Phylogeography of Sarmarutilus rubilio (Cypriniformes: Leuciscidae): Complex Genetic Structure, Clues to a New Cryptic Species and Further Insights into Roaches Phylogeny
Abstract
:1. Introduction
2. Materials and Methods
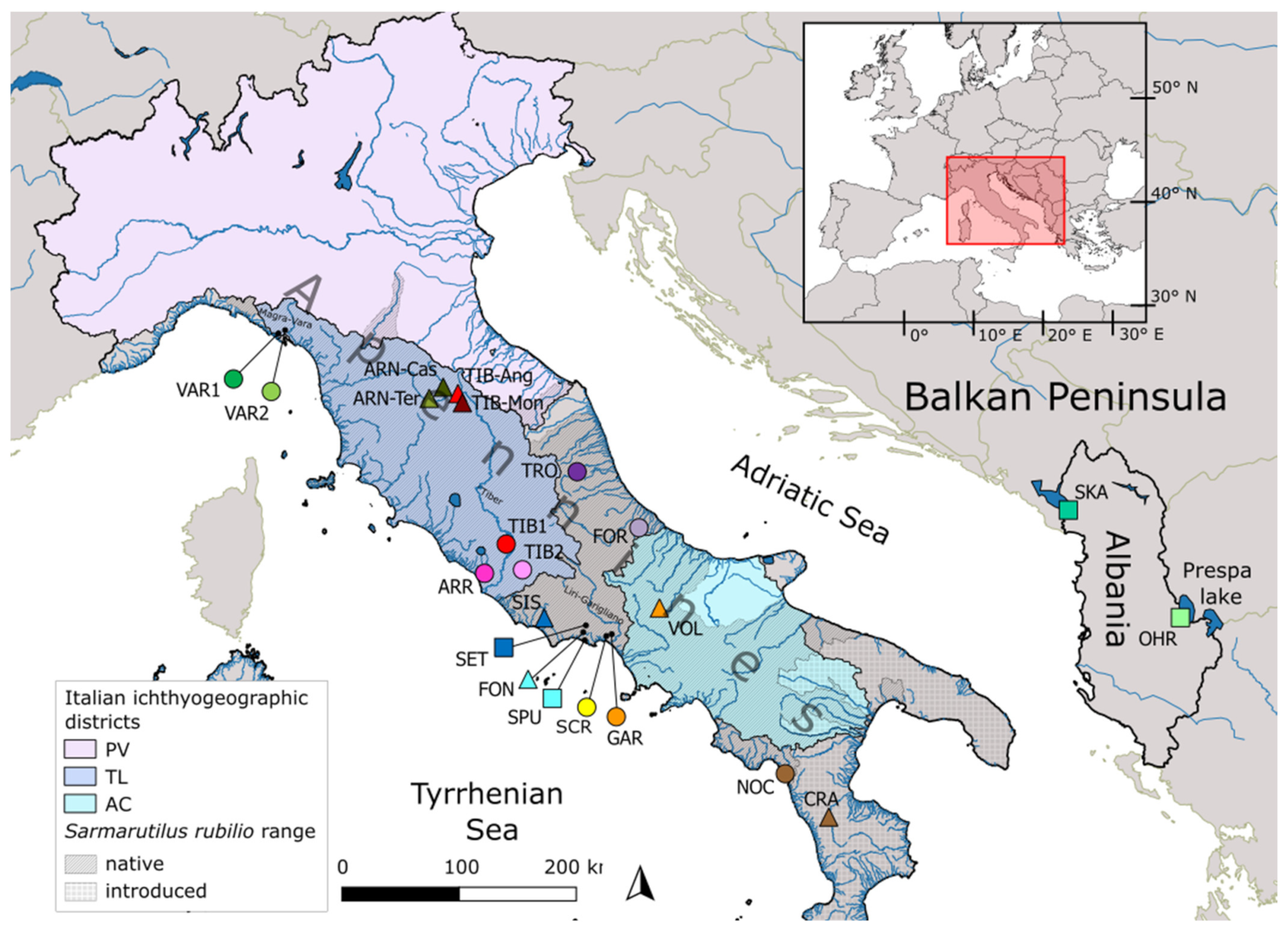
2.1. Sampling, Morphological Identification and DNA Extraction
2.2. PCR Amplification
2.3. Data Analysis
3. Results
3.1. Molecular Identification and Phylogeny
3.2. Genetic Variability and Demographic History
3.3. Genetic Structure and Phylogeography
4. Discussion
4.1. Molecular Identification and Phylogeny
4.2. Divergence between Sarmarutilus Lineages: Cryptic Species?
4.3. Phylogeography of Italian Sarmarutilus
4.3.1. Haplogroup C
4.3.2. Haplogroup A
4.3.3. Haplogroup B
4.4. Implication for Conservation and Management
- (a).
- New research is necessary to precisely map the distribution of lineage C. This new putative cryptic species is endemic to the Magra-Vara basin and is found in tributaries not included in protected areas; therefore, it could be particularly exposed to threats. This underlines the importance of also protecting small river course habitats, as they might represent refugia of relict native fish populations [119].
- (b).
- (c).
