Identification of Recurrent Chromosome Breaks Underlying Structural Rearrangements in Mammary Cancer Cell Lines
Abstract
:1. Introduction
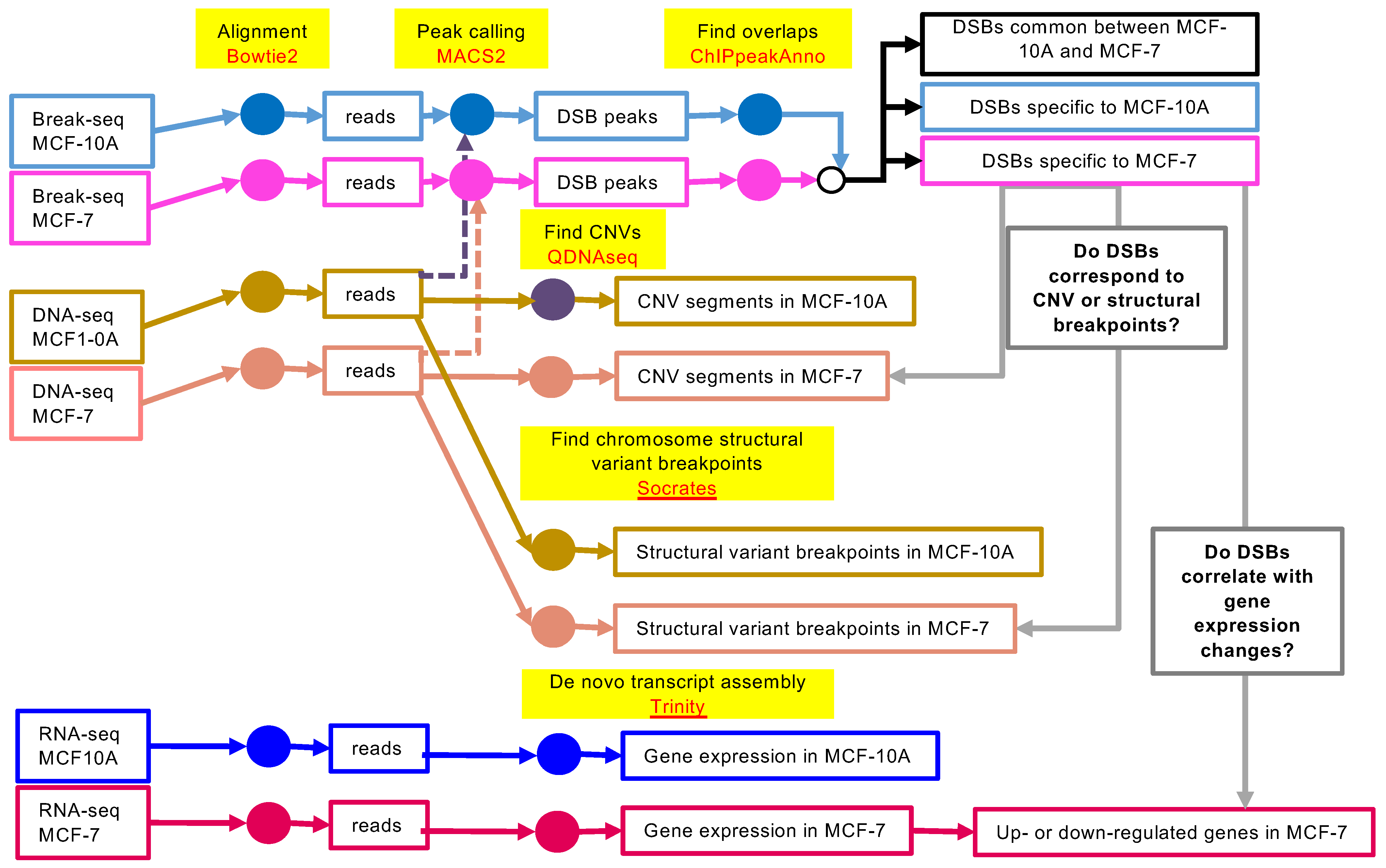
2. Materials and Methods
2.1. Cell Lines and Growth Conditions
2.2. Break-Seq
2.3. DNA-Seq
2.4. RNA-Seq
2.5. Survival Prediction Analysis
2.6. Gene Ontology
3. Results
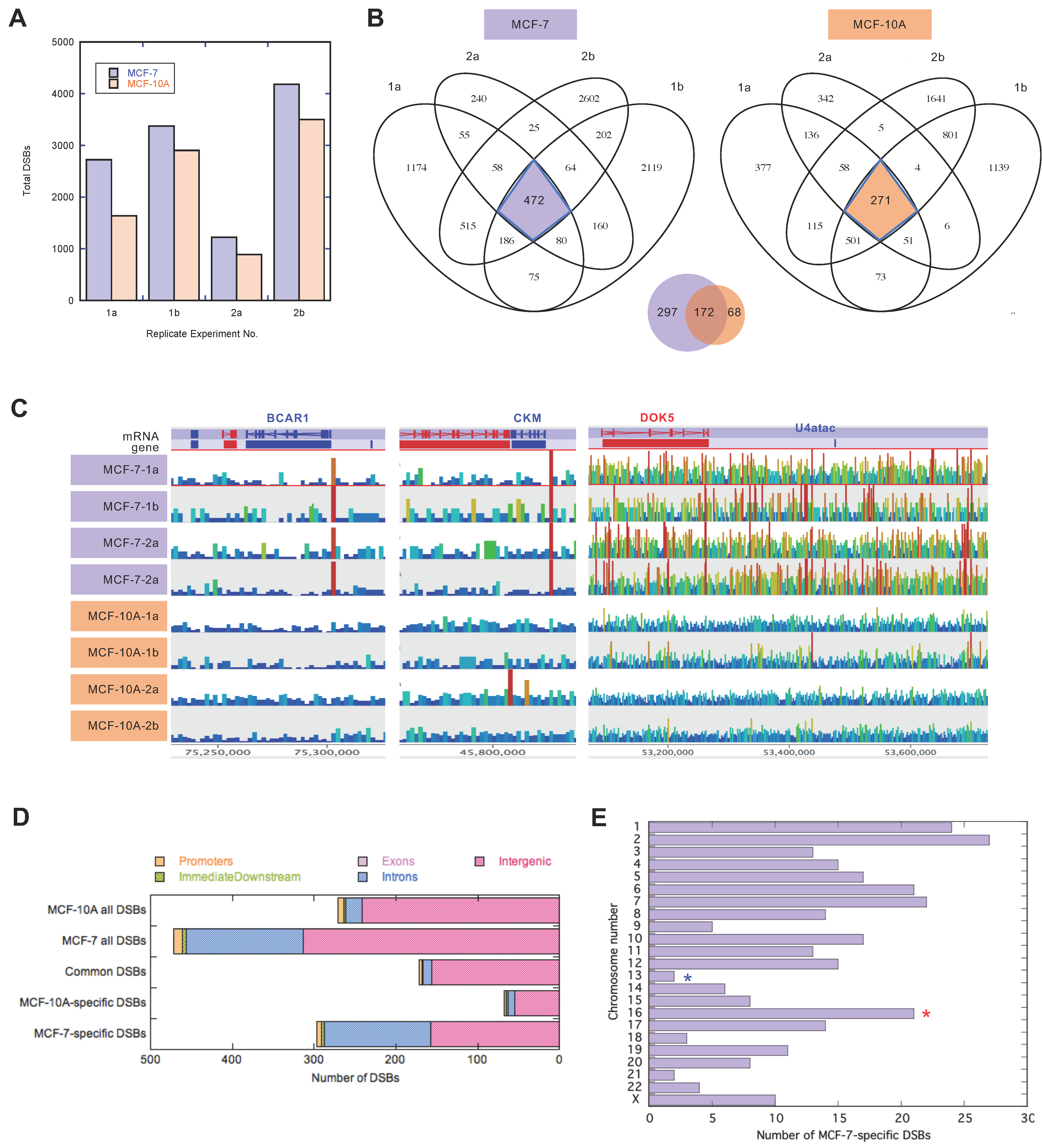
3.1. High Level of Gene-Associated Spontaneous Chromosome Breakage in the MCF-7 Cell Line
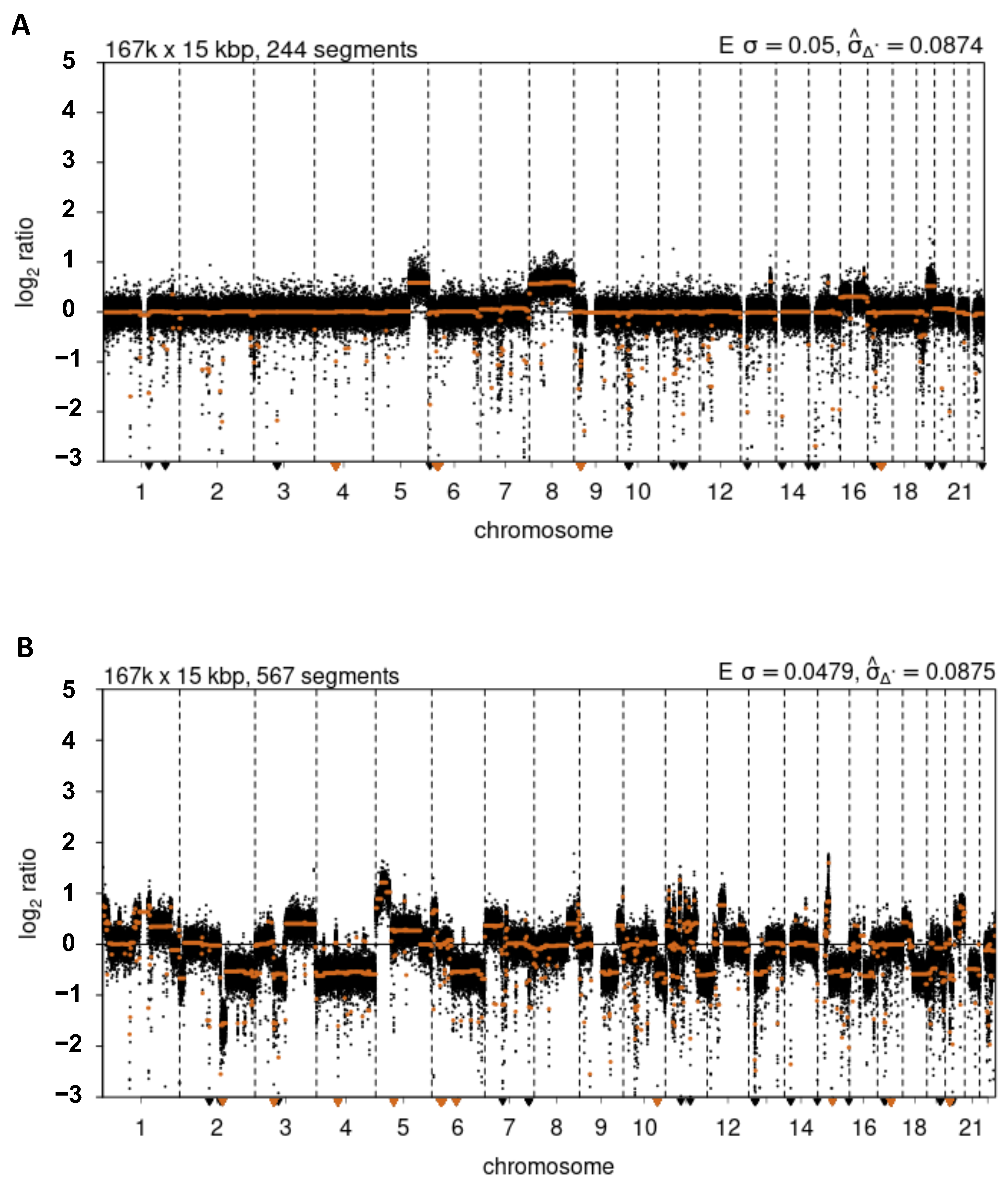
3.2. Concurrent Cancer-Specific Spontaneous DSBs and Structural Variation Breakpoints on the Pericentromere of 16q
3.3. Genes Immediately Downstream from the Pericentromere of 16q Showed High Expression in MCF-7 Cells
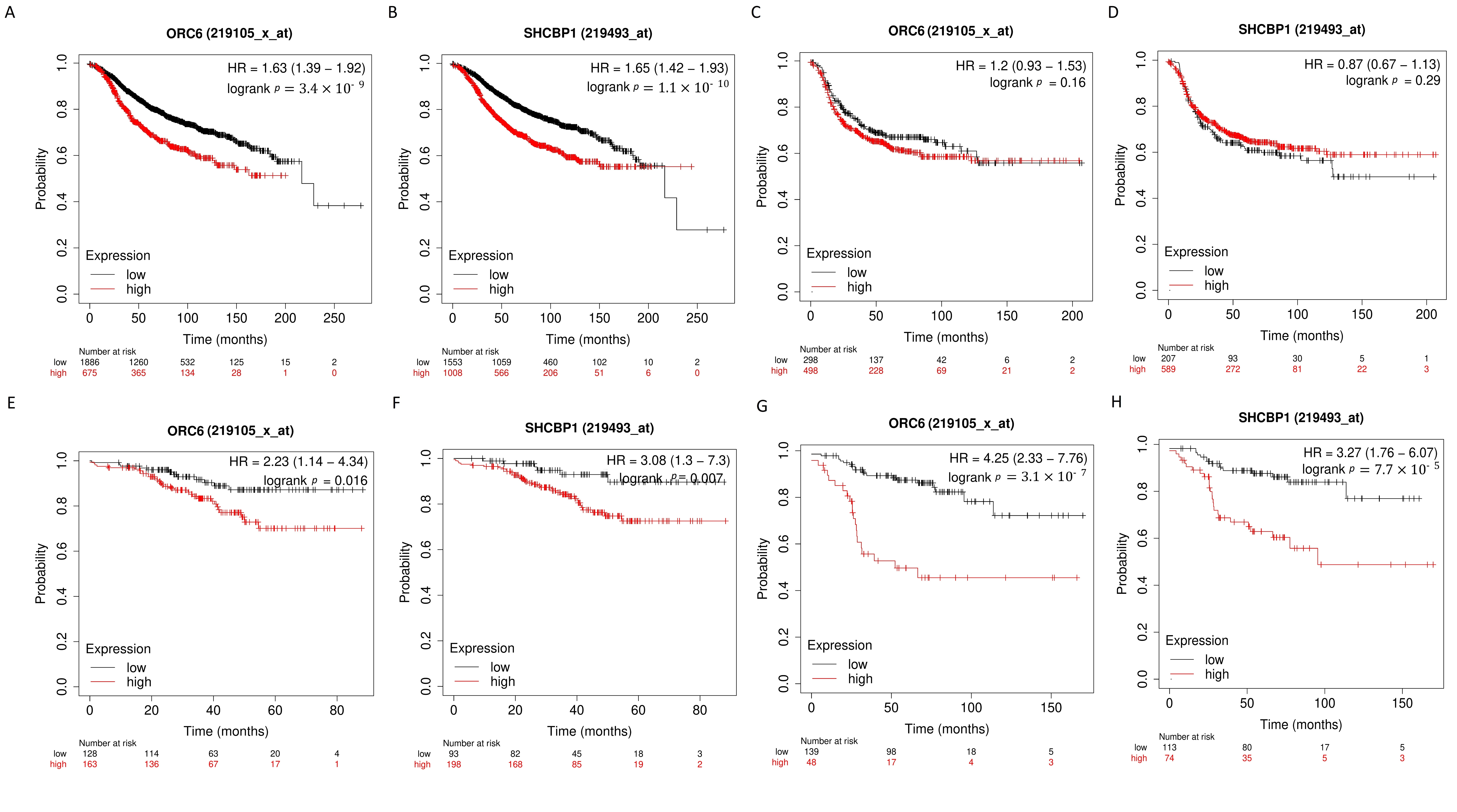
3.4. SHCBP1 and ORC6 Are Effective Predictive and Poor Prognosis Markers for (ER)-Positive Breast Cancer Patients
3.5. Genes Associated with MCF-7-Specific DSBs Were Enriched in Biological Pathways including the ER Signaling Pathway
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lengauer, C.; Kinzler, K.W.; Vogelstein, B. Genetic instabilities in human cancers. Nature 1998, 396, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Pino, M.S.; Chung, D.C. The chromosomal instability pathway in colon cancer. Gastroenterology 2010, 138, 2059–2072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, D.S.; Wersto, R.P.; Zhou, W.; Chrest, F.J.; Garrett, E.S.; Kwon, T.K.; Gabrielson, E. Variable levels of chromosomal instability and mitotic spindle checkpoint defects in breast cancer. Am. J. Pathol. 2002, 161, 391–397. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, A.; Jenjaroenpun, P.; Li, J.; El Hilali, S.; McCulley, A.; Haarer, B.; Hoffman, E.A.; Belak, A.; Thorland, A.; Hehnly, H.; et al. Replication Stress Induces Global Chromosome Breakage in the Fragile X Genome. Cell Rep. 2020, 32, 108179. [Google Scholar] [CrossRef] [PubMed]
