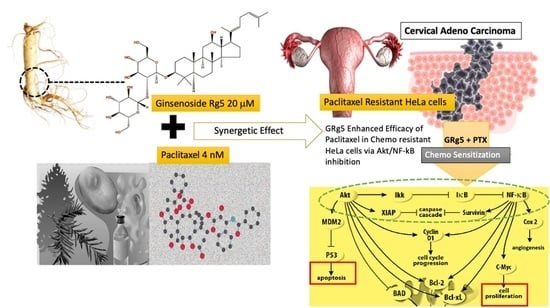
Ginsenoside Rg5 Sensitizes Paclitaxel—Resistant Human Cervical-Adeno-Carcinoma Cells to Paclitaxel—And Enhances the Anticancer Effect of Paclitaxel
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Cell Culture and Treatment
2.3. MTT Assay
2.4. Colony Formation Assay (CFA)
2.5. DAPI Staining Assay
2.6. Annexin V-FITC Staining for Apoptosis Assay Using Flow Cytometry
2.7. Determination of Gene Expression by the RT-PCR
2.8. Western Blot Analysis
2.9. Statistical Analysis
3. Results
3.1. Efficacy of GRg5 Combined Paclitaxel Treatment on Paclitaxel Chemo Resistant Cervical Cancer Cells (HeLa PTX—R Cells)
3.1.1. Cytotoxic Effect of GRg5, PTX, and Combination of GRg5 and PTX in HeLa-PTX-R Cells
3.1.2. Effect of GRg5 in PTX Induced Apoptosis in HeLa-PTX-R Cervical Cancer Cells
3.1.3. Effect of GRg5 on mRNA Expression of Apoptotic Regulatory Genes in PTX Treated HeLa-PTX-R Cell Lines
3.1.4. Effect of GRg5 on Apoptotic Regulatory Proteins in PTX Treated HeLa-PTX-R Cell Lines
3.1.5. Effect of GRg5 on Cell Cycle Regulatory Proteins in PTX Treated HeLa-PTX-R Cell Lines
3.1.6. Effect of GRg5-PTX Combination Treatment on Akt/IKK-α/NF-κB Signaling Pathways
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Konoshima, T.; Takasaki, M.; Tokuda, H. Anti-carcinogenic activity of the roots of Panax notoginseng II. Biol. Pharm. Bull. 1999, 22, 1150–1152. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liu, Q.Z.; Xing, S.P.; Zhang, J.L. Inhibiting effect of Endostar combined with ginsenoside Rg3 on breast cancer tumor growth in tumor-bearing mice. Asian Pac. J. Trop. Med. 2016, 9, 180–183. [Google Scholar] [CrossRef] [Green Version]
- Seo, Y.S.; Shon, M.Y.; Kong, R.; Kang, O.H.; Zhou, T.; Kim, D.Y.; Kwon, D.Y. Black ginseng extract exerts anti-hyperglycemic effect via modulation of glucose metabolism in liver and muscle. J. Ethnopharmacol. 2016, 190, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wang, A.; Feng, J.; Zhang, Q.; Liu, L.; Ren, H. Ginsenoside GRg5 induces apoptosis in human esophageal cancer cells through the phosphoinositide3 kinase/protein kinase B signalling pathway. Mol. Med. Rep. 2019, 19, 4019–4026. [Google Scholar] [PubMed]
- Kim, S.J.; Kim, A.K. Anti-breast cancer activity of Fine Black ginseng (Panax ginseng Meyer) and ginsenoside GRg5. J. Ginseng Res. 2015, 39, 125–134. [Google Scholar] [CrossRef] [Green Version]
- Cho, Y.L.; Hur, S.M.; Kim, J.Y.; Kim, J.H.; Lee, D.K.; Choe, J.; Won, M.H.; Ha, K.S.; Jeoung, D.; Han, S.; et al. Specific activation of insulin-like growth factor-1 receptor by ginsenoside GRg5 promotes angiogenesis and vasorelaxation. J. Biol. Chem. 2015, 290, 467–477. [Google Scholar] [CrossRef] [Green Version]
- Hyung, J.K.; Tae, K.O.; Yoon, H.K.; Jaesun, L.; Joo, M.M.; Yong, S.P.; Chang, M.S. Pharmacokinetics of Ginsenoside Rb1, Rg3, Rk1, GRg5, F2, and Compound K from Red Ginseng Extract in Healthy Korean Volunteers. Evid.-Based Complementary Altern. Med. 2022, 202, 8427519. [Google Scholar]
