Indirect DNA Transfer and Forensic Implications: A Literature Review
Abstract
:1. Introduction
2. Materials and Methods
2.1. Database Search Terms and Timeline
2.2. Inclusion and Exclusion Criteria
2.3. Quality Assessment and Data Extraction
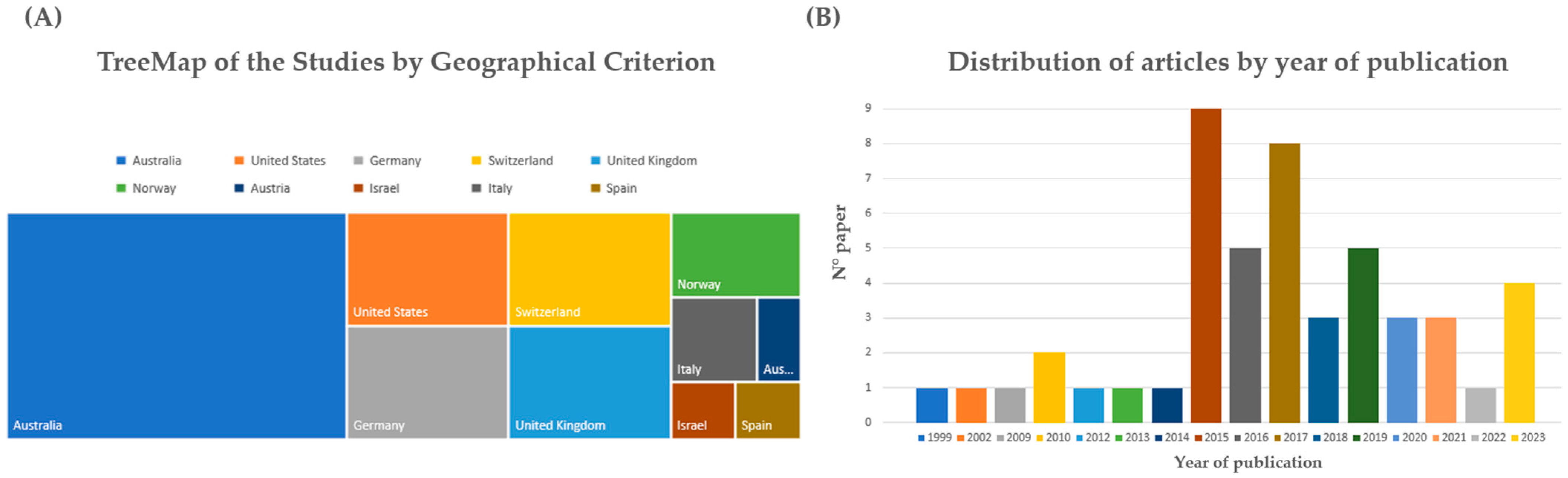
2.4. Characteristics of Eligible Studies
3. Results
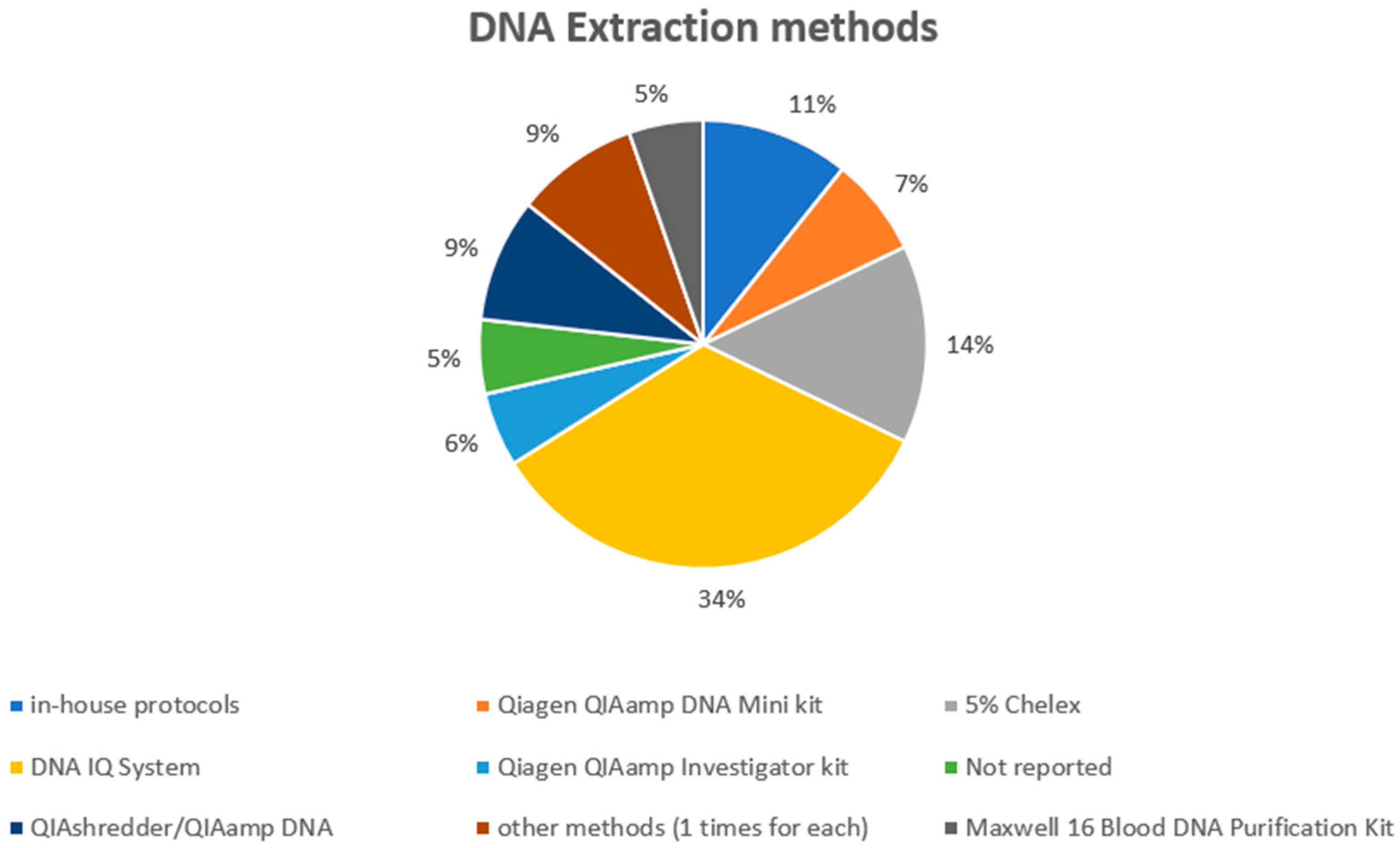
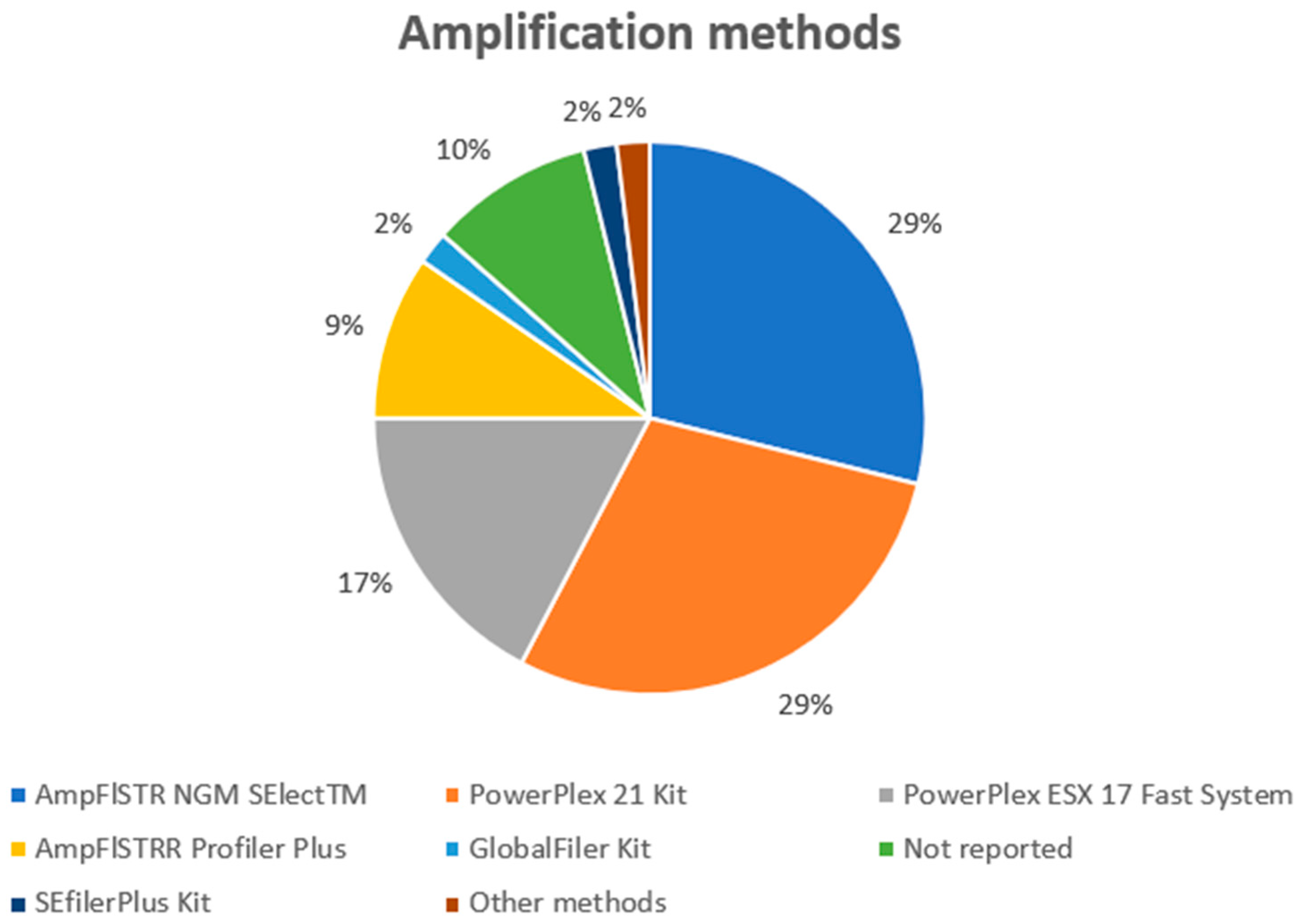
3.1. Technical Results
3.2. Main Findings
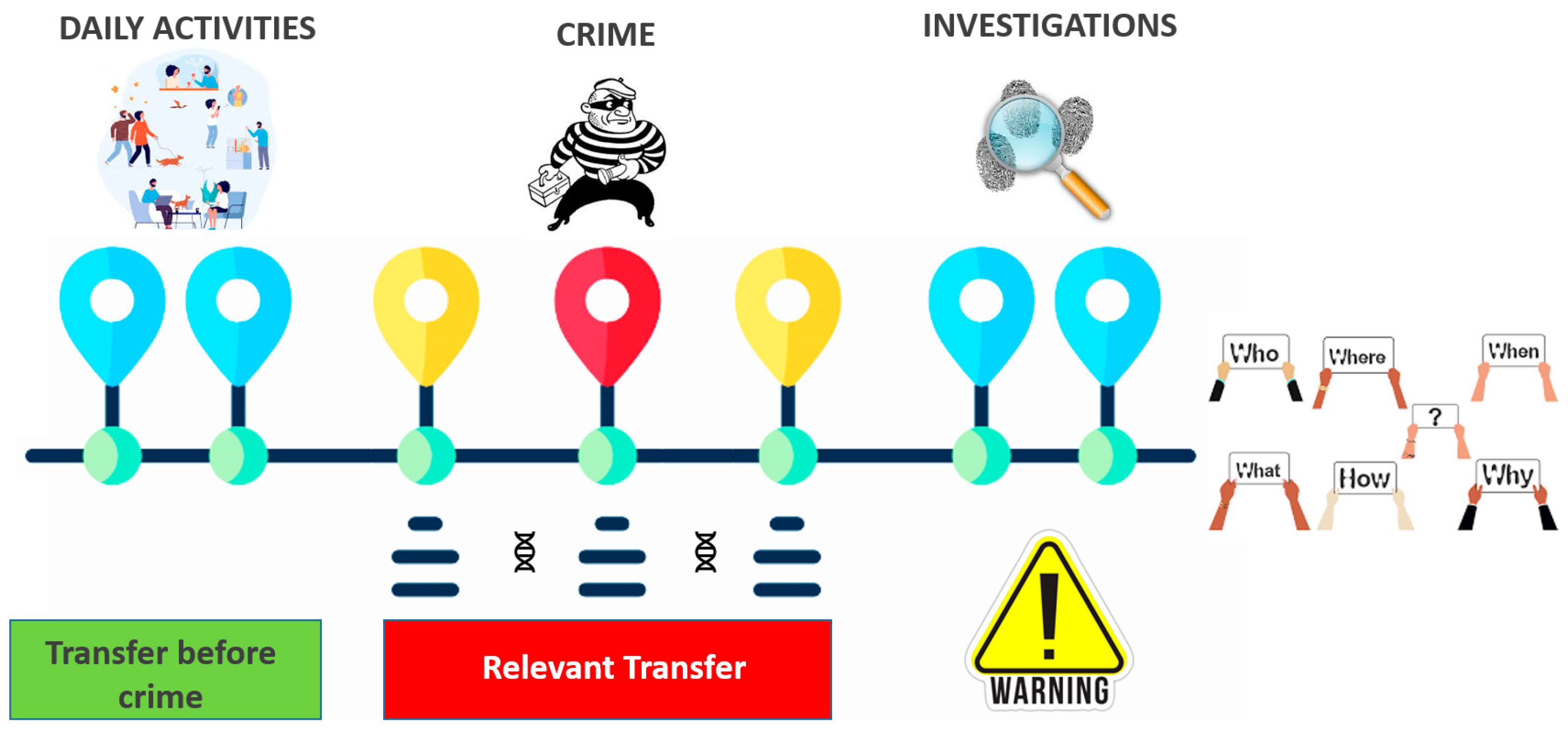
4. Discussion
- The subject’s characteristics: These include age, sex, shedder status (good or bad), and lifestyle habits. Some people tend to shed more DNA than others, which can affect the amount of DNA transferred and detected. Age and sex can also influence the quality and quantity of DNA, as well as lifestyle habits such as smoking, drinking, or using cosmetics [24,49,50,56,58,62].
- The type and duration of the contact: The type of contact can be direct (touching) or indirect (through an intermediary). The duration of contact can range from seconds to hours. Generally, direct and longer contacts are more likely to result in secondary DNA transfer than indirect and shorter contacts [24,29,32,36,40,43,55,57,63,69].
