute heat stress on heat shock protein 70 and its corresponding mRNA expression in the heart, liver, and kidney of broilers. Asian Austral. J. Anim. 2008, 21, 1116–1126. [Google Scholar] [CrossRef]
- Bhusari, S.; Liu, Z.; Hearne, L.B.; Spiers, D.E.; Lamberson, W.R.; Antoniou, E. Expression profiling of heat stress effects on mice fed ergot alkaloids. Toxicol. Sci. 2007, 95, 89–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Purohit, G.K.; Mahanty, A.; Suar, M.; Sharma, A.P.; Mohanty, B.P.; Mohanty, S. Investigating hsp gene expression in liver of Channa striatus under heat stress for understanding the upper thermal acclimation. Biomed Res. Int. 2014, 2014, 381719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabergh, C.M.; Airaksinen, S.; Soitamo, A.; Bjorklund, H.V.; Johansson, T.; Nikinmaa, M.; Sistonen, L. Tissue-specific expression of zebrafish (Danio rerio) heat shock factor 1 mRNAs in response to heat stress. J. Exp. Biol. 2000, 203, 1817–1824. [Google Scholar] [CrossRef]
- Cui, Y.; Hao, Y.; Li, J.; Bao, W.; Li, G.; Gao, Y.; Gu, X. Chronic Heat Stress Induces Immune Response, Oxidative Stress Response, and Apoptosis of Finishing Pig Liver: A Proteomic Approach. Int. J. Mol. Sci. 2016, 17, 393. [Google Scholar] [CrossRef] [Green Version]
- Ronchi, B.; Bernabucci, U.; Lacetera, N.; Verini Supplizi, A.; Nardone, A. Distinct and common effects of heat stress and restricted feeding on metabolic status of Holstein heifers. Zootec. E Nutr. Anim. 1999, 25, 11–20. [Google Scholar]
- Rakesh, V.; Stallings, J.D.; Helwig, B.G.; Leon, L.R.; Jackson, D.A.; Reifman, J. A 3-D mathematical model to identify organ-specific risks in rats during thermal stress. J Appl. Physiol. 2013, 115, 1822–1837. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.; Decuypere, E.; Buyse, J. Acute heat stress induces oxidative stress in broiler chickens. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2006, 144, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Kew, M.; Bersohn, I.; Seftel, H.; Kent, G. Liver damage in heatstroke. Am. J. Med. 1970, 49, 192–202. [Google Scholar] [CrossRef]
- Zhu, G.; Li, W.; Zhang, F.; Guo, W. RNA-seq analysis reveals alternative splicing under salt stress in cotton, Gossypium davidsonii. BMC Genom. 2018, 19, 73. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Li, J.; Gao, Z.; Lu, Y.; Yu, J.; Zheng, Q.; Yan, S.; Zhang, W.; He, H.; Ma, L.; et al. SKIP Confers Osmotic Tolerance during Salt Stress by Controlling Alternative Gene Splicing in Arabidopsis. Mol. Plant 2015, 8, 1038–1052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Syed, N.H.; Kalyna, M.; Marquez, Y.; Barta, A.; Brown, J.W. Alternative splicing in plants--coming of age. Trends Plant Sci. 2012, 17, 616–623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calixto, C.P.G.; Guo, W.; James, A.B.; Tzioutziou, N.A.; Entizne, J.C.; Panter, P.E.; Knight, H.; Nimmo, H.G.; Zhang, R.; Brown, J.W.S. Rapid and Dynamic Alternative Splicing Impacts the Arabidopsis Cold Response Transcriptome. Plant Cell 2018, 30, 1424–1444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, F.; Cui, P.; Wang, Z.; Zhang, S.; Ali, S.; Xiong, L. Genome-wide analysis of alternative splicing of pre-mRNA under salt stress in Arabidopsis. BMC Genom. 2014, 15, 431. [Google Scholar] [CrossRef] [Green Version]