- Joshi, I.; DeRycke, J.; Palmowski, M.; LeSuer, R.; Feng, W. Genome-wide mapping of DNA double-strand breaks from eukaryotic cell cultures using Break-seq. STAR Protoc. 2021, 2, 100554. [Google Scholar] [CrossRef] [PubMed]
- Prat, A.; Karginova, O.; Parker, J.S.; Fan, C.; He, X.; Bixby, L.; Harrell, J.C.; Roman, E.; Adamo, B.; Troester, M.; et al. Characterization of cell lines derived from breast cancers and normal mammary tissues for the study of the intrinsic molecular subtypes. Breast Cancer Res. Treat. 2013, 142, 237–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hampton, O.A.; Den Hollander, P.; Miller, C.A.; Delgado, D.A.; Li, J.; Coarfa, C.; Harris, R.A.; Richards, S.; Scherer, S.E.; Muzny, D.M.; et al. A sequence-level map of chromosomal breakpoints in the MCF-7 breast cancer cell line yields insights into the evolution of a cancer genome. Genome Res. 2009, 19, 167–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kytölä, S.; Rummukainen, J.; Nordgren, A.; Karhu, R.; Farnebo, F.; Isola, J.; Larsson, C. Chromosomal alterations in 15 breast cancer cell lines by comparative genomic hybridization and spectral karyotyping. Genes Chromosomes Cancer 2000, 28, 308–317. [Google Scholar] [CrossRef]