- The main conservation efforts should be focused on avoiding the introduction of allochthonous species, especially those that can compete or hybridize with Sarmarutilus. In addition, the translocation of Sarmarutilus populations from different districts and within the AC district (where subclusters are present) should be avoided, as these roaches represent different genetic entities.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pereira, L.H.G.; Hanner, R.; Foresti, F.; Oliveira, C. Can DNA Barcoding Accurately Discriminate Megadiverse Neotropical Freshwater Fish Fauna? BMC Genet. 2013, 14, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perea, S.; Böhme, M.; Zupančič, P.; Freyhof, J.; Šanda, R.; Özulu, M.; Abdoli, A.; Doadrio, I. Phylogenetic Relationships and Biogeographical Patterns in Circum-Mediterranean Subfamily Leuciscinae (Teleostei, Cyprinidae) Inferred from Both Mitochondrial and Nuclear Data. BMC Evol. Biol. 2010, 10, 265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.Z.; Gan, X.N.; Li, J.B.; Mayden, R.L.; He, S.P. Cyprinid Phylogeny Based on Bayesian and Maximum Likelihood Analyses of Partitioned Data: Implications for Cyprinidae Systematics. Sci. China Life Sci. 2012, 55, 761–773. [Google Scholar] [CrossRef] [Green Version]
- Tancioni, L.; Russo, T.; Cataudella, S.; Milana, V.; Hett, A.K.; Corsi, E.; Rossi, A.R. Testing Species Delimitations in Four Italian Sympatric Leuciscine Fishes in the Tiber River: A Combined Morphological and Molecular Approach. PLoS ONE 2013, 8, e60392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramirez, J.L.; Carvalho-Costa, L.F.; Venere, P.C.; Carvalho, D.C.; Troy, W.P.; Galetti, P.M. Testing Monophyly of the Freshwater Fish Leporinus (Characiformes, Anostomidae) through Molecular Analysis. J. Fish Biol. 2016, 88, 1204–1214. [Google Scholar] [CrossRef] [PubMed]
- Mangit, F.; Yerli, S.V. Systematic Evaluation of the Genus Alburnus (Cyprinidae) with Description of a New Species. Hydrobiologia 2018, 807, 297–312. [Google Scholar] [CrossRef]
- Lucentini, L.; Puletti, M.E.; Ricciolini, C.; Gigliarelli, L.; Fontaneto, D.; Lanfaloni, L.; Bilò, F.; Natali, M.; Panara, F. Molecular and Phenotypic Evidence of a New Species of Genus Esox (Esocidae, Esociformes, Actinopterygii): The Southern Pike, Esox flaviae. PLoS ONE 2011, 6, e25218. [Google Scholar] [CrossRef]
- Palandačić, A.; Bravničar, J.; Zupančič, P.; Šanda, R.; Snoj, A. Molecular Data Suggest a Multispecies Complex of Phoxinus (Cyprinidae) in the Western Balkan Peninsula. Mol. Phylogenet. Evol. 2015, 92, 118–123. [Google Scholar] [CrossRef]
- Artaev, O.N.; Ermakov, O.A.; Vekhov, D.A.; Konovalov, A.F.; Levina, M.A.; Pozdeev, I.V.; Ruchin, A.B.; Alyushin, I.V.; Iljin, V.Y.; Levin, B.A. Genetic Screening of Distribution Pattern of Roaches Rutilus rutilus and R. lacustris (Cyprinidae) in Broad Range of Secondary Contact (Volga Basin). Inl. Water Biol. 2021, 14, 205–214. [Google Scholar] [CrossRef]
- De Santis, V.; Delmastro, G.B.; Vanetti, I.; Britton, J.R.; Zaccara, S. Species Composition of Introduced and Natural Minnow Populations of the Phoxinus Cryptic Complex in the Westernmost Part of the Po River Basin (North Italy). Biol. Invasions 2021, 23, 657–668. [Google Scholar] [CrossRef]
- Milana, V.; Šanda, R.; Vukić, J.; Ciccotti, E.; Riccato, F.; Petrosino, G.; Rossi, A.R. Far from Home: Genetic Variability of Knipowitschia Sp. from Italy Revealed Unexpected Species in Coastal Lagoons of the Tyrrhenian Coast. Estuar. Coast. Shelf Sci. 2021, 251, 107260. [Google Scholar] [CrossRef]
- Rossi, A.R.; Milana, V.; Pulcini, D.; Cataudella, S.; Martinoli, M.; Tancioni, L. An Integrated Genetic and Morphological Approach to Clarify the Conservation Status of the Threatened Italian Endemic Species Alburnus albidus (Cypriniformes: Cyprinidae). Hydrobiologia 2016, 770, 73–87. [Google Scholar] [CrossRef]
- Bernatchez, L. The Evolutionary History of Brown Trout (Salmo trutta L.) Inferred from Phylogeographic, Nested Clade, and Mismatch Analyses of Mitochondrial DNA Variation. Evolution 2001, 55, 351–379. [Google Scholar] [CrossRef] [PubMed]
- Buonerba, L.; Zaccara, S.; Delmastro, G.B.; Lorenzoni, M.; Salzburger, W.; Gante, H.F. Intrinsic and Extrinsic Factors Act at Different Spatial and Temporal Scales to Shape Population Structure, Distribution and Speciation in Italian Barbus (Osteichthyes: Cyprinidae). Mol. Phylogenet. Evol. 2015, 89, 115–129. [Google Scholar] [CrossRef] [PubMed]
- Buj, I.; Marčić, Z.; Caleta, M.; Sanda, R.; Geiger, M.F.; Freyhof, J.; Machordom, A.; Vukić, J. Ancient Connections among the European Rivers and Watersheds Revealed from the Evolutionary History of the Genus Telestes (Actinopterygii; Cypriniformes). PLoS ONE 2017, 12, e0187366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Myers, N.; Mittermeler, R.A.; Mittermeler, C.G.; da Fonseca, G.A.B.; Kent, J. Biodiversity Hotspots for Conservation Priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef]