- The characteristics of the item: These include material, usage, size, shape, texture, and cleanliness. Different materials have different affinities for DNA, such as cotton being more absorbent than plastic. Usage can affect the amount of DNA background on an item, such as a frequently used phone having more DNA than a rarely used pen. Size, shape, and texture can affect the surface area and roughness of an item, which can influence the amount of contact and friction between the item and the DNA source. Cleanliness can affect the presence of contaminants or inhibitors that can degrade or interfere with DNA analysis [7,25,26,31,32,33,37,42,45,46,50,51,53,55,57,58,60,64,66,69].
- Trace type: This refers to whether the trace is fresh or dry, visible or invisible, single-source or mixed-source. Fresh traces are more likely to contain viable cells that can be amplified by PCR than dry traces. Visible traces are easier to locate and collect than invisible traces. Single-source traces are easier to interpret than mixed-source traces that contain DNA from multiple contributors [27,28,29,34,38,39,47,50,51,53,65,66,69].
- The activities made before contact: These include washing hands, wearing gloves, handling other items, or performing other actions that can affect the amount and quality of DNA on the hands or other body parts. Washing hands can reduce the amount of DNA available for transfer. Wearing gloves can prevent direct contact between the source and the target of DNA transfer. Handling other items can introduce additional sources of DNA or contaminants that can affect the analysis [30,33,34,38,40,59,61,63,68].
- Time: The period of time between the primary and secondary contact and the interval between the secondary contact and the sampling of the evidence can affect the amount and quality of DNA transferred. Generally, the longer the time gap, the lower the chance of detecting secondary transfer DNA. However, there is no clear consensus on how long DNA can persist on different surfaces or objects after secondary transfer [36,38,40,41,42,46,54,56,63].
- Environmental conditions: The temperature, humidity, presence of microbial contamination, and other environmental factors can influence the degradation and persistence of DNA after secondary transfer. For example, high temperature and humidity can accelerate DNA degradation, while low temperature and humidity can preserve DNA for longer periods. Microbial contamination can also degrade DNA or interfere with its detection [32,33,37,41,44,47,52,53,60,65,68].
- Technical methods: The sampling methods, extraction methods, and profiling techniques used in forensic analysis can also affect the detection and interpretation of secondary transfer DNA. For example, different sampling methods (such as swabbing, taping, or cutting) can yield different amounts of DNA from the same surface or object. Different extraction methods (such as organic, Chelex, or silica-based) can be more efficient in isolating DNA from complex mixtures. Different profiling techniques (such as STRs, SNPs, or NGS) can have different sensitivities and specificities in amplifying and analyzing DNA from low-template DNA or degraded samples [38,39,42,48,52].
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bukyya, J.L.; Tejasvi, M.L.A.; Avinash, A.; Chanchala, H.P.; Talwade, P.; Afroz, M.M.; Pokala, A.; Neela, P.K.; Shyamilee, T.K.; Srisha, V. DNA Profiling in Forensic Science: A Review. Glob. Med. Genet. 2021, 8, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Machado, H.; Granja, R. DNA Technologies in Criminal Investigation and Courts BT—Forensic Genetics in the Governance of Crime; Machado, H., Granja, R., Eds.; Springer Singapore: Singapore, 2020; pp. 45–56. ISBN 978-981-15-2429-5. [Google Scholar]
- Latham, K.E.; Miller, J.J. DNA Recovery and Analysis from Skeletal Material in Modern Forensic Contexts. Forensic Sci. Res. 2019, 4, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Haddrill, P.R. Developments in Forensic Dna Analysis. Emerg. Top. Life Sci. 2021, 5, 381–393. [Google Scholar] [CrossRef] [PubMed]
- Jordan, D.; Mills, D. Past, Present, and Future of DNA Typing for Analyzing Human and Non-Human Forensic Samples. Front. Ecol. Evol. 2021, 9, 646130. [Google Scholar] [CrossRef]
- Alessandrini, F.; Onofri, V.; Turchi, C.; Buscemi, L.; Pesaresi, M.; Tagliabracci, A. Past, Present and Future in Forensic Human Identification; Springer: Berlin/Heidelberg, Germany, 2020; ISBN 9783030338329. [Google Scholar]
- Carrara, L.; Hicks, T.; Samie, L.; Taroni, F.; Castella, V. DNA Transfer When Using Gloves in Burglary Simulations. Forensic Sci. Int. Genet. 2023, 63, 102823. [Google Scholar] [CrossRef] [PubMed]
- Buckingham, A.K.; Harvey, M.L.; van Oorschot, R.A.H. The Origin of Unknown Source DNA from Touched Objects. Forensic Sci. Int. Genet. 2016, 25, 26–33. [Google Scholar] [CrossRef]
- Ferrara, M.; Sessa, F.; Rendine, M.; Spagnolo, L.; De Simone, S.; Riezzo, I.; Ricci, P.; Pascale, N.; Salerno, M.; Bertozzi, G.; et al. A Multidisciplinary Approach Is Mandatory to Solve Complex Crimes: A Case Report. Egypt J. Forensic Sci. 2019, 9, 11. [Google Scholar] [CrossRef]
- Pomara, C.; Gianpaolo, D.P.; Monica, S.; Maglietta, F.; Sessa, F.; Guglielmi, G.; Turillazzi, E. “Lupara Bianca” a Way to Hide Cadavers after Mafia Homicides. A Cemetery of Italian Mafia. A Case Study. Leg. Med. 2015, 17, 192–197. [Google Scholar] [CrossRef]
- Van Oorschot, R.A.H.; Jones, M.K.K. DNA Fingerprints from Fingerprints. Nature 1997, 387, 767. [Google Scholar] [CrossRef]
- Daly, D.J.; Murphy, C.; McDermott, S.D. The Transfer of Touch DNA from Hands to Glass, Fabric and Wood. Forensic Sci. Int. Genet. 2012, 6, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Ladd, C.; Adamowicz, M.S.; Bourke, M.T.; Scherczinger, C.A.; Lee, H.C. A Systematic Analysis of Secondary DNA Transfer. J. Forensic Sci. 1999, 44, 1270–1272. [Google Scholar] [CrossRef] [PubMed]
- Locard, E. The Analysis of Dust Traces. Am. J. Police Sci. 1930, 1, 276–298. [Google Scholar] [CrossRef]
- Burrill, J.; Daniel, B.; Frascione, N. A Review of Trace “Touch DNA” Deposits: Variability Factors and an Exploration of Cellular Composition. Forensic Sci. Int. Genet. 2019, 39, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Tozzo, P.; Mazzobel, E.; Marcante, B.; Delicati, A.; Caenazzo, L. Touch DNA Sampling Methods: Efficacy Evaluation and Systematic Review. Int. J. Mol. Sci. 2022, 23, 15541. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Ostrander, J.; Wickenheiser, R.; Hall, A. Touch DNA in Forensic Science: The Use of Laboratory-Created Eccrine Fingerprints to Quantify DNA Loss. Forensic Sci. Int. 2020, 2, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Sessa, F.; Salerno, M.; Bertozzi, G.; Messina, G.; Ricci, P.; Ledda, C.; Rapisarda, V.; Cantatore, S.; Turillazzi, E.; Pomara, C. Touch DNA: Impact of Handling Time on Touch Deposit and Evaluation of Different Recovery Techniques: An Experimental Study. Sci. Rep. 2019, 9, 9542. [Google Scholar] [CrossRef]
- Jansson, L.; Swensson, M.; Gifvars, E.; Hedell, R.; Forsberg, C.; Ansell, R.; Hedman, J. Individual Shedder Status and the Origin of Touch DNA. Forensic Sci. Int. Genet. 2022, 56, 102626. [Google Scholar] [CrossRef]