- Volik, S.; Zhao, S.; Chin, K.; Brebner, J.H.; Herndon, D.R.; Tao, Q.; Kowbel, D.; Huang, G.; Lapuk, A.; Kuo, W.L.; et al. End-sequence profiling: Sequence-based analysis of aberrant genomes. Proc. Natl. Acad. Sci. USA 2003, 100, 7696–7701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dutrillaux, B.; Gerbault-Seureau, M.; Zafrani, B. Characterization of chromosomal anomalies in human breast cancer. A comparison of 30 paradiploid cases with few chromosome changes. Cancer Genet. Cytogenet. 1990, 49, 203–217. [Google Scholar] [CrossRef]
- Dutrillaux, B.; Gerbault-Seureau, M.; Remvikos, Y.; Zafrani, B.; Prieur, M. Breast cancer genetic evolution: I. Data from cytogenetics and DNA content. Breast Cancer Res. Treat. 1991, 19, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Kokalj-Vokac, N.; Alemeida, A.; Gerbault-Seureau, M.; Malfoy, B.; Dutrillaux, B. Two-color FISH characterization of i(1q) and der(1;16) in human breast cancer cells. Genes Chromosomes Cancer 1993, 7, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Pandis, N.; Heim, S.; Bardi, G.; Limon, J.; Mandahl, N.; Mitelman, F. Improved technique for short-term culture and cytogenetic analysis of human breast cancer. Genes Chromosomes Cancer 1992, 5, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Pandis, N.; Jin, Y.; Gorunova, L.; Petersson, C.; Bardi, G.; Idvall, I.; Johansson, B.; Ingvar, C.; Mandahl, N.; Mitelman, F.; et al. Chromosome analysis of 97 primary breast carcinomas: Identification of eight karyotypic subgroups. Genes Chromosomes Cancer 1995, 12, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, H.; Takarabe, T.; Susumu, N.; Inazawa, J.; Okada, S.; Hirohashi, S. Detection of numerical and structural alterations and fusion of chromosomes 16 and 1 in low-grade papillary breast carcinoma by fluorescence in situ hybridization. Am. J. Pathol. 1997, 151, 1027–1034. [Google Scholar]