- Tierno de Figueroa, J.M.; López-Rodríguez, M.J.; Fenoglio, S.; Sánchez-Castillo, P.; Fochetti, R. Freshwater Biodiversity in the Rivers of the Mediterranean Basin. Hydrobiologia 2013, 719, 137–186. [Google Scholar] [CrossRef]
- Seifertová, M.; Bryja, J.; Vyskočilová, M.; Martínková, N.; Šimková, A. Multiple Pleistocene Refugia and Post-Glacial Colonization in the European Chub (Squalius cephalus) Revealed by Combined Use of Nuclear and Mitochondrial Markers. J. Biogeogr. 2012, 39, 1024–1040. [Google Scholar] [CrossRef]
- Lévêque, C.; Oberdorff, T.; Paugy, D.; Stiassny, M.L.J.; Tedesco, P.A. Global Diversity of Fish (Pisces) in Freshwater. Hydrobiologia 2008, 595, 545–567. [Google Scholar] [CrossRef]
- Geiger, M.F.; Herder, F.; Monaghan, M.T.; Almada, V.; Barbieri, R.; Bariche, M.; Berrebi, P.; Bohlen, J.; Casal-Lopez, M.; Delmastro, G.B.; et al. Spatial Heterogeneity in the Mediterranean Biodiversity Hotspot Affects Barcoding Accuracy of Its Freshwater Fishes. Mol. Ecol. Resour. 2014, 14, 1210–1221. [Google Scholar] [CrossRef] [Green Version]
- Bianco, P.G. An Update on the Status of Native and Exotic Freshwater Fishes of Italy. J. Appl. Ichthyol. 2014, 30, 62–77. [Google Scholar] [CrossRef]
- Bianco, P.G. Factors Affecting the Distribution of Freshwater Fishes Especially in Italy. Cybium 1995, 19, 241–259. [Google Scholar]
- AIIAD. Principi Guida Riguardanti Le Immissioni Di Fauna Ittica Nelle Acque Interne Italiane. 2021. Available online: http://www.aiiad.it/sito/images/docs/sistematica/AIIAD-Principi_guida_immissioni_fauna_ittica_05032021.pdf (accessed on 21 March 2022).
- Rondinini, C.; Battistoni, A.; Peronace, V.; Teofili, C. Lista Rossa Dei Vertebrati Italiani. Comitato Italiano IUCN e Ministero dell’Ambiente e della Tutela del Territorio e del Mare, 2013. Available online: http://www.iucn.it/pdf/Comitato_IUCN_Lista_Rossa_dei_vertebrati_italiani.pdf (accessed on 22 March 2022).
- Schönhuth, S.; Vukić, J.; Šanda, R.; Yang, L.; Mayden, R.L. Phylogenetic Relationships and Classification of the Holarctic Family Leuciscidae (Cypriniformes: Cyprinoidei). Mol. Phylogenet. Evol. 2018, 127, 781–799. [Google Scholar] [CrossRef] [PubMed]
- Marchetto, F.; Zaccara, S.; Muenzel, F.M.; Salzburger, W. Phylogeography of the Italian Vairone (Telestes muticellus, Bonaparte 1837) Inferred by Microsatellite Markers: Evolutionary History of a Freshwater Fish Species with a Restricted and Fragmented Distribution. BMC Evol. Biol. 2010, 10, 111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaccara, S.; Quadroni, S.; de Santis, V.; Vanetti, I.; Carosi, A.; Britton, R.; Lorenzoni, M. Genetic and Morphological Analyses Reveal a Complex Biogeographic Pattern in the Endemic Barbel Populations of the Southern Italian Peninsula. Ecol. Evol. 2019, 9, 10185–10197. [Google Scholar] [CrossRef] [Green Version]
- Rossi, A.R.; Petrosino, G.; Crescenzo, S.; Milana, V.; Talarico, L.; Martinoli, M.; Rakaj, A.; Lorenzoni, M.; Carosi, A.; Ciuffardi, L. Phylogeography and Population Structure of Squalius lucumonis: A Baseline for Conservation of an Italian Endangered Freshwater Fish. J. Nat. Conserv. 2021, 64, 126085. [Google Scholar] [CrossRef]
- Nonnis Marzano, F.; Lorenzoni, M.; Tancioni, L. PESCI (Ciclostomi e Osteitti). In Manuali per il Monitoraggio di Specie e Habitat di Interesse Comunitario (Direttiva 92/43/CEE) in Italia: Specie Animali; Stoch, P.G.F., Ed.; ISPRA: Rome, Italy, 2016; pp. 129–190. [Google Scholar]
- Bianco, P.G.; Ketmaier, V. A Revision of the Rutilus Complex from Mediterranean Europe with Description of a New Genus, Sarmarutilus, and a New Species, Rutilus stoumboudae (Teleostei: Cyprinidae). Zootaxa 2014, 3841, 379–402. [Google Scholar] [CrossRef] [Green Version]
- Djait, H.; Bahri-Sfar, L.; Laouar, H.; Missaoui, N.; Ben Hassine, O.K. Dietary Comparison of Pike-Perch, Sander lucioperca (Linnaeus, 1758) and Catfish, Silurus glanis Linnaeus, 1758 in Sidi Salem Dam Reservoir (Tunisia). Cybium 2019, 43, 61–69. [Google Scholar] [CrossRef]
- Keskin, E.; Unal, E.M.; Atar, H.H. Detection of Rare and Invasive Freshwater Fish Species Using EDNA Pyrosequencing: Lake Iznik Ichthyofauna Revised. Biochem. Syst. Ecol. 2016, 67, 29–36. [Google Scholar] [CrossRef]
- Bianco, P.G.; Caputo, V.; Ferrito, V.; Lorenzoni, M.; Nonnis Marzano, F.; Stefani, F.; Sabatini, A.; Tancioni, L. Rutilus rubilio. In Lista Rossa IUCN dei Vertebrati Italiani; Rondinini, C., Battistoni, A., Peronace, V., Teofili, C., Eds.; Comitato Italiano IUCN e Ministero dell’Ambiente e della Tutela del Territorio e del Mare, 2013. Available online: http://www.iucn.it/scheda.php?id=-251974278 (accessed on 21 February 2022).