- Alessandrini, F.; Cecati, M.; Pesaresi, M.; Turchi, C.; Carle, F.; Tagliabracci, A. Fingerprints as Evidence for a Genetic Profile: Morphological Study on Fingerprints and Analysis of Exogenous and Individual Factors Affecting DNA Typing. J. Forensic Sci. 2003, 48, 586–592. [Google Scholar] [CrossRef]
- Johannessen, H.; Gill, P.; Roseth, A.; Fonneløp, A.E. Determination of Shedder Status: A Comparison of Two Methods Involving Cell Counting in Fingerprints and the DNA Analysis of Handheld Tubes. Forensic Sci. Int. Genet. 2021, 53, 102541. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.Y.C.; Tan, J.; Lee, Y.S.; Syn, C.K.C. Shedder Status—An Analysis over Time and Assessment of Various Contributing Factors. J. Forensic Sci. 2023, 68, 1292–1301. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Lowe, A.; Murray, C.; Whitaker, J.; Tully, G.; Gill, P. The Propensity of Individuals to Deposit DNA and Secondary Transfer of Low Level DNA from Individuals to Inert Surfaces. Forensic Sci. Int. 2002, 129, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Goray, M.; Eken, E.; Mitchell, R.J.; van Oorschot, R.A.H. Secondary DNA Transfer of Biological Substances under Varying Test Conditions. Forensic Sci. Int. Genet. 2010, 4, 62–67. [Google Scholar] [CrossRef]
- Goray, M.; Mitchell, R.J.; van Oorschot, R.A. Investigation of Secondary DNA Transfer of Skin Cells under Controlled Test Conditions. Leg. Med. 2010, 12, 117–120. [Google Scholar] [CrossRef]
- Wiegand, P.; Heimbold, C.; Klein, R.; Immel, U.; Stiller, D.; Klintschar, M. Transfer of Biological Stains from Different Surfaces. Int. J. Leg. Med. 2011, 125, 727–731. [Google Scholar] [CrossRef] [PubMed]
- Warshauer, D.H.; Marshall, P.; Kelley, S.; King, J.; Budowle, B. An Evaluation of the Transfer of Saliva-Derived DNA. Int. J. Leg. Med. 2012, 126, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, V.J.; Mitchell, R.J.; Ballantyne, K.N.; van Oorschot, R.A.H. Following the Transfer of DNA: How Far Can It Go? Forensic Sci. Int. Genet. Suppl. Ser. 2013, 4, e53–e54. [Google Scholar] [CrossRef]
- Zoppis, S.; Muciaccia, B.; D’Alessio, A.; Ziparo, E.; Vecchiotti, C.; Filippini, A. DNA Fingerprinting Secondary Transfer from Different Skin Areas: Morphological and Genetic Studies. Forensic Sci. Int. Genet. 2014, 11, 137–143. [Google Scholar] [CrossRef]
- Fonneløp, A.E.; Johannessen, H.; Gill, P. Persistence and Secondary Transfer of DNA from Previous Users of Equipment. Forensic Sci. Int. Genet. Suppl. Ser. 2015, 5, e191–e192. [Google Scholar] [CrossRef]
- Fonneløp, A.E.; Egeland, T.; Gill, P. Secondary and Subsequent DNA Transfer during Criminal Investigation. Forensic Sci. Int. Genet. 2015, 17, 155–162. [Google Scholar] [CrossRef]
- Goray, M.; van Oorschot, R.A.H. The Complexities of DNA Transfer during a Social Setting. Leg. Med. 2015, 17, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Kamphausen, T.; Fandel, S.B.; Gutmann, J.S.; Bajanowski, T.; Poetsch, M. Everything Clean? Transfer of DNA Traces between Textiles in the Washtub. Int. J. Leg. Med. 2015, 129, 709–714. [Google Scholar] [CrossRef] [PubMed]
- Montpetit, S.; O’Donnell, P. An Optimized Procedure for Obtaining DNA from Fired and Unfired Ammunition. Forensic Sci. Int. Genet. 2015, 17, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Oldoni, F.; Castella, V.; Hall, D. Exploring the Relative DNA Contribution of First and Second Object’s Users on Mock Touch DNA Mixtures. Forensic Sci. Int. Genet. Suppl. Ser. 2015, 5, e300–e301. [Google Scholar] [CrossRef]
- Szkuta, B.; Harvey, M.L.; Ballantyne, K.N.; Van Oorschot, R.A.H. DNA Transfer by Examination Tools—A Risk for Forensic Casework? Forensic Sci. Int. Genet. 2015, 16, 246–254. [Google Scholar] [CrossRef]
- Szkuta, B.; Harvey, M.L.; Ballantyne, K.N.; van Oorschot, R.A.H. Residual DNA on Examination Tools Following Use. Forensic Sci. Int. Genet. Suppl. Ser. 2015, 5, e495–e497. [Google Scholar] [CrossRef]
- Verdon, T.J.; Mitchell, R.J.; van Oorschot, R.A.H. Preliminary Investigation of Differential Tapelifting for Sampling Forensically Relevant Layered Deposits. Leg. Med. 2021, 17, 553–559. [Google Scholar] [CrossRef]
- Cale, C.M.; Earll, M.E.; Latham, K.E.; Bush, G.L. Could Secondary DNA Transfer Falsely Place Someone at the Scene of a Crime? J. Forensic Sci. 2016, 61, 196–203. [Google Scholar] [CrossRef]
- Jones, S.; Scott, K.; Lewis, J.; Davidson, G.; Allard, J.E.; Lowrie, C.; McBride, B.M.; McKenna, L.; Teppett, G.; Rogers, C.; et al. DNA Transfer through Nonintimate Social Contact. Sci. Justice 2016, 56, 90–95. [Google Scholar] [CrossRef]
- Oldoni, F.; Castella, V.; Hall, D. Shedding Light on the Relative DNA Contribution of Two Persons Handling the Same Object. Forensic Sci. Int. Genet. 2016, 24, 148–157. [Google Scholar] [CrossRef]
- Samie, L.; Hicks, T.; Castella, V.; Taroni, F. Stabbing Simulations and DNA Transfer. Forensic Sci. Int. Genet. 2016, 22, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.; Abarno, D.; Rowe, E.; Rask-Nielsen, L. Observations of DNA Transfer within an Operational Forensic Biology Laboratory. Forensic Sci. Int. Genet. 2016, 23, 33–49. [Google Scholar] [CrossRef]
- Fonneløp, A.E.; Ramse, M.; Egeland, T.; Gill, P. The Implications of Shedder Status and Background DNA on Direct and Secondary Transfer in an Attack Scenario. Forensic Sci. Int. Genet. 2017, 29, 48–60. [Google Scholar] [CrossRef] [PubMed]
- McColl, D.L.; Harvey, M.L.; van Oorschot, R.A.H. DNA Transfer by Different Parts of a Hand. Forensic Sci. Int. Genet. Suppl. Ser. 2017, 6, e29–e31. [Google Scholar] [CrossRef]
- Meakin, G.E.; Butcher, E.V.; van Oorschot, R.A.H.; Morgan, R.M. Trace DNA Evidence Dynamics: An Investigation into the Deposition and Persistence of Directly- and Indirectly-Transferred DNA on Regularly-Used Knives. Forensic Sci. Int. Genet. 2017, 29, 38–47. [Google Scholar] [CrossRef]
- Neuhuber, F.; Kreindl, G.; Kastinger, T.; Dunkelmann, B.; Zahrer, W.; Cemper-Kiesslich, J.; Grießner, I. Police Officer’s DNA on Crime Scene Samples—Indirect Transfer as a Source of Contamination and Its Database-Assisted Detection in Austria. Forensic Sci. Int. Genet. Suppl. Ser. 2017, 6, e608–e609. [Google Scholar] [CrossRef]
- Pfeifer, C.M.; Wiegand, P. Persistence of Touch DNA on Burglary-Related Tools. Int. J. Leg. Med. 2017, 131, 941–953. [Google Scholar] [CrossRef] [PubMed]
- Szkuta, B.; Ballantyne, K.N.; van Oorschot, R.A.H. Transfer and Persistence of DNA on the Hands and the Influence of Activities Performed. Forensic Sci. Int. Genet. 2017, 28, 10–20. [Google Scholar] [CrossRef]
- Szkuta, B.; Van Oorschot, R.A.H.; Ballantyne, K.N. DNA Decontamination of Fingerprint Brushes. Forensic Sci. Int. 2017, 277, 41–50. [Google Scholar] [CrossRef]
- Taylor, D.; Biedermann, A.; Samie, L.; Pun, K.-M.; Hicks, T.; Champod, C. Helping to Distinguish Primary from Secondary Transfer Events for Trace DNA. Forensic Sci. Int. Genet. 2017, 28, 155–177. [Google Scholar] [CrossRef]
- Ruan, T.; Barash, M.; Gunn, P.; Bruce, D. Investigation of DNA Transfer onto Clothing during Regular Daily Activities. Int. J. Leg. Med. 2018, 132, 1035–1042. [Google Scholar] [CrossRef]
- Szkuta, B.; Ballantyne, K.N.; Kokshoorn, B.; van Oorschot, R.A.H. Transfer and Persistence of Non-Self DNA on Hands over Time: Using Empirical Data to Evaluate DNA Evidence given Activity Level Propositions. Forensic Sci. Int. Genet. 2018, 33, 84–97. [Google Scholar] [CrossRef]