- Aswad, L.; Yenamandra, S.P.; Ow, G.S.; Grinchuk, O.; Ivshina, A.V.; Kuznetsov, V.A. Genome and transcriptome delineation of two major oncogenic pathways governing invasive ductal breast cancer development. Oncotarget 2015, 6, 36652–36674. [Google Scholar] [CrossRef] [Green Version]
- Scheinin, I.; Sie, D.; Bengtsson, H.; van de Wiel, M.A.; Olshen, A.B.; van Thuijl, H.F.; van Essen, H.F.; Eijk, P.P.; Rustenburg, F.; Meijer, G.A.; et al. DNA copy number analysis of fresh and formalin-fixed specimens by shallow whole-genome sequencing with identification and exclusion of problematic regions in the genome assembly. Genome Res. 2014, 24, 2022–2032. [Google Scholar] [CrossRef]
- Schröder, J.; Hsu, A.; Boyle, S.E.; Macintyre, G.; Cmero, M.; Tothill, R.W.; Johnstone, R.W.; Shackleton, M.; Papenfuss, A.T. Socrates: Identification of genomic rearrangements in tumour genomes by re-aligning soft clipped reads. Bioinformatics 2014, 30, 1064–1072. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Jenjaroenpun, P.; Pillai, A.M.; Ivshina, A.V.; Ow, G.S.; Efthimios, M.; Zhiqun, T.; Tan, T.Z.; Lee, S.C.; Rogers, K.; et al. Transposon insertional mutagenesis in mice identifies human breast cancer susceptibility genes and signatures for stratification. Proc. Natl. Acad. Sci. USA 2017, 114, E2215–E2224. [Google Scholar] [CrossRef] [Green Version]
- Motakis, E.; Ivshina, A.V.; Kuznetsov, V.A. Data-driven approach to predict survival of cancer patients: Estimation of microarray genes’ prediction significance by Cox proportional hazard regression model. IEEE Eng. Med. Biol. Mag. Q. Mag. Eng. Med. Biol. Soc. 2009, 28, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.X.; Li, X.Q.; Chang, Y. Signature Gene Identification of Cancer Occurrence and Pattern Recognition. J. Comput. Biol. A J. Comput. Mol. Cell Biol. 2018, 25, 907–916. [Google Scholar] [CrossRef] [PubMed]