- Cui, P.; Zhang, S.; Ding, F.; Ali, S.; Xiong, L. Dynamic regulation of genome-wide pre-mRNA splicing and stress tolerance by the Sm-like protein LSm5 in Arabidopsis. Genome Biol. 2014, 15, R1. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.F.; Imam, J.S.; Wilkinson, M.F. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007, 76, 51–74. [Google Scholar] [CrossRef] [Green Version]
- Cartegni, L.; Chew, S.L.; Krainer, A.R. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat. Rev. Genet. 2002, 3, 285–298. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arroyo, J.D.; Jourdain, A.A.; Calvo, S.E.; Ballarano, C.A.; Doench, J.G.; Root, D.E.; Mootha, V.K. A Genome-wide CRISPR Death Screen Identifies Genes Essential for Oxidative Phosphorylation. Cell Metab. 2016, 24, 875–885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oslowski, C.M.; Urano, F. Measuring ER stress and the unfolded protein response using mammalian tissue culture system. Methods Enzym. 2011, 490, 71–92. [Google Scholar] [CrossRef] [Green Version]
- Alemu, T.W.; Pandey, H.O.; Salilew, W.D.; Gebremedhn, S.; Neuhof, C.; Tholen, E.; Holker, M.; Schellander, K.; Tesfaye, D. Oxidative and endoplasmic reticulum stress defense mechanisms of bovine granulosa cells exposed to heat stress. Theriogenology 2018, 110, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Wheelock, J.B.; Rhoads, R.P.; Vanbaale, M.J.; Sanders, S.R.; Baumgard, L.H. Effects of heat stress on energetic metabolism in lactating Holstein cows. J. Dairy Sci. 2010, 93, 644–655. [Google Scholar] [CrossRef]
- Pearce, S.C.; Gabler, N.K.; Ross, J.W.; Escobar, J.; Patience, J.F.; Rhoads, R.P.; Baumgard, L.H. The effects of heat stress and plane of nutrition on metabolism in growing pigs. J. Anim. Sci. 2013, 91, 2108–2118. [Google Scholar] [CrossRef] [Green Version]
- Torlinska, T.; Banach, R.; Paluszak, J.; Gryczka-Dziadecka, A. Hyperthermia effect on lipolytic processes in rat blood and adipose tissue. Acta Physiol. Pol. 1987, 38, 361–366. [Google Scholar]
- Geiger, P.C.; Gupte, A.A. Heat shock proteins are important mediators of skeletal muscle insulin sensitivity. Exerc. Sport Sci. Rev. 2011, 39, 34–42. [Google Scholar] [CrossRef]
- Pearce, N.J.; Yates, J.W.; Berkhout, T.A.; Jackson, B.; Tew, D.; Boyd, H.; Camilleri, P.; Sweeney, P.; Gribble, A.D.; Shaw, A.; et al. The role of ATP citrate-lyase in the metabolic regulation of plasma lipids. Hypolipidaemic effects of SB-204990, a lactone prodrug of the potent ATP citrate-lyase inhibitor SB-201076. Biochem. J. 1998, 334 Pt 1, 113–119. [Google Scholar] [CrossRef]