- Voskoboinik, L.; Amiel, M.; Reshef, A.; Gafny, R.; Barash, M. Laundry in a Washing Machine as a Mediator of Secondary and Tertiary DNA Transfer. Int. J. Leg. Med. 2018, 132, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Butcher, E.V.; van Oorschot, R.A.H.; Morgan, R.M.; Meakin, G.E. Opportunistic Crimes: Evaluation of DNA from Regularly-Used Knives after a Brief Use by a Different Person. Forensic Sci. Int. Genet. 2019, 42, 135–140. [Google Scholar] [CrossRef]
- Champion, J.; van Oorschot, R.A.H.; Linacre, A. Visualising DNA Transfer: Latent DNA Detection Using Diamond Dye. Forensic Sci. Int. Genet. Suppl. Ser. 2019, 7, 229–231. [Google Scholar] [CrossRef]
- Otten, L.; Banken, S.; Schürenkamp, M.; Schulze-Johann, K.; Sibbing, U.; Pfeiffer, H.; Vennemann, M. Secondary DNA Transfer by Working Gloves. Forensic Sci. Int. Genet. 2019, 43, 102126. [Google Scholar] [CrossRef] [PubMed]
- Romero-García, C.; Rosell-Herrera, R.; Revilla, C.J.; Baeza-Richer, C.; Gomes, C.; Palomo-Díez, S.; Arroyo-Pardo, E.; López-Parra, A.M. Effect of the Activity in Secondary Transfer of DNA Profiles. Forensic Sci. Int. Genet. Suppl. Ser. 2019, 7, 578–579. [Google Scholar] [CrossRef]
- Szkuta, B.; Ansell, R.; Boiso, L.; Connolly, E.; Kloosterman, A.D.; Kokshoorn, B.; McKenna, L.G.; Steensma, K.; van Oorschot, R.A.H. Assessment of the Transfer, Persistence, Prevalence and Recovery of DNA Traces from Clothing: An Inter-Laboratory Study on Worn Upper Garments. Forensic Sci. Int. Genet. 2019, 42, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Gosch, A.; Euteneuer, J.; Preuß-Wössner, J.; Courts, C. DNA Transfer to Firearms in Alternative Realistic Handling Scenarios. Forensic Sci. Int. Genet. 2020, 48, 102355. [Google Scholar] [CrossRef]
- Samie, L.; Taroni, F.; Champod, C. Estimating the Quantity of Transferred DNA in Primary and Secondary Transfers. Sci. Justice 2020, 60, 128–135. [Google Scholar] [CrossRef]
- Szkuta, B.; Ansell, R.; Boiso, L.; Connolly, E.; Kloosterman, A.D.; Kokshoorn, B.; McKenna, L.G.; Steensma, K.; van Oorschot, R.A.H. DNA Transfer to Worn Upper Garments during Different Activities and Contacts: An Inter-Laboratory Study. Forensic Sci. Int. Genet. 2020, 46, 102268. [Google Scholar] [CrossRef] [PubMed]
- Tanzhaus, K.; Reiß, M.-T.; Zaspel, T. “I’ve Never Been at the Crime Scene!”—Gloves as Carriers for Secondary DNA Transfer. Int. J. Leg. Med. 2021, 135, 1385–1393. [Google Scholar] [CrossRef] [PubMed]
- Thornbury, D.; Goray, M.; van Oorschot, R.A.H. Indirect DNA Transfer without Contact from Dried Biological Materials on Various Surfaces. Forensic Sci. Int. Genet. 2021, 51, 102457. [Google Scholar] [CrossRef] [PubMed]
- Thornbury, D.; Goray, M.; van Oorschot, R.A.H. Transfer of DNA without Contact from Used Clothing, Pillowcases and Towels by Shaking Agitation. Sci. Justice 2021, 61, 797–805. [Google Scholar] [CrossRef] [PubMed]
- Reither, J.B.; van Oorschot, R.A.H.; Szkuta, B. DNA Transfer between Worn Clothing and Flooring Surfaces with Known Histories of Use. Forensic Sci. Int. Genet. 2022, 61, 102765. [Google Scholar] [CrossRef] [PubMed]
- McCrane, S.M.; Mulligan, C.J. An Innovative Transfer DNA Experimental Design and QPCR Assay: Protocol and Pilot Study. J. Forensic Sci. 2023, 68, 990–1000. [Google Scholar] [CrossRef] [PubMed]
- Onofri, M.; Altomare, C.; Severini, S.; Tommolini, F.; Lancia, M.; Carlini, L.; Gambelunghe, C.; Carnevali, E. Direct and Secondary Transfer of Touch DNA on a Credit Card: Evidence Evaluation Given Activity Level Propositions and Application of Bayesian Networks. Genes 2023, 14, 996. [Google Scholar] [CrossRef]
- Monkman, H.; Szkuta, B.; van Oorschot, R.A.H. Presence of Human DNA on Household Dogs and Its Bi-Directional Transfer. Genes 2023, 14, 1486. [Google Scholar] [CrossRef]
- Kallupurackal, V.; Kummer, S.; Voegeli, P.; Kratzer, A.; Dørum, G.; Haas, C.; Hess, S. Sampling Touch DNA from Human Skin Following Skin-to-Skin Contact in Mock Assault Scenarios—A Comparison of Nine Collection Methods. J. Forensic Sci. 2021, 66, 1889–1900. [Google Scholar] [CrossRef]
- Tozzo, P.; Giuliodori, A.; Rodriguez, D.; Caenazzo, L. Effect of Dactyloscopic Powders on DNA Profiling from Enhanced Fingerprints: Results from an Experimental Study. Am. J. Forensic Med. Pathol. 2014, 35, 68–72. [Google Scholar] [CrossRef]
- Sessa, F.; Esposito, M.; Roccuzzo, S.; Cocimano, G.; Li Rosi, G.; Sablone, S.; Salerno, M. DNA Profiling from Fired Cartridge Cases: A Literature Review. Minerva Forensic Med. 2023, 143, 34–43. [Google Scholar] [CrossRef]
- Sessa, F.; Maglietta, F.; Asmundo, A.; Pomara, C. Forensic Genetics and Genomic. In Forensic and Clinical Forensic Autopsy; Pomara, C., Fineschi, V., Eds.; CRC Press: Boca Raton, FL, USA, 2021; pp. 177–192. [Google Scholar]
- Turchi, C.; Previderè, C.; Bini, C.; Carnevali, E.; Grignani, P.; Manfredi, A.; Melchionda, F.; Onofri, V.; Pelotti, S.; Robino, C.; et al. Assessment of the Precision ID Identity Panel Kit on Challenging Forensic Samples. Forensic Sci. Int. Genet. 2020, 49, 102400. [Google Scholar] [CrossRef] [PubMed]
- Turchi, C.; Onofri, V.; Melchionda, F.; Fattorini, P.; Tagliabracci, A. Development of a Forensic DNA Phenotyping Panel Using Massive Parallel Sequencing. Forensic Sci. Int. Genet. Suppl. Ser. 2019, 7, 177–179. [Google Scholar] [CrossRef]
- Tozzo, P.; Politi, C.; Delicati, A.; Gabbin, A.; Caenazzo, L. External Visible Characteristics Prediction through SNPs Analysis in the Forensic Setting: A Review. Front. Biosci. Landmark 2021, 26, 828–850. [Google Scholar] [CrossRef]
- Kayser, M.; De Knijff, P. Improving Human Forensics through Advances in Genetics, Genomics and Molecular Biology. Nat. Rev. Genet. 2011, 12, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Sijen, T.; Harbison, S. On the Identification of Body Fluids and Tissues: A Crucial Link in the Investigation and Solution of Crime. Genes 2021, 12, 1728. [Google Scholar] [CrossRef] [PubMed]
- Lech, K.; Liu, F.; Ackermann, K.; Revell, V.L.; Lao, O.; Skene, D.J.; Kayser, M. Evaluation of MRNA Markers for Estimating Blood Deposition Time: Towards Alibi Testing from Human Forensic Stains with Rhythmic Biomarkers. Forensic Sci. Int. Genet. 2016, 21, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Gondhali, U.; Choudhary, S. Potential of DNA Technique-Based Body Fluid; Springer: Singapore, 2022; ISBN 9789811643187. [Google Scholar]
- Lynch, C.; Fleming, R. Partial Validation of Multiplexed Real-Time Quantitative PCR Assays for Forensic Body Fluid Identification. Sci. Justice 2023, 63, 724–735. [Google Scholar] [CrossRef]
- Weber, A.R.; Lednev, I.K. Crime Clock—Analytical Studies for Approximating Time since Deposition of Bloodstains. Forensic Chem. 2020, 19, 100248. [Google Scholar] [CrossRef]
- Gosch, A.; Bhardwaj, A.; Courts, C. TrACES of Time: Transcriptomic Analyses for the Contextualization of Evidential Stains—Identification of RNA Markers for Estimating Time-of-Day of Bloodstain Deposition. Forensic Sci. Int. Genet. 2023, 67, 102915. [Google Scholar] [CrossRef]
- van Oorschot, R.A.H.; Szkuta, B.; Meakin, G.E.; Kokshoorn, B.; Goray, M. DNA Transfer in Forensic Science: A Review. Forensic Sci. Int. Genet. 2019, 38, 140–166. [Google Scholar] [CrossRef] [PubMed]