- Tan Ide, A.; Ricciardelli, C.; Russell, D.L. The metalloproteinase ADAMTS1: A comprehensive review of its role in tumorigenic and metastatic pathways. Int. J. Cancer 2013, 133, 2263–2276. [Google Scholar] [CrossRef] [PubMed]
- Porter, S.; Scott, S.D.; Sassoon, E.M.; Williams, M.R.; Jones, J.L.; Girling, A.C.; Ball, R.Y.; Edwards, D.R. Dysregulated expression of adamalysin-thrombospondin genes in human breast carcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2004, 10, 2429–2440. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.D.; Yuan, Y.L.; Yu, H.B.; Tian, G.J.; Li, D.Y. SHCBP1 is a novel target and exhibits tumor-promoting effects in gastric cancer. Oncol. Rep. 2019, 41, 1649–1657. [Google Scholar] [CrossRef]
- Liu, L.; Yang, Y.; Liu, S.; Tao, T.; Cai, J.; Wu, J.; Guan, H.; Zhu, X.; He, Z.; Li, J.; et al. EGF-induced nuclear localization of SHCBP1 activates β-catenin signaling and promotes cancer progression. Oncogene 2019, 38, 747–764. [Google Scholar] [CrossRef]
- Mo, M.; Tong, S.; Yin, H.; Jin, Z.; Zu, X.; Hu, X. SHCBP1 regulates STAT3/c-Myc signaling activation to promote tumor progression in penile cancer. Am. J. Cancer Res. 2020, 10, 3138–3156. [Google Scholar]
- Peng, C.; Zhao, H.; Chen, W.; Song, Y.; Wang, X.; Li, J.; Qiao, Y.; Wu, D.; Ma, S.; Wang, X.; et al. Identification of SHCBP1 as a novel downstream target gene of SS18-SSX1 and its functional analysis in progression of synovial sarcoma. Oncotarget 2016, 7, 66822–66834. [Google Scholar] [CrossRef] [Green Version]