- Di Tizio, L.; di Felice, P.L. Rutilus rubilio (Bonaparte, 1837) (Rovella). In Manuali per il Monitoraggio di Specie e Habitat di Interesse Comunitario (Direttiva 92/43/CEE) in Italia: Specie Animali; Stoch, F., Genovesi, P., Eds.; ISPRA: Rome, Italy, 2016; pp. 164–165. [Google Scholar]
- EEC. Council Directive 92/43/ECC. Off. J. Eur. Union 1992. [Google Scholar]
- Marić, D. Rutilus albus Sp. n. (Teleostei: Cyprinidae) from Lake Skadar. Period. Biol. 2010, 112, 153–158. [Google Scholar]
- Milošević, D.; Winkler, K.A.; Marić, D.; Weiss, S. Genotypic and Phenotypic Evaluation of Rutilus Spp. from Skadar, Ohrid and Prespa Lakes Supports Revision of Endemic as Well as Taxonomic Status of Several Taxa. J. Fish Biol. 2011, 79, 1094–1110. [Google Scholar] [CrossRef] [PubMed]
- Stoch, F.; Genovesi, P. Manuali per Il Monitoraggio Di Specie e Habitat Di Interesse Comunitario (Direttiva 92/43/CEE) in Italia: Habitat; ISPRA: Rome, Italy, 2016. [Google Scholar]
- Kottelat, M.; Freyhof, J. Handbook of European Freshwater Fishes; Publications Kottelat: Cornol, Switzerland, 2007. [Google Scholar]
- Aljanabi, S.M.; Martinez, I. Universal and Rapid Salt-Extraction of High Quality Genomic DNA for PCR-Based Techniques. Nucleic Acids Res. 1997, 25, 4692–4693. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.N.; Ratnasingham, S.; DeWaard, J.R. Barcoding Animal Life: Cytochrome c Oxidase Subunit 1 Divergences among Closely Related Species. Proc. R. Soc. B Biol. Sci. 2003, 270 (Suppl. 1), 96–99. [Google Scholar] [CrossRef] [Green Version]
- Lieckfeldt, D.; Hett, A.K.; Ludwig, A.; Freyhof, J. Detection, Characterization and Utility of a New Highly Variable Nuclear Marker Region in Several Species of Cyprinid Fishes (Cyprinidae). Eur. J. Wildl. Res. 2006, 52, 63–65. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The Barcode of Life Data System: Barcoding. Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X Version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A Computer Program to Estimate Gene Genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef] [Green Version]
- Leigh, J.W.; Bryant, D. POPART: Full-Feature Software for Haplotype Network Construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucl. Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. Mrbayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Bio. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. JModelTest 2: More Models, New Heuristics and Parallel Computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [Green Version]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin (Version 3.0): An Integrated Software Package for Population Genetics Data Analysis. Evol. Bioinforma. 2005, 1, 117693430500100. [Google Scholar] [CrossRef] [Green Version]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: Mew York, NY, USA, 1987. [Google Scholar]
- Kimura, M. A Simple Method for Estimating Evolutionary Rates of Base Substitutions through Comparative Studies of Nucleotide Sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Adamack, A.T.; Gruber, B. PopGenReport: Simplifying Basic Population Genetic Analyses in R. Methods Ecol. Evol. 2014, 5, 384–387. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. 2020. Available online: https://www.R-project.org/ (accessed on 1 February 2022).
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian Phylogenetic and Phylodynamic Data Integration Using BEAST 1.10. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef] [Green Version]
- Tipton, M.L.; Gignoux-Wolfsohn, S.; Stonebraker, P.; Chernoff, B. Postglacial Recolonization of Eastern Blacknose Dace, Rhinichthys atratulus (Teleostei: Cyprinidae), through the Gateway of New England. Ecol. Evol. 2011, 1, 343–358. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Harris, A.J.; He, X. S-DIVA (Statistical Dispersal-Vicariance Analysis): A Tool for Inferring Biogeographic Histories. Mol. Phylogenet. Evol. 2010, 56, 848–850. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Harris, A.J.; Blair, C.; He, X. RASP (Reconstruct Ancestral State in Phylogenies): A Tool for Historical Biogeography. Mol. Phylogenet. Evol. 2015, 87, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Harpending, H.C. Signature of Ancient Population Growth in a Low-Resolution Mitochondrial DNA Mismatch Distribution. Human Biol. 1994, 66, 591–600. [Google Scholar]
- Schneider, S.; Excoffier, L. Estimation of Past Demographic Parameters from the Distribution of Pairwise Differences When the Mutation Rates Vary among Sites: Application to Human Mitochondrial DNA. Genetics 1999, 152, 1079–1089. [Google Scholar] [CrossRef]
- Tajima, F. Statistical Method for Testing the Neutral Mutation Hypothesis by DNA Polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Fu, Y. Statistical Tests of Neutrality of Mutations Against Population Growth, Hitchhiking and Background Selection. Genetics 1997, 147, 915–925. [Google Scholar] [CrossRef]
- Ramos-Onsins, S.E.; Rozas, J. Statistical Properties of New Neutrality Tests against Population Growth. Mol. Biol. Evol. 2002, 19, 2092–2100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogers, A.R.; Harpending, H. Population Growth Makes Waves in the Distribution of Pairwise Genetic Differences. Mol. Biol. Evol. 1992, 9, 552–569. [Google Scholar] [CrossRef] [PubMed]
- Hashemzadeh Segherloo, I.; Ghojoghi, F.; Tabatabaei, S.N.; Normandeau, E.; Hernandez, C.; Hallerman, E.; Boyle, B.; Bernatchez, L. Population Genomics of the Southern Caspian Sea Vobla Rutilus Lacustris. Hydrobiologia 2021, 848, 345–361. [Google Scholar] [CrossRef]
- Fricke, R.; Eschmeyer, W.N.; van der Laan, R. Eschmeyer’s Catalog of Fishes: Genera, Species, References; Electronic Version. Available online: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (accessed on 25 April 2022).