- Xin, M.; Qiao, Z.; Li, J.; Liu, J.; Song, S.; Zhao, X.; Miao, P.; Tang, T.; Wang, L.; Liu, W.; et al. miR-22 inhibits tumor growth and metastasis by targeting ATP citrate lyase: Evidence in osteosarcoma, prostate cancer, cervical cancer and lung cancer. Oncotarget 2016, 7, 44252–44265. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Yin, L.; Wei, J.; Yang, Z.; Jiang, G. ATP citrate lyase is increased in human breast cancer, depletion of which promotes apoptosis. Tumour. Biol. 2017, 39, 1393391326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatzivassiliou, G.; Zhao, F.; Bauer, D.E.; Andreadis, C.; Shaw, A.N.; Dhanak, D.; Hingorani, S.R.; Tuveson, D.A.; Thompson, C.B. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell 2005, 8, 311–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flees, J.; Rajaei-Sharifabadi, H.; Greene, E.; Beer, L.; Hargis, B.M.; Ellestad, L.; Porter, T.; Donoghue, A.; Bottje, W.G.; Dridi, S. Effect of Morinda citrifolia (Noni)-Enriched Diet on Hepatic Heat Shock Protein and Lipid Metabolism-Related Genes in Heat Stressed Broiler Chickens. Front. Physiol. 2017, 8, 919. [Google Scholar] [CrossRef] [Green Version]
- Yadav, S.K.; Pandey, A.; Kumar, L.; Devi, A.; Kushwaha, B.; Vishvkarma, R.; Maikhuri, J.P.; Rajender, S.; Gupta, G. The thermo-sensitive gene expression signatures of spermatogenesis. Reprod. Biol. Endocrinol. 2018, 16, 56. [Google Scholar] [CrossRef] [PubMed]
- Eslamizad, M.; Albrecht, D.; Kuhla, B. The effect of chronic, mild heat stress on metabolic changes of nutrition and adaptations in rumen papillae of lactating dairy cows. J. Dairy Sci. 2020, 103, 8601–8614. [Google Scholar] [CrossRef]
- Moon, Y.A.; Kim, K.S.; Park, S.W.; Kim, Y.S. Cloning and identification of exon-intron organization of the rat ATP-citrate lyase gene. Biochim. Biophys. Acta 1996, 1307, 280–284. [Google Scholar] [CrossRef]
- Xu, N.; Chen, C.Y.; Shyu, A.B. Versatile role for hnRNP D isoforms in the differential regulation of cytoplasmic mRNA turnover. Mol. Cell. Biol. 2001, 21, 6960–6971. [Google Scholar] [CrossRef] [Green Version]
- Latorre, E.; Torregrossa, R.; Wood, M.E.; Whiteman, M.; Harries, L.W. Mitochondria-targeted hydrogen sulfide attenuates endothelial senescence by selective induction of splicing factors HNRNPD and SRSF2. Aging 2018, 10, 1666–1681. [Google Scholar] [CrossRef]
- Hayakawa, H.; Fujikane, A.; Ito, R.; Matsumoto, M.; Nakayama, K.I.; Sekiguchi, M. Human proteins that specifically bind to 8-oxoguanine-containing RNA and their responses to oxidative stress. Biochem. Biophys. Res. Commun. 2010, 403, 220–224. [Google Scholar] [CrossRef]