- Peng, C.; Zhao, H.; Song, Y.; Chen, W.; Wang, X.; Liu, X.; Zhang, C.; Zhao, J.; Li, J.; Cheng, G.; et al. SHCBP1 promotes synovial sarcoma cell metastasis via targeting TGF-β1/Smad signaling pathway and is associated with poor prognosis. Mol. Carcinog. 2017, 36, 141. [Google Scholar] [CrossRef]
- Ren, C.; Zhou, Z.; Wang, X.; Hua, X.; Zou, M.; Zhang, X. SHCBP1 Promotes the Progression of Esophageal Squamous Cell Carcinoma Via the TGFβ Pathway. J. Cancer Res. Clin. Oncol. 2021, 29, 136–143. [Google Scholar] [CrossRef]
- Tao, H.C.; Wang, H.X.; Dai, M.; Gu, C.Y.; Wang, Q.; Han, Z.G.; Cai, B. Targeting SHCBP1 inhibits cell proliferation in human hepatocellular carcinoma cells. Asian Pac. J. Cancer Prev. APJCP 2013, 14, 5645–5650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.; Li, Y.; Zhang, Z.; Wang, J.; Wang, J. SHCBP1 regulates apoptosis in lung cancer cells through phosphatase and tensin homolog. Oncol. Lett. 2019, 18, 1888–1894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, N.; Wu, Y.P.; Yin, H.B.; Chen, S.H.; Li, X.D.; Xue, X.Y.; Gou, X. SHCBP1 promotes tumor cell proliferation, migration, and invasion, and is associated with poor prostate cancer prognosis. J. Cancer Res. Clin. Oncol. 2020, 146, 1953–1969. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Hu, J.F.; Zhan, Q.; Wang, Z.W.; Li, G.; Pan, J.J.; Huang, L.; Liao, C.Y.; Huang, Y.; Tian, Y.F.; et al. SHCBP1 interacting with EOGT enhances O-GlcNAcylation of NOTCH1 and promotes the development of pancreatic cancer. Genomics 2021, 113, 827–842. [Google Scholar] [CrossRef]
- Zhang, G.Y.; Ma, Z.J.; Wang, L.; Sun, R.F.; Jiang, X.Y.; Yang, X.J.; Long, B.; Ye, H.L.; Zhang, S.Z.; Yu, Z.Y.; et al. The Role of Shcbp1 in Signaling and Disease. Nat. Commun. 2019, 19, 854–862. [Google Scholar] [CrossRef]