- Nag, S.A.; Qin, J.J.; Wang, W.; Wang, M.H.; Wang, H.; Zhang, R. Ginsenosides as Anticancer Agents, In vitro and in vivo Activities, Structure-Activity Relationships, and Molecular Mechanisms of Action. Front. Pharmacol. 2012, 3, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, S.L.; Luo, H.B.; Cai, L.; Zhang, J.; Wang, D.; Chen, Y.J.; Zhan, H.X.; Jiang, Z.H.; Xie, Y. Ginsenoside GRg5 overcomes chemotherapeutic multidrug resistance mediated by ABCB1 transporter, in vitro and in vivo study. J. Ginseng Res. 2020, 44, 247–257. [Google Scholar] [CrossRef]
- Liu, Y.; Fan, D. Ginsenoside GRg5 induces apoptosis and autophagy via the inhibition of the PI3K/Akt pathway against breast cancer in a mouse model. Food Funct. 2018, 9, 5513–5527. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.D.; He, T.; Du, T.W.; Fan, Y.G.; Chen, D.S.; Wang, Y. GinsenosideGRg5 induces apoptosis and DNA damage in human cervical cancer cells. Mol. Med. Rep. 2015, 11, 940–946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christensen, L.P. Ginsenosides chemistry, biosynthesis, analysis, and potential health effects. Adv. Nutr. Res. 2009, 55, 1–99. [Google Scholar]
- Walboomers, J.M.; Jacobs, M.V.; Manos, M.M.; Bosch, F.X.; Kummer, J.A.; Shah, K.V.; Snijders, P.J.; Peto, J.; Meijer, C.J.; Muñoz, N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999, 189, 12–19. [Google Scholar] [CrossRef]
- Sreedevi, A.; Javed, R.; Dinesh, A. Epidemiology of cervical cancer with special focus on India. Int. J. Women’s Health 2015, 7, 405–414. [Google Scholar]
- Shen, Y.; Yang, L.; Wang, Z. Treatment of early bulky cervical cancer with neoadjuvant paclitaxel, carboplatin and cisplatin prior to laparoscopical radical hysterectomy and pelvic lymphadenectomy. Oncol. Lett. 2012, 3, 641–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schnell, F.M. Chemotherapy-induced nausea and vomiting, the importance of acute antiemetic control. Oncologist 2003, 8, 187–198. [Google Scholar] [CrossRef]
- Schwab, C.L.; English, D.P.; Roque, D.M.; Santin, A.D. Taxanes, their impact on gynecologic malignancy. Anticancer. Drugs 2014, 25, 522–535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.H.; Hu, S.L.; Shen, G.D.; Shen, G. Tumor suppressor genes and their underlying interactions in paclitaxel resistance in cancer therapy. Cancer Cell Int. 2016, 16, 13. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.; Cang, S.; Ma, Y.; Chiao, J.W. Synergistic effect of paclitaxel and epigenetic agent phenethyl isothiocyanate on growth inhibition, cell cycle arrest and apoptosis in breast cancer cells. Cancer Cell Int. 2013, 13, 10. [Google Scholar] [CrossRef] [Green Version]
- Yusuf, R.Z.; Duan, Z.; Lamendola, D.E.; Penson, R.T.; Seiden, M.V. Paclitaxel resistance, molecular mechanisms and pharmacologic manipulation. Curr. Cancer Drug Targets 2013, 3, 1–19. [Google Scholar] [CrossRef]
- Engelman, J.A. Targeting PI3K signalling in cancer, opportunities, challenges and limitations. Nat. Rev. Cancer 2009, 9, 550–562. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wu, J.; Ling, M.T.; Zhao, L.; Zhao, K.N. The role of the PI3K/Akt/mTOR signalling pathway in human cancers induced by infection with human papillomaviruses. Mol. Cancer 2015, 14, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, H.; Lin, L.; Zhang, Z.; Zhang, H.; Hu, H. Targeting NF-κB pathway for the therapy of diseases, mechanism and clinical study. Signal Transduct. Target. Ther. 2020, 5, 209. [Google Scholar] [CrossRef] [PubMed]