- Lee, Y.; Rio, D.C. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu. Rev. Biochem. 2015, 84, 291–323. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Chen, K.L.; Zheng, X.M.; Li, H.X.; Wang, G.L. Identification and bioinformatics analysis of microRNAs associated with stress and immune response in serum of heat-stressed and normal Holstein cows. Cell Stress Chaperones 2014, 19, 973–981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Yang, C.; Du, J.; Zhang, B.; He, Y.; Hu, Q.; Li, M.; Zhang, Y.; Wang, C.; Zhong, J. Characterization of miRNA profiles in the mammary tissue of dairy cattle in response to heat stress. BMC Genom. 2018, 19, 975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, L.; Liao, R.; Wu, N.; Zhu, G.; Tu, Y.; Yang, C. Integrating miRNA and mRNA expression profiles in plasma of laying hens associated with heat stress. Mol. Biol. Rep. 2019, 46, 2779–2789. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Wang, H.; Wang, A.H.; Zhang, L.Y.; Bai, J. MicroRNA-532 and microRNA-3064 inhibit cell proliferation and invasion by acting as direct regulators of human telomerase reverse transcriptase in ovarian cancer. PLoS ONE 2017, 12, e173912. [Google Scholar] [CrossRef]
- Zhang, P.; Ha, M.; Li, L.; Huang, X.; Liu, C. MicroRNA-3064-5p sponged by MALAT1 suppresses angiogenesis in human hepatocellular carcinoma by targeting the FOXA1/CD24/Src pathway. FASEB J. 2020, 34, 66–81. [Google Scholar] [CrossRef] [Green Version]
- Donato, L.; Bramanti, P.; Scimone, C.; Rinaldi, C.; Giorgianni, F.; Beranova-Giorgianni, S.; Koirala, D.; D’Angelo, R.; Sidoti, A. miRNA expression profile of retinal pigment epithelial cells under oxidative stress conditions. FEBS Open Bio 2018, 8, 219–233. [Google Scholar] [CrossRef] [Green Version]





| A3SS | A5SS | MXE | RI | SE | AS | AE | Unknown | |
|---|---|---|---|---|---|---|---|---|
| Control | 28 | 32 | 8 | 32 | 47 | 53 | 26 | 6 |
| H120 | 31 | 35 | 11 | 31 | 57 | 76 | 28 | 8 |
| Increase (%) | 10.71 | 9.38 | 37.50 | −3.12 | 21.28 | 43.40 | 7.69 | 33.33 |
| Gene Symbol | Start | End | Type | Transcription | q-Value | dPSI |
|---|---|---|---|---|---|---|
| Strn4 | chr1:78756007:+ | chr1:78756355:+ | A3SS:P | NM_001107480 | 0.0481 | 0.20 |
| Strn4 | chr1:78756007:+ | chr1:78756355:+ | A3SS:D | NM_001161809 | 0.0494 | −0.20 |
| Ngrn | chr1:142050672:+ | chr1:142051084:+ | RI:E | NM_001033900 | 0.0031 | −0.16 |
| Ngrn | chr1:142050672:+ | chr1:142051084:+ | RI:R | NR_028055 | 0.0031 | 0.16 |
| Wbp1l | chr1:266401987:+ | chr1:266409945:+ | A5SS:D | NM_001127484 | 0.0024 | −0.10 |
| Wbp1l | chr1:266401987:+ | chr1:266409945:+ | A5SS:P | NM_001313908 | 0.0024 | 0.10 |
| Shc1 | chr2:188745503:+ | chr2:188748894:+ | AS | NM_053517 | 0.0048 | −0.17 |
| Shc1 | chr2:188748359:+ | chr2:188748894:+ | AS | NM_001164060 | 0.0048 | 0.17 |
| Ensa | chr2:197756356:+ | chr2:197759882:+ | AE | NM_021842 | 0.0000 | −0.24 |
| Ensa | chr2:197756356:+ | chr2:197758162:+ | AE | NM_001033974 | 0.0000 | 0.24 |
| Cast | chr2:1506234:− | chr2:1501715:− | SE:S | NM_001033716 | 0.0000 | 0.39 |
| Cast | chr2:1506234:− | chr2:1501715:− | SE:I | NM_001033715 NM_053295 | 0.0000 | −0.39 |
| Hnrnpf | chr4:149957206:+ | chr4:149970689:+ | AS | NM_001037287 | 0.0051 | −0.31 |
| Hnrnpf | chr4:149970237:+ | chr4:149970689:+ | AS | NM_001037285 NM_022397 | 0.6048 | 0.08 |