- Zhou, Y.; Tan, Z.; Chen, K.; Wu, W.; Zhu, J.; Wu, G.; Cao, L.; Zhang, X.; Zeng, X.; Li, J.; et al. Overexpression of SHCBP1 promotes migration and invasion in gliomas by activating the NF-κB signaling pathway. Mol. Carcinog. 2018, 57, 1181–1190. [Google Scholar] [CrossRef]
- Feng, W.; Li, H.C.; Xu, K.; Chen, Y.F.; Pan, L.Y.; Mei, Y.; Cai, H.; Jiang, Y.M.; Chen, T.; Feng, D.X. SHCBP1 is over-expressed in breast cancer and is important in the proliferation and apoptosis of the human malignant breast cancer cell line. Oncogene 2016, 587, 91–97. [Google Scholar] [CrossRef]
- Shi, W.; Zhang, G. Hyperactivation of HER2-SHCBP1-PLK1 axis promotes tumor cell mitosis and impairs trastuzumab sensitivity to gastric cancer. Nature Commun. 2021, 12, 2812. [Google Scholar] [CrossRef]
- Cheng, L.; Tan, Z.; Huang, Z.; Pan, Y.; Zhang, W.; Wang, J. Expression Profile and Prognostic Values of Mini-Chromosome Maintenance Families (MCMs) in Breast Cancer. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2020, 26, e923673. [Google Scholar] [CrossRef]
- Hu, Y.; Wang, L.; Li, Z.; Wan, Z.; Shao, M.; Wu, S.; Wang, G. Potential Prognostic and Diagnostic Values of CDC6, CDC45, ORC6 and SNHG7 in Colorectal Cancer. OncoTargets Ther. 2019, 12, 11609–11621. [Google Scholar] [CrossRef] [Green Version]
- Idilli, A.I.; Pagani, F.; Kerschbamer, E.; Berardinelli, F.; Bernabe, M.; Cayuela, M.L.; Piazza, S.; Poliani, P.L.; Cusanelli, E.; Mione, M.C. Changes in the Expression of Pre-Replicative Complex Genes in hTERT and ALT Pediatric Brain Tumors. Cancers 2020, 12, 1028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, R.; Wang, Z.; Zhang, Y.; Chen, Y.; Liu, Q.; Zhang, T.; Liu, Y. Development and validation of a novel prognostic signature in gastric adenocarcinoma. OncoTargets Ther. 2020, 12, 22233–22252. [Google Scholar] [CrossRef]