- Hayden, M.S.; Ghosh, S. Shared principles in NF-kappaB signaling. Cell 2008, 132, 344–362. [Google Scholar] [CrossRef] [Green Version]
- Israel, A. The IKK complex, a central regulator of NF-κB activation. Cold Spring Harb. Perspect. Biol. 2010, 2, a000158. [Google Scholar] [CrossRef] [Green Version]
- Rajendran, P.; Ben, A.R.; Al-Saeedi, F.J.; Elsayed, M.M.; Islam, M.; Al-Ramadan, S.Y. Thidiazuron decreases epithelial-mesenchymal transition activity through the NF-kB and PI3K/AKT signalling pathways in breast cancer. J. Cell. Mol. Med. 2020, 24, 14525–14538. [Google Scholar] [CrossRef]
- Sikka, S.; Shanmugam, M.K.; Siveen, K.S.; Ong, T.H.; Yang, M.H.; Lee, J.H.; Rajendran, P.; Chinnathambi, A.; Alharbi, S.A.; Alahmadi, T.A.; et al. Diosgenin attenuates tumor growth and metastasis in transgenic prostate cancer mouse model by negatively regulating both NF-κB/STAT3 signaling cascades. Eur. J. Pharmacol. 2021, 906, 174274. [Google Scholar] [CrossRef]
- He, H.; Ni, J.; Huang, J. Molecular mechanisms of chemoresistance in osteosarcoma (Review). Oncol. Lett. 2014, 7, 1352–1362. [Google Scholar] [CrossRef] [Green Version]
- Stanton, R.A.; Gernert, K.M.; Nettles, J.H.; Aneja, R. Drugs that target dynamic microtubules, a new molecular perspective. Med. Res. Rev. 2011, 31, 443–481. [Google Scholar] [CrossRef] [Green Version]
- Zhou, M.; Zhao, Y.; Ding, Y.; Liu, H.; Liu, Z.; Fodstad, O.; Riker, A.I.; Kamarajugadda, S.; Lu, J.; Owen, L.B. Warburg effect in chemosensitivity, targeting lactate dehydrogenase-A re-sensitizes taxol-resistant cancer cells to taxol. Mol. Cancer 2010, 9, 33. [Google Scholar] [CrossRef] [Green Version]
- Rose, P.G.; Blessing, J.A.; Gershenson, D.M.; McGehee, R. Paclitaxel and cisplatin as first-line therapy in recurrent or advanced squamous cell carcinoma of the cervix, a gynecologic oncology group study. J. Clin. Oncol. 1999, 17, 2676–2680. [Google Scholar] [CrossRef] [PubMed]
- Bayat, M.R.; Homayouni, T.S.; Baluch, N.; Morgatskaya, E.; Kumar, S.; Das, B.; Yeger, H. Combination therapy in combating cancer. Oncotarget 2017, 8, 38022–38043. [Google Scholar]
- Semenza, G.L.; Jiang, B.H.; Leung, S.W.; Passantino, R.; Concordet, J.P.; Maire, P.; Giallongo, A. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 1996, 271, 32529–32537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choudhari, A.S.; Mandave, P.C.; Deshpande, M.; Ranjekar, P.; Prakash, O. Phytochemicals in Cancer Treatment, From Preclinical Studies to Clinical Practice. Front. Pharmacol. 2019, 10, 1614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pratheeshkumar, P.; Sreekala, C.; Zhang, Z.; Budhraja, A.; Ding, S.; Son, Y.O.; Wang, X.; Hitron, A.; Hyun-Jung, K.; Wang, L. Cancer prevention with promising natural products, mechanisms of action and molecular targets. Anti-Cancer Agents Med. Chem. 2012, 12, 1159–1184. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Huang, J.; Yu, C.; Xiang, L.; Li, L.; Shi, D.; Lin, F. Quercetin Enhanced Paclitaxel Therapeutic Effects Towards PC-3 Prostate Cancer Through ER Stress Induction and ROS Production. OncoTargets Ther. 2020, 13, 513–523. [Google Scholar] [CrossRef] [Green Version]