- Oikonomou, A.; Leprieur, F.; Leonardos, I.D. Biogeography of Freshwater Fishes of the Balkan Peninsula. Hydrobiologia 2014, 738, 205–220. [Google Scholar] [CrossRef]
- Tougard, C.; Justy, F.; Guinand, B.; Douzery, E.J.P.; Berrebi, P. Salmo macrostigma (Teleostei, Salmonidae): Nothing More than a Brown Trout (S. Trutta) Lineage? J. Fish Biol. 2018, 93, 302–310. [Google Scholar] [CrossRef] [Green Version]
- Ketmaier, V.; Bianco, P.G.; Durand, J.D. Molecular Systematics, Phylogeny and Biogeography of Roaches (Rutilus, Teleostei, Cyprinidae). Mol. Phylogenet. Evol. 2008, 49, 362–367. [Google Scholar] [CrossRef]
- Jurajda, P.; Pavlov, I. Rediscovery of Rutilus virgo in the River Dyje, Czech Republic. Folia Zool. 2016, 65, 98–100. [Google Scholar] [CrossRef]
- Tsoumani, M.; Georgiadis, A.; Giantsis, I.A.; Leonardos, I.; Apostolidis, A.P. Phylogenetic Relationships among Southern Balkan Rutilus Species Inferred from Cytochrome b Sequence Analysis: Micro-Geographic Resolution and Taxonomic Implications. Biochem. Syst. Ecol. 2014, 54, 172–178. [Google Scholar] [CrossRef]
- Levin, B.A.; Simonov, E.P.; Ermakov, O.A.; Levina, M.A.; Interesova, E.A.; Kovalchuk, O.M.; Malinina, Y.A.; Mamilov, N.S.; Mustafayev, N.J.; Pilin, D.V.; et al. Phylogeny and Phylogeography of the Roaches, Genus Rutilus (Cyprinidae), at the Eastern Part of Its Range as Inferred from MtDNA Analysis. Hydrobiologia 2017, 788, 33–46. [Google Scholar] [CrossRef]
- Ward, R.D. DNA Barcode Divergence among Species and Genera of Birds and Fishes. Mol. Ecol. Resour. 2009, 9, 1077–1085. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, Y.P.; Yang, Z.Y.; Liu, Z.; Du, Y.Y. DNA Barcoding Reveals Cryptic Diversity in the Underestimated Genus Triplophysa (Cypriniformes: Cobitidae, Nemacheilinae) from the Northeastern Qinghai-Tibet Plateau. BMC Evol. Biol. 2020, 20, 151. [Google Scholar] [CrossRef] [PubMed]
- Pereira, L.H.G.; Castro, J.R.C.; Vargas, P.M.H.; Gomez, J.A.M.; Oliveira, C. The Use of an Integrative Approach to Improve Accuracy of Species Identification and Detection of New Species in Studies of Stream Fish Diversity. Genetica 2021, 149, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Guimarães, K.L.A.; Lima, M.P.; Santana, D.J.; de Souza, M.F.B.; Barbosa, R.S.; Rodrigues, L.R.R. DNA Barcoding and Phylogeography of the Hoplias malabaricus Species Complex. Sci. Rep. 2022, 12, 5288. [Google Scholar] [CrossRef] [PubMed]
- Tsoupas, A.; Papavasileiou, S.; Minoudi, S.; Gkagkavouzis, K.; Petriki, O.; Bobori, D.; Sapounidis, A.; Koutrakis, E.; Leonardos, I.; Karaiskou, N.; et al. DNA Barcoding Identification of Greek Freshwater Fishes. PLoS ONE 2022, 17, e0263118. [Google Scholar] [CrossRef] [PubMed]
- Valić, D.; Vardić Smrzlić, I.; Kapetanović, D.; Teskeredžić, Z.; Pleše, B.; Teskeredžić, E. Identification, Phylogenetic Relationships and a New Maximum Size of Two Rudd Populations (Scardinius, Cyprinidae) from the Adriatic Sea Drainage, Croatia. Biologia 2013, 68, 539–545. [Google Scholar] [CrossRef]
- Scribner, K.T.; Page, K.S.; Bartron, M.L. Hybridization in Freshwater Fishes: A Review of Case Studies and Cytonuclear Methods of Biological Inference. Rev. Fish Biol. Fish. 2000, 10, 293–323. [Google Scholar] [CrossRef]
- Hayden, B.; Pulcini, D.; Kelly-Quinn, M.; O’Grady, M.; Caffrey, J.; McGrath, A.; Mariani, S. Hybridisation between Two Cyprinid Fishes in a Novel Habitat: Genetics, Morphology and Life-History Traits. BMC Evol. Biol. 2010, 10, 169. [Google Scholar] [CrossRef] [Green Version]
- Sousa-Santos, C.; Matono, P.; Silva, J.D.A.; Ilhéu, M. Evaluation of Potential Hybridization between Native Fishes and the Invasive Bleak, Alburnus alburnus (Actinopterygii: Cypriniformes: Cyprinidae). Acta Ichthyol. Piscat. 2018, 48, 109–122. [Google Scholar] [CrossRef] [Green Version]
- Freyhof, J.; Lieckfeldt, D.; Pitra, C.; Ludwig, A. Molecules and Morphology: Evidence for Introgression of Mitochondrial DNA in Dalmatian Cyprinids. Mol. Phylogenet. Evol. 2005, 37, 347–354. [Google Scholar] [CrossRef]