| Hnrnpf | chr4:149970567:+ | chr4:149970689:+ | AS | NM_001037286 | 0.0168 | 0.23 |
| Ak2 | chr5:147200851:+ | chr5:147204050:+ | AE | NM_001033967 | 0.0165 | −0.10 |
| Ak2 | chr5:147200851:+ | chr5:147201014:+ | AE | NM_030986 | 0.0165 | 0.10 |
| Ppm1b | chr6:8261060:+ | chr6:8271055:+ | AE | NM_001270619 | 0.0165 | 0.14 |
| Ppm1b | chr6:8261060:+ | chr6:8273549:+ | AE | NM_033096 | 0.8322 | 0.04 |
| Ppm1b | chr6:8261060:+ | chr6:8280124:+ | AE | NM_001270620 | 0.0000 | −0.17 |
| Srsf5 | chr6:104611145:+ | chr6:104612019:+ | A5SS:D | NM_001195506 | 0.0000 | −0.21 |
| Srsf5 | chr6:104611145:+ | chr6:104612019:+ | A5SS:P | NM_019257 NM_001195505 | 0.0000 | 0.21 |
| Tcf3 | chr7:12164343:+ | chr7:12167727:+ | Unknown | NM_001035237 | 0.0013 | 0.15 |
| Tcf3 | chr7:12164343:+ | chr7:12167727:+ | Unknown | NM_133524 | 0.0013 | −0.15 |
| Nabp2 | chr7:2825608:− | chr7:2825124:− | AS | NM_001244819 | 0.0261 | −0.18 |
| Nabp2 | chr7:2825498:− | chr7:2825124:− | AS | NM_001034939 | 0.0261 | 0.18 |
| Sqstm1 | chr10:35713296:− | chr10:35704728:− | AE | NM_175843 | 0.0000 | −0.24 |
| Sqstm1 | chr10:35713296:− | chr10:35713103:− | AE | NM_181550 | 0.0000 | 0.24 |
| Srebf1 | chr10:46582854:− | chr10:46579444:− | AS | NM_001276708 | 0.0168 | −0.14 |
| Srebf1 | chr10:46593009:− | chr10:46579444:− | AS | NM_001276707 | 0.0172 | 0.14 |
| Acly | chr10:88419161:− | chr10:88413611:− | SE:S | NM_001111095 | 0.0000 | 0.33 |
| Acly | chr10:88419161:− | chr10:88413611:− | SE:I | NM_016987 | 0.0000 | −0.33 |
| Ddx5 | chr10:94988461:− | chr10:94982051:− | AS | NM_001007613 | 0.0000 | −0.34 |
| Mir3064 | chr10:94982117:− | chr10:94982051:− | AS | NR_128673 | 0.0000 | 0.34 |
| Zcchc2 | chr13:26000532:+ | chr13:26014926:+ | AE | NM_001122677 | 0.0013 | −0.29 |
| Zcchc2 | chr13:26000532:+ | chr13:26000769:+ | AE | NM_001271042 | 0.0015 | 0.29 |
| Tor1aip2 | chr13:73708912:+ | chr13:73718239:+ | SE:S | NM_001165897 | 0.0000 | 0.39 |
| Tor1aip2 | chr13:73708912:+ | chr13:73718239:+ | SE:I | NM_001165896 | 0.0000 | −0.39 |
| Ephx1 | chr13:99287887:− | chr13:99284094:− | AS | NM_012844 | 0.0000 | 0.18 |
| Ephx1 | chr13:99300580:− | chr13:99284094:− | AS | NM_001034090 | 0.0000 | −0.18 |
| Hnrnpd | chr14:11256779:+ | chr14:11268562:+ | SE:S | NM_001082539 NM_001082541 | 0.0403 | 0.23 |
| Hnrnpd | chr14:11256779:+ | chr14:11268562:+ | SE:I | NM_001082540 NM_024404 | 0.0403 | −0.23 |
| Abcg3l4 | chr14:5794507:− | chr14:5607839:− | MXE | NM_001037205 | 0.0022 | 0.51 |
| Abcg3l2 | chr14:5794507:− | chr14:5607839:− | MXE | NM_001014133 | 0.0022 | −0.51 |
| Tmem33 | chr14:42540006:− | chr14:42539079:− | RI:E | NM_001034198 | 0.0037 | −0.14 |
| Tmem33 | chr14:42540006:− | chr14:42539079:− | RI:R | NM_021671 | 0.0037 | 0.14 |
| Crem | chr17:57082726:+ | chr17:57088162:+ | A3SS:P | NM_001110860 NM_001271247 NM_001271246 NM_001271102 | 0.0024 | 0.18 |
| Crem | chr17:57082726:+ | chr17:57088162:+ | A3SS:D | NM_001271101 NM_001271245 NM_017334 NM_001271248 | 0.0024 | −0.18 |