- Georgaki, S.; Skopeliti, M.; Tsiatas, M.; Nicolaou, K.A.; Ioannou, K.; Husband, A.; Bamias, A.; Dimopoulos, M.A.; Constantinou, A.I.; Tsitsilonis, O.E. Phenoxodiol, an anticancer isoflavene, induces immunomodulatory effects in vitro and in vivo. J. Cell. Mol. Med. 2009, 13, 3929–3938. [Google Scholar] [CrossRef] [Green Version]
- Xu, T.M.; Xin, Y.; Cui, M.H.; Jiang, X.; Gu, L.P. Inhibitory effect of ginsenoside Rg3 combined with cyclophosphamide on growth and angiogenesis of ovarian cancer. Chin. Med. J. 2007, 120, 584–588. [Google Scholar] [CrossRef]
- Noh, S.; Choi, E.; Hwang, C.H.; Jung, J.H.; Kim, S.H.; Kim, B. Dietary Compounds for Targeting Prostate Cancer. Nutrients 2019, 11, 2401. [Google Scholar] [CrossRef] [Green Version]
- Lowe, S.W.; Lin, A.W. Apoptosis in cancer. Carcinogenesis 2000, 21, 485–495. [Google Scholar] [CrossRef] [Green Version]
- Cheng, E.H.; Kirsch, D.G.; Clem, R.J.; Ravi, R.; Kastan, M.B.; Bedi, A.; Ueno, K.; Hardwick, J.M. Conversion of Bcl-2 to a Bax-like death effector by caspases. Science 1997, 278, 1966–1968. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.H.; Reynolds, C.P. Bcl-2 inhibitors, targeting mitochondrial apoptotic pathways in cancer therapy. Clin. Cancer Res. 2009, 15, 1126–1132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chipuk, J.E.; Green, D.R. How do BCL-2 proteins induce mitochondrial outer membrane permeabilization. Trends Cell Biol. 2008, 18, 157–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, G.; O’Rourke, K.; Dixit, V.M. Caspase-9, Bcl-XL, and Apaf-1 form a ternary complex. J. Biol. Chem. 1998, 273, 5841–5845. [Google Scholar] [CrossRef] [Green Version]
- Whitfield, M.L.; Sherlock, G.; Saldanha, A.J.; Murray, J.I.; Ball, C.A.; Alexander, K.E.; Matese, J.C.; Perou, C.M.; Hurt, M.M.; Brown, P.O.; et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell 2002, 13, 1977–2000. [Google Scholar] [CrossRef] [Green Version]
- Tan, M.L.; Ooi, J.P.; Ismail, N.; Moad, A.I.; Muhammad, T.S. Programmed cell death pathways and current antitumor targets. Pharm. Res. 2009, 26, 1547–1560. [Google Scholar] [CrossRef]
- Yoshinouchi, M.; Hongo, A.; Takamoto, N.; Ono, Y.; Nagao, S.; Miyagi, Y.; Kudo, T.; Kodama, J. Alteration of the CDKN2/p16 gene is not required for HPV-positive uterine cervical cancer cell lines. Int. J. Oncol. 2000, 16, 537–541. [Google Scholar] [CrossRef]
- Dirks, P.B.; Rutka, J.T. Current concepts in neuro-oncology, the cell cycle—A review. Neurosurgery 1997, 40, 1000–1015. [Google Scholar] [CrossRef]
- Clarke, B.; Chetty, R. Cell cycle aberrations in the pathogenesis of squamous cell carcinoma of the uterine cervix. Gynecol. Oncol. 2001, 82, 238–246. [Google Scholar] [CrossRef]
- Cho, N.H.; Lim, S.Y.; Kim, Y.T.; Kim, D.; Kim, Y.S.; Kim, J.W. G2 checkpoint in uterine cervical cancer with HPV 16 E6 according to p53 polymorphism and its screening value. Gynecol. Oncol. 2003, 90, 15–22. [Google Scholar] [CrossRef]
- Xu, W.; Yang, Z.; Lu, N. A new role for the PI3K/Akt signaling pathway in the epithelial-mesenchymal transition. Cell Adh. Migr. 2015, 9, 317–324. [Google Scholar] [CrossRef] [Green Version]