- Carranza, S.; Romano, A.; Arnold, E.N.; Sotgiu, G. Biogeography and Evolution of European Cave Salamanders, Hydromantes (Urodela: Plethodontidae), Inferred from MtDNA Sequences. J. Biogeogr. 2008, 35, 724–738. [Google Scholar] [CrossRef]
- Mattoccia, M.; Marta, S.; Romano, A.; Sbordoni, V. Phylogeography of an Italian Endemic Salamander (Genus Salamandrina): Glacial Refugia, Postglacial Expansions, and Secondary Contact. Biol. J. Linn. Soc. 2011, 104, 903–992. [Google Scholar] [CrossRef] [Green Version]
- Canestrelli, D.; Salvi, D.; Maura, M.; Bologna, M.A.; Nascetti, G. One Species, Three Pleistocene Evolutionary Histories: Phylogeography of the Italian Crested Newt, Triturus carnifex. PLoS ONE 2012, 7, e41754. [Google Scholar] [CrossRef] [PubMed]
- Maura, M.; Salvi, D.; Bologna, M.A.; Nascetti, G.; Canestrelli, D. Northern Richness and Cryptic Refugia: Phylogeography of the Italian Smooth Newt Lissotriton vulgaris meridionalis. Biol. J. Linn. Soc. 2014, 113, 590–603. [Google Scholar] [CrossRef] [Green Version]
- Schultze, N.; Spitzweg, C.; Corti, C.; Delaugerre, M.; di Nicola, M.R.; Geniez, P.; Lapini, L.; Liuzzi, C.; Lunghi, E.; Novarini, N.; et al. Mitochondrial Ghost Lineages Blur Phylogeography and Taxonomy of Natrix helvetica and N. natrix in Italy and Corsica. Zool. Scr. 2020, 49, 395–411. [Google Scholar] [CrossRef] [Green Version]
- Stefani, F.; Galli, P.; Crosa, G.; Zaccara, S.; Calamari, D. Alpine and Apennine Barriers Determining the Differentiation of the Rudd (Scardinius erythrophthalmus L.) in the Italian Peninsula. Ecol. Freshw. Fish 2004, 13, 168–175. [Google Scholar] [CrossRef]
- Schmitt, T.; Fritz, U.; Delfino, M.; Ulrich, W.; Habel, J.C. Biogeography of Italy Revisited: Genetic Lineages Confirm Major Phylogeographic Patterns and a Pre-Pleistocene Origin of Its Biota. Front. Zool. 2021, 18, 34. [Google Scholar] [CrossRef] [PubMed]
- Lorenzoni, M.; Carosi, A.; Quadroni, S.; de Santis, V.; Vanetti, I.; Delmastro, G.B.; Zaccara, S. Cryptic Diversity within Endemic Italian Barbels: Revalidation and Description of New Barbus Species (Teleostei: Cyprinidae). J. Fish Biol. 2021, 98, 1433–1449. [Google Scholar] [CrossRef]
- Raggi, G. Neotettonica ed evoluzione paleogeografica plio-pleistocenica del bacino del fiume Magra. Mem. Della Soc. Geol. Ital. 1985, 30, 35–62. [Google Scholar]
- Ciuffardi, L.; Oneto, F.; Raineri, V. L’ittiofauna Delle Acque Interne Della Liguria: Aspetti Filogeografici e Distributivi Rilevanti Ai Fini Dell’applicazione Della Direttiva 2000/60/CE. Ann. Mus. Civ. Stor. Nat. Giacomo Doria 2015, 107, 213–283. [Google Scholar]
- Zaccara, S.; Delmastro, G.B. Tyrrhenian Basins of Ligury as a New Peri-Mediterranean Ichthyogeographic District? Population Structure of Telestes muticellus (Osteichthyes, Cyprinidae), a Primary Freshwater Fish. Hydrobiologia 2009, 632, 285–295. [Google Scholar] [CrossRef]
- Bianco, P.G. L’Ittiofauna Continentale Dell’Appennino Umbro-Marchigiano, Barriera Semipermeabile Allo Scambio Di Componenti Primarie Tra Gli Opposti Versanti Dell’Italia Centrale. Biogeogr.–J. Integr. Biogeogr. 1994, 17, 427–485. [Google Scholar] [CrossRef] [Green Version]
- Piacentini, T.; Miccadei, E. The Role of Drainage Systems and Intermontane Basins in the Quaternary Landscape of the Central Apennines Chain (Italy). Rend. Lincei 2014, 25, 139–150. [Google Scholar] [CrossRef]
- Splendiani, A.; Giovannotti, M.; Cerioni, P.N.; Caniglia, M.L.; Caputo, V. Phylogeographic Inferences on the Native Brown Trout MtDNA Variation in Central Italy. Ital. J. Zool. 2006, 73, 179–189. [Google Scholar] [CrossRef] [Green Version]
- Rossi, A.R.; Talarico, L.; Petrosino, G.; Crescenzo, S.; Tancioni, L. Conservation Genetics of Mediterranean Brown Trout in Central Italy (Latium): A Multi-Marker Approach Anna. Water 2022, 14, 937. [Google Scholar] [CrossRef]
- Taviani, S.; Henriksen, H.J. The Application of a Groundwater/Surface-Water Model to Test the Vulnerability of Bracciano Lake (near Rome, Italy) to Climatic and Water-Use Stresses. Hydrogeol. J. 2015, 23, 1481–1498. [Google Scholar] [CrossRef]