- Gosch, A.; Courts, C. On DNA Transfer: The Lack and Difficulty of Systematic Research and How to Do It Better. Forensic Sci. Int. Genet. 2019, 40, 24–36. [Google Scholar] [CrossRef] [PubMed]
- van Oorschot, R.A.H.; Meakin, G.E.; Kokshoorn, B.; Goray, M.; Szkuta, B. DNA Transfer in Forensic Science: Recent Progress towards Meeting Challenges. Genes 2021, 12, 1766. [Google Scholar] [CrossRef] [PubMed]
- Meakin, G.E.; Kokshoorn, B.; van Oorschot, R.A.H.; Szkuta, B. Evaluating Forensic DNA Evidence: Connecting the Dots. WIREs Forensic Sci. 2021, 3, e1404. [Google Scholar] [CrossRef]
- Esposito, M.; Sessa, F.; Cocimano, G.; Zuccarello, P.; Roccuzzo, S.; Salerno, M. Advances in Technologies in Crime Scene Investigation. Diagnostics 2023, 13, 3169. [Google Scholar] [CrossRef] [PubMed]
- Bini, C.; Giorgetti, A.; Fazio, G.; Amurri, S.; Pelletti, G.; Pelotti, S. Impact on Touch DNA of an Alcohol-Based Hand Sanitizer Used in COVID-19 Prevention. Int. J. Leg. Med. 2023, 137, 645–653. [Google Scholar] [CrossRef]
- Taylor, D.; Samie, L.; Champod, C. Using Bayesian Networks to Track DNA Movement through Complex Transfer Scenarios. Forensic Sci. Int. Genet. 2019, 42, 69–80. [Google Scholar] [CrossRef]
- Taroni, F.; Garbolino, P.; Aitken, C. A Generalised Bayes’ Factor Formula for Evidence Evaluation under Activity Level Propositions: Variations around a Fibres Scenario. Forensic Sci. Int. 2021, 322, 110750. [Google Scholar] [CrossRef]
- Sessa, F.; Salerno, M.; Pomara, C. The Interpretation of Mixed DNA Samples BT—Handbook of DNA Profiling; Dash, H.R., Shrivastava, P., Lorente, J.A., Eds.; Springer Singapore: Singapore, 2020; pp. 1–22. ISBN 978-981-15-9364-2. [Google Scholar]
- Butler, J.M.; Kline, M.C.; Coble, M.D. NIST Interlaboratory Studies Involving DNA Mixtures (MIX05 and MIX13): Variation Observed and Lessons Learned. Forensic Sci. Int. Genet. 2018, 37, 81–94. [Google Scholar] [CrossRef]
- Gill, P.; Hicks, T.; Butler, J.M.; Connolly, E.; Gusmão, L.; Kokshoorn, B.; Morling, N.; van Oorschot, R.A.H.; Parson, W.; Prinz, M.; et al. DNA Commission of the International Society for Forensic Genetics: Assessing the Value of Forensic Biological Evidence—Guidelines Highlighting the Importance of Propositions. Part II: Evaluation of Biological Traces Considering Activity Level Propositions. Forensic Sci. Int. Genet. 2020, 44, 102186. [Google Scholar] [CrossRef]
- Kotsoglou, K.N.; McCartney, C. To the Exclusion of All Others? DNA Profile and Transfer Mechanics—R v Jones (William Francis) [2020] EWCA Crim 1021 (03 Aug 2020). Int. J. Evid. Proof 2021, 25, 135–140. [Google Scholar] [CrossRef]
- Veldhuis, M.S.; Ariëns, S.; Ypma, R.J.F.; Abeel, T.; Benschop, C.C.G. Explainable Artificial Intelligence in Forensics: Realistic Explanations for Number of Contributor Predictions of DNA Profiles. Forensic Sci. Int. Genet. 2022, 56, 102632. [Google Scholar] [CrossRef] [PubMed]
- Morgan, R.M. Conceptualising Forensic Science and Forensic Reconstruction. Part I: A Conceptual Model. Sci. Justice 2017, 57, 455–459. [Google Scholar] [CrossRef]
- Earwaker, H.; Nakhaeizadeh, S.; Smit, N.M.; Morgan, R.M. A Cultural Change to Enable Improved Decision-Making in Forensic Science: A Six Phased Approach. Sci. Justice 2020, 60, 9–19. [Google Scholar] [CrossRef]
- Margiotta, G.; Tasselli, G.; Tommolini, F.; Lancia, M.; Massetti, S.; Carnevali, E. Risk of DNA Transfer by Gloves in Forensic Casework. Forensic Sci. Int. Genet. Suppl. Ser. 2015, 5, e527–e529. [Google Scholar] [CrossRef]
- Wienroth, M.; Granja, R.; McCartney, C.; Lipphardt, V.; Amoako, E.N. Ethics as Lived Practice. Anticipatory Capacity and Ethical Decision-Making in Forensic Genetics. Genes 2021, 12, 1868. [Google Scholar] [CrossRef]
- Amankwaa, A.O.; McCartney, C. The Effectiveness of the Current Use of Forensic DNA in Criminal Investigations in England and Wales. WIREs Forensic Sci. 2021, 3, e1414. [Google Scholar] [CrossRef]



| Reference, Year, and Nationality | Experimental Model | Main Findings |
|---|---|---|
| Ladd et al., 1999, United States [13] | The researchers examined two forms of secondary transfer, which included skin to skin contact through handshaking and skin to object to skin contact. | Secondary transfer was not observed in this experimental model. |
| Lowe et al., 2002, United Kingdom [24] | In the first scenario, participants categorized as good and poor shedders were asked to hold hands for 1 min. Following this, poor shedders were instructed to hold a plastic 50 mL tube for 10 s. | Secondary transfer occurred when the DNA from the hand of the good shedder was transferred to an object through the poor shedder. The authors concluded that secondary transfer under optimized conditions is possible and may result in a single full profile. |
| In the second scenario, there was a 30 min delay between the human contact and the poor shedders gripping the tube during the experiment. | In both the second and third scenarios, it was observed that the first shedder pairing led to secondary transfer. The recovered DNA profiles were mixed and included between 80 and 100% of the good shedder’s profile in every instance of secondary transfer. | |
| In the third scenario, there was a 1 h gap between the human contact and the tube gripping for the poor shedders in the experiment. | ||
| Goray et al., 2009, Australia [25] | In this experimental model, three biological materials (pure DNA, blood, and saliva) were tested, evaluating the transfer between two different substrates: plastic (hard/non-porous) and cotton and wool (soft/porous). Wet samples were handled by depositing the biological fluid onto the primary substrate and then applying the secondary substrate within a time frame of 10–60 s. In the case of dry samples, the biological fluid was deposited onto the primary substrate and allowed to dry for 18–24 h (at room temperature) before the secondary substrate was applied. Contact was established through three modes: passive, pressure, and friction. | In this experimental model, it was found that the secondary transfer is significantly influenced by the moisture content (i.e., in the case of wet substrate), item substrate type, and manner of contact (passive and pressure contact). Although the experiment involved testing three distinct biological sources (pure DNA, saliva, and blood), there were no significant differences detected in the secondary transfer rates. |
| Goray et al., 2010, Australia [26] | The experiment involved creating a primary deposit of touch DNA by rubbing the skin over the designated area for 10–15 s. The researchers then evaluated the effects of different moisture levels (fresh and dry), primary and secondary substrates (soft porous cotton and hard non-porous plastic), and modes of contact (passive, pressure, and friction). | According to the experiment, the type of substrate used for the primary transfer plays a crucial role in the secondary transfer evaluation. If skin cells are deposited on cotton, the retrieved amount is approximately 20 times greater than when deposited on plastic. The secondary transfer of skin cells was found to occur more easily when the primary substrate was non-porous than porous. Contact through friction was also observed to significantly increase the rate of transfer. |