- Wang, X.K.; Wang, Q.Q.; Huang, J.L.; Zhang, L.B.; Zhou, X.; Liu, J.Q.; Chen, Z.J.; Liao, X.W.; Huang, R.; Yang, C.K.; et al. Novel candidate biomarkers of origin recognition complex 1, 5 and 6 for survival surveillance in patients with hepatocellular carcinoma. Cancers 2020, 11, 1869–1882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Bao, L.; Hu, J.; Wu, D.; Tong, X. ORC6, Negatively Regulated by miR-1-3p, Promotes Proliferation, Migration, and Invasion of Hepatocellular Carcinoma Cells. Front. Cell Dev. Biol. 2021, 9, 652292. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, K.M.; Jones, R.M.; Petermann, E.; Jeggo, P.A. Diminished origin-licensing capacity specifically sensitizes tumor cells to replication stress. Mol. Cancer Res. MCR 2013, 11, 370–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Senter, N.C.; McCulley, A.; Kuznetsov, V.A.; Feng, W. Identification of Recurrent Chromosome Breaks Underlying Structural Rearrangements in Mammary Cancer Cell Lines. Genes 2022, 13, 1228. https://doi.org/10.3390/genes13071228
Senter NC, McCulley A, Kuznetsov VA, Feng W. Identification of Recurrent Chromosome Breaks Underlying Structural Rearrangements in Mammary Cancer Cell Lines. Genes. 2022; 13(7):1228. https://doi.org/10.3390/genes13071228
Chicago/Turabian StyleSenter, Natalie C., Andrew McCulley, Vladimir A. Kuznetsov, and Wenyi Feng. 2022. "Identification of Recurrent Chromosome Breaks Underlying Structural Rearrangements in Mammary Cancer Cell Lines" Genes 13, no. 7: 1228. https://doi.org/10.3390/genes13071228