- Plati, J.; Bucur, O.; Khosravi-Far, R. Dysregulation of apoptotic signaling in cancer, molecular mechanisms and therapeutic opportunities. J. Cell. Biochem. 2008, 104, 1124–1149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riggio, M.; Perrone, M.C.; Polo, M.L.; Rodriguez, M.J.; May, M.; Abba, M.; Lanari, C.; Novaro, V. AKT1 and AKT2 isoforms play distinct roles during breast cancer progression through the regulation of specific downstream proteins. Sci. Rep. 2017, 7, 44244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinod, B.S.; Nair, H.H.; Vijayakurup, V.; Shabna, A.; Shah, S.; Krishna, A.; Pillai, K.S.; Thankachan, S.; Anto, R.J. Resveratrol chemosensitizes HER-2-overexpressing breast cancer cells to docetaxel chemoresistance by inhibiting docetaxel-mediated activation of HER-2-Akt axis. Cell Death Discov. 2015, 1, 1506. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.J.; Chen, G.; Hu, X.; Zhang, W.; Liu, Y.; Zhu, L.X.; Zhou, Q.; Zhao, Y.F. Activation of PI3K/Akt/IKK-α/NF-κB signaling pathway is required for the apoptosis-evasion in human salivary adenoid cystic carcinoma, its inhibition by quercetin. Apoptosis 2010, 15, 850–863. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.; Edwards, J.; Pepper, C.; Mackay, S. Inhibitory-κB Kinase (IKK) α and Nuclear Factor-κB (NFκB)-Inducing Kinase (NIK) as Anti-Cancer Drug Targets. Cells 2018, 7, 176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.J.; Ho, J.Y.; Lee, H.W.; Baik, M.W.; Kim, O.; Choi, Y.J.; Hur, S.Y. Inhibition of Phosphatidylinositol 3-kinase (PI3K) Signaling Synergistically Potentiates Antitumor Efficacy of Paclitaxel and Overcomes Paclitaxel-Mediated Resistance in Cervical Cancer. Int. J. Mol. Sci. 2019, 20, 3383. [Google Scholar] [CrossRef] [Green Version]











| Gene | Forward Sequence | Reverse Sequence |
|---|---|---|
| BCL-2 | 5′ GATGTGATGCCTCTGCGAAG 3′ | 5′ CATGCTGATGTCTCTGGAATCT 3′ |
| BCL-xL | 5′AACATCCCAGCTTCACATAACCCC 3′ | 5′ GCGACCCCAGTTTACTCCATCC 3′ |
| c-IAP-1 | 5′ GTTTTAAAACCAGCTTGGGTTATATTG 3′ | 5′ GTTCCTCACCCAACCAGTCTACTTAG 3′ |
| MCL-1 | 5′ CCAAGAAAGCTGCATCGAACCAT 3′ | 5′ CAGCACATTCCTGATGCCACCT 3′ |
| BAX | 5′ GGTTGTCGCCCTTTTCTA 3′ | 5′ CGGAGGAAGTCCAATGTC 3′ |
| BID | 5′ AAATAGTTTGGGGATTTTGAAT 3′ | 5′ AATACACTCACCACCCTCC 3′ |
| BAK | 5′ GGCAGGGTATGGTATGGTTG 3′ | 5′ TCCCGACTGCCTGGTTACTG 3′ |
| PUMA | 5′ AGTACATCCTCTGGGCTCTGC3′ | 5′ CGGACAAGTCAGGACTTGCAGG 3′ |
| GAPDH | 5′ GCGAGAAGATGACCCAGAT 3′ | 5′ GAGGCGTACAGGGATAGC 3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramesh, J.; Thilakan, R.C.; Gopalakrishnan, R.M.; Vijayapoopathi, S.; Dorschel, A.; Venugopal, B. Ginsenoside Rg5 Sensitizes Paclitaxel—Resistant Human Cervical-Adeno-Carcinoma Cells to Paclitaxel—And Enhances the Anticancer Effect of Paclitaxel. Genes 2022, 13, 1142. https://doi.org/10.3390/genes13071142
Ramesh J, Thilakan RC, Gopalakrishnan RM, Vijayapoopathi S, Dorschel A, Venugopal B. Ginsenoside Rg5 Sensitizes Paclitaxel—Resistant Human Cervical-Adeno-Carcinoma Cells to Paclitaxel—And Enhances the Anticancer Effect of Paclitaxel. Genes. 2022; 13(7):1142. https://doi.org/10.3390/genes13071142
Chicago/Turabian StyleRamesh, Janani, Rejani Chalikkaran Thilakan, Raja Mohan Gopalakrishnan, Singaravel Vijayapoopathi, Arianna Dorschel, and Bhuvarahamurthy Venugopal. 2022. "Ginsenoside Rg5 Sensitizes Paclitaxel—Resistant Human Cervical-Adeno-Carcinoma Cells to Paclitaxel—And Enhances the Anticancer Effect of Paclitaxel" Genes 13, no. 7: 1142. https://doi.org/10.3390/genes13071142