| Wiegand et al., 2011, Germany [27] | To conduct the experiment, 50 µL of saliva or venous blood from male donors were placed on paper, cotton cloth, and plastic surfaces, covering an area of 2 × 2 cm with equal moisture. The stains were left to air dry overnight and then subjected to two different scenarios for evaluation. The first scenario involved pressing the thumb on the stain for 2 s, while the second scenario involved rubbing the surface of a paper with the thumb for 10 s before swabbing the contact area to assess secondary transfer. | The transfer of saliva stains through rubbing and pressing onto paper only yielded 50% of detectable stains with very low levels of DNA (ranging from 0–1 pg/μL). On the other hand, the secondary transfer of blood stains resulted in relevant values. However, it was found that only a few instances of DNA concentrations were sufficient for complete DNA profiles, and these instances were transfers from stains on plastic. |
| Warshauer et al., 2012, United States [28] | First scenario: thumbs were licked, and subjects grasped sterilized plastic conical tubes after each drying time. | According to this study, when DNA is transferred from saliva, the genetic material of the original contributor can make up most of the resulting mixture. The amount of moisture present during the transfer, along with the texture and surface area of the object(s) involved, are important factors that affect the transfer. It is important to consider these factors when analyzing DNA transfers. |
| Second scenario: saliva was deposited on pens; subsequently, subjects were required to pass the pens to their designated partners after each drying time; pens were gripped in the same way as the tubes; the partners’ palms were then swabbed. | ||
| Third scenario: thumbs covered by gloves were licked, and subjects grasped sterilized plastic conical tubes after each drying time; tubes were then swabbed. | ||
| In the fourth scenario of this study, pens were contaminated with saliva and then passed between subjects after different drying times. The pens were held like tubes and the subjects’ palms were moistened before they grasped them; the pens were swabbed for DNA analysis. | ||
| In the fifth scenario of this study, the subjects moistened their thumbs after each drying time by licking them. Following that, they grasped sterilized plastic conical tubes, which were then swabbed to collect any DNA. | ||
| Lehmann et al., 2013, Australia [29] | This study involved depositing blood onto the substrate in 15 μL amounts and transferring touch DNA through various methods such as rubbing hands over cotton or repeatedly placing hands onto glass. Wet blood was immediately transferred while dry blood was allowed to dry completely. Touch DNA was deposited on the primary substrate and then transferred onto subsequent substrates within an hour. The primary substrate with DNA was flipped over and placed on top of the second substrate for deposition. | According to this study, the transfer of DNA from saliva is influenced by the substrate and the biological source types. The researchers confirmed the evidence of secondary and subsequent DNA transfer in their experimental model. They found that DNA transferred more readily to and from glass than it did to and from cotton. Additionally, the transfer of touch DNA was found to be less significant than wet or dry blood on either cotton or glass. Notably, the study found that wet blood transferred more effectively than dry blood on both cotton and glass surfaces. |
| Zoppis et al., 2014, Italy [30] | Three different scenarios were evaluated. First scenario: before handwashing, 8 subjects were asked to rub a fingertip on a typical sebaceous skin area of another individual (i.e., back of the hand and back of the forearm). Subsequently, they pressed on a glass slide. Second scenario: the same deposition was provided 10 min after conventional handwashing. Third scenario: the same deposition was provided 10 min after handwashing with antiseptic soap and air drying. | In evaluating genetic results, it is important to consider the specific previously touched cutaneous area, whether it is sebaceous or non-sebaceous skin areas, as DNA secondary transfer is a significant phenomenon. |
| Fonneløp et al., 2015, Norway [31] | In this study, each participant’s computer keyboard and mouse were exchanged with those of another participant, and the new user used the equipment for the entire duration of the study. | According to the findings of this study, it is feasible for the DNA of the first user to be transferred into the hands of a new user even eight days after the latter has touched the computer equipment. |
| Fonneløp et al., 2015, Norway [32] | The first substrate, either a piece of wood or a plastic tube, was picked up by the donors and held for 30 s with moderate pressure and friction. After this, the substrate was placed on a clean bench paper. The “investigator”, who wore personal protective equipment, then picked up the same substrate and held it in their right-hand glove for 30 s. Finally, the substrate was placed back onto the bench paper and the right-hand glove was held against new pre-cleaned items with moderate pressure and friction. | Based on the results of this study, DNA was readily transferred to wood and plastic, while less was transferred to a metal door handle, demonstrating that a second and tertiary transfer is a possible event. |
| Goray and van Oorschot, 2015, Australia [33] | During a 20 min social interaction, three individuals were invited to participate in a blind experiment where they had a drink of juice and chatted while being video recorded. The experiment involved collecting samples from various segments such as the table, chair arms (top only), jug handle, remaining surface of the jug, entire outer surface of the glasses, and left and right hands of each participant. | DNA that can be measured was discovered on numerous surfaces and objects during the testing. The lowest number of contributors necessary to account for the findings was recorded for each tested surface. In addition, some of the tested surfaces and objects exhibited unidentified DNA profiles. |
| Kamphausen et al., 2015, Germany [34] | The authors washed two pieces of clothing, one with skin cells and the other one with blood, either using a washing machine or hand-washing techniques. | According to the findings of the research, blood cells were consistently observed to transfer from one object to another. Combining buccal swabs and clothes for washing did not yield complete STR profiles. Lastly, the transmission of enough epithelial skin cells from one fabric to another during washing for a reliable STR analysis (i.e., a full profile) is highly unlikely. |
| Montpetit and O’Donnell, 2015, United States [35] | The goal of this study was to investigate the collection and profiling of DNA from both fired and unfired ammunition, which are frequently discovered during searches of individuals. To simulate various contact scenarios, DNA testing was conducted on casings and cartridges. | According to the findings of the current research, a combined profile was typically detected. However, in approximately 97% of cases, the individual operating the weapon’s ammunition loader was identified as the source of the profile. |