- Cavinato, G.P.; Miccadei, E.; Parotto, M. Stato Dell’arte Delle Conoscenze Sulla Geologia Plio-Quaternaria Dell’Italia Centrale (Settore Laziale). Studi Geol. Camerti Vol. Spec. 1992, 1, 27–31. [Google Scholar]
- Aucelli, P.P.C.; D’Argenio, B.; della Seta, M.; Giano, S.I.; Schiattarella, M. Foreword: Intermontane Basins: Quaternary Morphoevolution of Central-Southern Italy. Rend. Lincei 2014, 25, 107–110. [Google Scholar] [CrossRef] [Green Version]
- Bernatchez, L.; Wilson, C.C. Comparative Phylogeography of Nearctic and Palearctic Fishes. Mol. Ecol. 1998, 7, 431–452. [Google Scholar] [CrossRef] [Green Version]
- Tschá, M.K.; Bachmann, L.; Abilhoa, V.; Boeger, W.A. Past Connection and Isolation of Catchments: The Sea-hanges Affect the Distribution and Genetic Variability of Coastal Freshwater Fishes. Estuar. Coast. Shelf Sci. 2017, 190, 31–39. [Google Scholar] [CrossRef]
- Lambeck, K.; Antonioli, F.; Purcell, A.; Silenzi, S. Sea-Level Change along the Italian Coast for the Past 10,000 Yr. Quat. Sci. Rev. 2004, 23, 1567–1598. [Google Scholar] [CrossRef]
- Vai, G.B.; Cantelli, L. Litho-Palaeoenvironmental Maps of Italy During the Last Two Climatic Extremes; Museo Geologico Giovanni Capellini: Bologna, Italy, 2004. [Google Scholar]
- Grant, W.S.; Bowen, B.W. Shallow Population Histories in Deep Evolutionary Lineages of Marine Fishes: Insights from Sardines and Anchovies and Lessons for Conservation. J. Hered. 1998, 89, 415–426. [Google Scholar] [CrossRef]
- Bianco, P.G. Mediterranean Endemic Freshwater Fishes of Italy. Biol. Conserv. 1995, 72, 159–170. [Google Scholar] [CrossRef]
- Chiesa, S.; Filonzi, L.; Vaghi, M.; Papa, R.; Marzano, F.N. Molecular Barcoding of an Atypical Cyprinid Population Assessed by Cytochrome b Gene Sequencing. Zoolog. Sci. 2013, 30, 408–413. [Google Scholar] [CrossRef] [PubMed]
- Celletti, P.; Giovinazzo, C.; Molinaro, A.; Biddittu, I.; Zarattini, A. A Middle Pleistocene Deposit with Elephas Antiquus Remains near Colleferro (Roma). In The World of Elephants. Proceedings of the First International Congress. Consiglio Nazionale delle Ricerche; The World of Elephants—International Congres: Roma, Italy, 2001; pp. 34–37. [Google Scholar]
- Bersani, P.; Corda, L. New Data on the Tuscolano-Artemisio Phase of the Alban Hills: Some Insights on Climatic Conditions. J. Mediterr. Earth Sci. 2011, 3, 25–32. [Google Scholar] [CrossRef]
- Aiello, G.; Marsella, E.; Sacchi, M. Quaternary Structural Evolution of Terracina and Gaeta Basins (Eastern Tyrrhenian Margin, Italy). Rend. Lincei 2000, 11, 41–58. [Google Scholar] [CrossRef]
- Bellucci, D.; Novaga, R.; Freyhof, J. New Data on the Distribution of the Volturno Spined Loach Cobitis zanandreai (Teleostei: Cobitidae). J. Appl. Ichthyol. 2021, 37, 885–892. [Google Scholar] [CrossRef]
- Carosi, A.; Ghetti, L.; Forconi, A.; Lorenzoni, M. Fish Community of the River Tiber Basin (Umbria-Italy): Temporal Changes and Possible Threats to Native Biodiversity. Knowl. Manag. Aquat. Ecosyst. 2015, 416, 22. [Google Scholar] [CrossRef] [Green Version]
- Allendorf, F.W.; Luikart, G.H.; Aitken, S.N. Conservation and the Genetics of Populations; Wiley-Blackwell: Oxford, UK, 2013. [Google Scholar]
- Ward, D.R.; Zemlak, T.S.; Innes, B.H.; Last, P.R.; Hebert, P.D.N. DNA barcoding Australia’s fish species. Philos. Trans. R. Soc. B 2005, 360, 1847–1857. [Google Scholar] [CrossRef]
- Gilles, A.; Lecointre, G.; Miquelis, A.; Loerstcher, M.; Chappaz, R.; Brun, G. Partial Combination Applied to Phylogeny of European Cyprinids Using the Mitochondrial Control Region. Mol. Phylogenet. Evol. 2001, 19, 22–33. [Google Scholar] [CrossRef] [Green Version]
- Chow, S.; Hazama, K. Universal primer for S7 ribosomal protein gene intron in fish. Mol. Ecol. 1998, 7, 1255–1256. [Google Scholar]




| CR | COI | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Drainage Basin | River/Lake | Pop ID | Lat (°N) | Lon (°E) | Protected Area | N | Hp (% Private) | Hp Rich | Hd (±s.e.) | π % (±s.e.) | N | Hp |
| Italy | ||||||||||||
| Magra-Vara | Riccò | VAR1 | 44.161 | 9.758 | 13 | 3 (0) | 3.00 | 0.51 (±0.14) | 1.51 (±0.81) | 11 | 4 | |