| Bicd2 | chr17:15677426:− | chr17:15675690:− | RI:E | NM_198765 | 0.0048 | −0.26 |
| Bicd2 | chr17:15677426:− | chr17:15675690:− | RI:R | NM_001033674 | 0.0048 | 0.26 |
| Zmynd11 | chr17:63831060:− | chr17:63830296:− | A5SS:P | NM_203367 NM_203369 | 0.0007 | 0.35 |
| Zmynd11 | chr17:63831060:− | chr17:63830296:− | A5SS:D | NM_203366 NM_203368 | 0.0007 | −0.35 |
| Slc39a7 | chr20:3822725:− | chr20:3822427:− | RI:E | NM_001164744 | 0.0000 | 0.19 |
| Slc39a7 | chr20:3822725:− | chr20:3822427:− | RI:R | NM_001008885 | 0.0000 | −0.19 |
| Group | ID | Description | p-Value | Count | Genes |
|---|---|---|---|---|---|
| BP | GO:0001889 | Liver development | 0.0000 | 5 | Cast, Ephx1, Ak2, Hnrnpd, Srsf5 |
| GO:0032868 | Response to insulin | 0.0072 | 3 | Srebf1, Shc1, Srsf5 | |
| GO:0032869 | Cellular response to insulin stimulus | 0.0094 | 3 | Srebf1, Shc1, Srsf5 | |
| GO:0000122 | Negative regulation of transcription from RNA polymerase II promoter | 0.0109 | 5 | Srebf1, Ddx5, Tcf3, Zmynd11, Sqstm1 | |
| GO:0008610 | Lipid biosynthetic process | 0.0178 | 2 | Srebf1, Acly | |
| GO:0014070 | Response to organic cyclic compound | 0.0415 | 3 | Srebf1, Shc1, Ephx1 | |
| GO:0050796 | Regulation of insulin secretion | 0.0480 | 2 | Srebf1, Ensa | |
| CC | GO:0005783 | Endoplasmic reticulum | 0.0051 | 6 | Cast, Srebf1, Tmem33, Tor1aip2, Slc39a7, Sqstm1 |
| GO:0005737 | Cytoplasm | 0.0152 | 14 | Cast, Srebf1, Shc1, Crem, Ak2, Bicd2, Acly, Hnrnpf, Ensa, Nabp2, Srsf5, Tcf3, Sqstm1, Zcchc2 | |
| GO:0005634 | Nucleus | 0.0219 | 13 | Cast, Srebf1, Ddx5, Shc1, Crem, Ak2, Ngrn, Hnrnpf, Nabp2, Hnrnpd, Srsf5, Tcf3, Zmynd11 | |
| GO:0005654 | Nucleoplasm | 0.0307 | 7 | Acly, Ddx5, Hnrnpf, Nabp2, Hnrnpd, Slc39a7, Zmynd11 | |
| GO:0005789 | Endoplasmic reticulum membrane | 0.0419 | 4 | Srebf1, Tmem33, Ephx1, Tor1aip2 | |
| MF | GO:0051721 | Protein phosphatase 2A binding | 0.0015 | 3 | Shc1, Ensa, Strn4 |
| GO:0005515 | Protein binding | 0.0073 | 8 | Cast, Ddx5, Shc1, Hnrnpd, Srsf5, Tcf3, Sqstm1, Strn4 | |
| GO:0044822 | Poly(A) RNA binding | 0.0246 | 6 | Cast, Ngrn, Ddx5, Hnrnpf, Hnrnpd, Srsf5 | |
| GO:0003682 | Chromatin binding | 0.0290 | 4 | Srebf1, Hnrnpd, Tcf3, Zmynd11 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, S.; Dou, J.; Li, Z.; Hu, L.; Yu, Y.; Wang, Y. Analysis of Genomic Alternative Splicing Patterns in Rat under Heat Stress Based on RNA-Seq Data. Genes 2022, 13, 358. https://doi.org/10.3390/genes13020358
Huang S, Dou J, Li Z, Hu L, Yu Y, Wang Y. Analysis of Genomic Alternative Splicing Patterns in Rat under Heat Stress Based on RNA-Seq Data. Genes. 2022; 13(2):358. https://doi.org/10.3390/genes13020358
Chicago/Turabian StyleHuang, Shangzhen, Jinhuan Dou, Zhongshu Li, Lirong Hu, Ying Yu, and Yachun Wang. 2022. "Analysis of Genomic Alternative Splicing Patterns in Rat under Heat Stress Based on RNA-Seq Data" Genes 13, no. 2: 358. https://doi.org/10.3390/genes13020358