| Graveglia | VAR2 | 44.191 | 9.790 | 20 | 4 (0) | 3.54 | 0.63 (±0.08) | 1.65 (±0.86) | 8 | 4 | ||
| Tronto | Tronto | TRO | 42.802 | 13.465 | SAC | 18 | 2 (0) | 1.72 | 0.11 (±0.10) | 0.01 (±0.02) | 3 | 2 |
| Foro | Foro | FOR | 42.246 | 14.186 | 16 | 2 (50) | 1.98 | 0.23 (±0.13) | 0.03 (±0.03) | 3 | 2 | |
| Arrone | Arrone | ARR | 41.914 | 12.265 | 15 | 4 (50) | 3.73 | 0.60 (±0.11) | 0.09 (±0.08) | 3 | 2 | |
| Tiber | Rio Martino | TIB1 | 42.173 | 12.545 | 33 | 4 (50) | 3.35 | 0.63 (±0.06) | 0.08 (±0.07) | 2 | 2 | |
| Fosso Passerano | TIB2 | 41.932 | 12.732 | 4 | 3 (0) | NA | 0.83 (±0.22) | 0.11 (±0.11) | 2 | 2 | ||
| Fondi | Settecannelle | SET | 41.368 | 13.421 | Regional Park | 20 | 5 (60) | 3.84 | 0.44 (±0.13) | 0.05 (±0.05) | 3 | 2 |
| San Puoto | San Puoto | SPU | 41.285 | 13.408 | SPA | 7 | 2 (0) | NA | 0.57 (±0.12) | 0.06 (±0.06) | 7 | 1 |
| Santa Croce | Santa Croce | SCR | 41.287 | 13.716 | SAC | 32 | 5 (40) | 3.47 | 0.60 (±0.07) | 0.21 (±0.13) | 3 | 2 |
| Liri-Garigliano | Ausentello | GAR | 41.303 | 13.743 | 24 | 3 (0) | 2.46 | 0.30 (±0.11) | 0.04 (±0.04) | 3 | 1 | |
| Noce | Pamafi | NOC | 39.934 | 15.752 | 6 | 2 (50) | NA | 0.33 (±0.22) | 0.58 (±0.38) | 6 | 2 | |
| Total | 208 | 21 | 54 | 14 | ||||||||
| Albania | ||||||||||||
| Skadar | Skadar | SKA | 42.059 | 19.455 | 8 | 8 (100) | NA | 1.00 (±0.06) | 0.39 (±0.25) | 8 | 4 | |
| Ohrid | Ohrid | OHR | 40.963 | 20.640 | 7 | 4 (100) | NA | 0.71 (±0.18) | 0.12 (±0.1) | 7 | 3 | |
| Total | 15 | 12 | 15 | 7 | ||||||||
| Haplogroup | N | Hp | Hd (±s.e.) | π % (±s.e.) | Tajima’s D | Fu’s F | R2 | SSD | Hri | τ | θ0 | θ1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 167 | 12 | 0.56 (±0.04) | 0.09 (±0.07) | −0.919 | −6.561 * | 0.052 | 0.001 | 0.056 | 0.926 | 0.100 | 3.196 |
| B | 29 | 7 | 0.55 (±0.10) | 0.13 (±0.09) | −1.722 * | −2.237 | 0.062 ** | 0.014 | 0.119 | 0.725 | 0.000 | 99,999.000 |
| C | 12 | 2 | 0.48 (±0.11) | 0.05 (±0.05) | 1.066 | 1.003 | 0.242 | 0.019 | 0.236 | 0.734 | 0.000 | 99,999.000 |
| Tot. | 208 | 21 | 0.71 (±0.03) | 0.75 (±0.39) | −0.107 | 1.541 | 0.081 | 0.035 | 0.040 | 0.072 | 1.816 | 99,999.000 |
| Among Groups | Among Populations within Groups | Within Populations | ||||
|---|---|---|---|---|---|---|
| N. of Groups and Group Composition | %var | ΦCT | %var | ΦSC | %var | ΦST |
| (1) no structure: 1 group | -- | -- | 57.68 | -- | 42.32 | 0.57682 *** |
| (2) ichthyogeographic districts: 3 groups (PV = TRO; TL = VAR1, VAR2, ARR, TIB1, TIB2; AC = FOR, SET, SPU, SCR, GAR, NOC) | −9.87 | −0.09873 | 66.04 | 0.60108 *** | 43.83 | 0.56169 *** |
| (3) Haplogroups: 3 groups (HpC = VAR1_C, VAR2_C; HpA = VAR1_A, VAR2_A, TRO, FOR, ARR, TIB1, TIB2, SCR_A, GAR, NOC_A; HpB = SET, SPU, SCR_B, NOC_B) | 95.06 | 0.951 *** | 2.07 | 0.418 *** | 2.88 | 0.971 *** |
| (4) NMDS, subgroups within haplogroups: 7 groups (VAR1_C, VAR2_C; VAR1_A, VAR2_A, TRO, FOR, TIB1, TIB2, SCR_A, GAR; ARR; NOC_A; SET, SPU; SCR_B; NOC_B) | 95.18 | 0.952 *** | 0.87 | 0.181 *** | 3.95 | 0.961 *** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrosino, G.; Tancioni, L.; Turani, M.; Rakaj, A.; Ciuffardi, L.; Rossi, A.R. Phylogeography of Sarmarutilus rubilio (Cypriniformes: Leuciscidae): Complex Genetic Structure, Clues to a New Cryptic Species and Further Insights into Roaches Phylogeny. Genes 2022, 13, 1071. https://doi.org/10.3390/genes13061071
Petrosino G, Tancioni L, Turani M, Rakaj A, Ciuffardi L, Rossi AR. Phylogeography of Sarmarutilus rubilio (Cypriniformes: Leuciscidae): Complex Genetic Structure, Clues to a New Cryptic Species and Further Insights into Roaches Phylogeny. Genes. 2022; 13(6):1071. https://doi.org/10.3390/genes13061071
Chicago/Turabian StylePetrosino, Gerardo, Lorenzo Tancioni, Martina Turani, Arnold Rakaj, Luca Ciuffardi, and Anna Rita Rossi. 2022. "Phylogeography of Sarmarutilus rubilio (Cypriniformes: Leuciscidae): Complex Genetic Structure, Clues to a New Cryptic Species and Further Insights into Roaches Phylogeny" Genes 13, no. 6: 1071. https://doi.org/10.3390/genes13061071