| Oldoni et al., 2015, Switzerland [36] | Various items, such as a computer mouse, pen, bracelet, necklace, key, watch, nurse cap, and nitrile gloves, were chosen for the study. The first participant used the objects frequently over a span of 8–10 days, while the second subject was asked to handle the same items for three separate simulation sessions of 5, 30, and 120 min each. | According to this study’s findings, the percentage contribution of the second user’s DNA profile increased significantly from 21% to 73% of the total DNA profile after 5 and 120 min, respectively, compared to the object’s owner on all objects examined. |
| Szkuta et al., 2015, Australia [37] | This study examined various situations to determine the extent and frequency of DNA transfer between simulated crime scene materials, such as cotton or glass, and high-risk vectors like scissors, forceps, and gloves. | According to this study, it was found that DNA-containing material could be transferred between exhibits through the use of scissors, forceps, and gloves. Touch DNA transfer was observed to be the highest when non-porous glass was used as the primary substrate, followed by porous cotton as the second substrate. These results demonstrate the potential for DNA transfer between different materials and objects and suggest that the source of the DNA profile may be identifiable even after transfer. |
| Szkuta et al., 2015, Australia [38] | In Experiment 1, dried blood or touch DNA was transferred from a primary substrate made of cotton or glass to a secondary substrate of DNA-free cotton or glass using scissors, forceps, or gloves. The researchers applied both heavy (multiple) and light (singular) contact in pairwise combinations. This implies that DNA can be transferred between exhibits through these common tools and that the source of the DNA profile may be identifiable even after transfer. | The authors concluded that a significant amount of DNA persisted on scissors, forceps, and gloves even after the transfer of dried blood from a primary cotton substrate to a DNA-free secondary cotton substrate. The nature of the contact did not impact the retention of dried blood on the vectors. However, the transfer of touch DNA from and to cotton resulted in fewer alleles remaining on these vectors. |
| Verdon et al., 2015, Australia [39] | Different scenarios were investigated (-touch/touch; -saliva/touch; -touch/saliva. | This study demonstrated that there is no clear preference for the sampling method, and the biological source is very important in order to determine the second transfer event. |
| Cale et al., 2016, United States [40] | The participants in this study wore gloves for 1.5 h before collecting samples to limit the presence of foreign DNA on their hands. Wearing gloves was also expected to promote the transfer of DNA by increasing the amount of sweat and oils on the participants’ hands. Once they removed the gloves, the participants shook hands vigorously for two minutes to simulate intimate contact, then immediately handled their assigned knife for two minutes. | The authors of this study were able to show that secondary DNA transfer, which refers to DNA belonging to an individual who did not directly touch the knife, was possible. Out of 20 instances, 16 were found to have alleles that could be attributed to this type of transfer. |
| Jones et al., 2016, United Kingdom [41] | In the first scenario, the male participant made physical contact with the female participant’s face for two minutes. Following that, both participants held hands and rubbed/massaged them together for three minutes. The male participant then proceeded to simulate urination by removing his penis from his underwear over the waistband and holding it with both hands for about 30 s. To increase the chances of DNA transfer, both hands were used to hold the penis before returning it to the underwear. Afterwards, the male volunteer removed his underwear while wearing gloves and swabbed the shaft of his penis. | On the underwear samples, DNA matching the female participant was detected. However, none of the penile samples taken 6 h after the staged non-intimate social contact events showed any matching female DNA. |
| In the second scenario, the male participant collected penile swabs after engaging in unprotected sexual intercourse with a female. The researchers collected the underwear that the participant wore immediately after the intercourse and recovered the samples from it. | The female participant’s DNA profile was found to match on all waistband samples, inside front samples, and on samples collected from the inside back, outside front, and outside back areas of the clothing. These findings suggest a possibility of DNA transfer through physical contact with the female participant. | |
| Oldoni et al., 2016, Switzerland [42] | The initial participant was instructed to interact with nine objects made of plastic, metal, nitrile, and fabric, which are typically present at crime scenes involving burglary or robbery. This interaction took place for at least 20 min per day, over a period of eight to ten consecutive days. Afterward, a second participant used the same objects in three different simulation sessions lasting 5, 30, or 120 min. | Indirectly transferred DNA accounted for only a small portion of the mixed DNA profiles observed, with the exception of 1 out of 234 cases. |
| Samie et al., 2016, Switzerland [43] | The objective of this study was to investigate the transfer of DNA from individuals with close connections to handlers. | Only a small percentage of the DNA profiles showed evidence of transfer from an unknown source, while the majority of profiles contained the DNA of the person who committed the stabbing. |
| Taylor et al., 2016, Australia [44] | These authors explore different work areas (laboratory areas, office areas, inaccessible areas, and common areas) in order to explore aspects of DNA transfer, including secondary and tertiary transfer. | Inaccessible areas were sampled to demonstrate secondary transfer. Moreover, these authors concluded that the detected profiles not always corresponded to the last person touching item. |
| Fonneløp et al., 2017, Norway [45] | Investigation on the secondary transfer using different scenarios in order to investigate possible occurrences of secondary transfer from co-workers (t-shirt used daily with investigation in order to detect an exogenous DNA profile) | These findings confirmed the possibility of obtaining a secondary transfer. |
| McColl et al., 2017, Australia [46] | The researchers utilized saliva from a male donor as a source of DNA that was manually transferred onto an object. To achieve this, four female participants pressed their dominant hand onto a plate coated with saliva for a period of 10 s and then immediately placed the same hand on a clean glass plate for another 10 s. | DNA transfer occurred in a different manner strictly related to the different parts of a hand. |
| Meakin et al., 2017, United Kingdom [47] | To mimic regular use, the researchers had each participant handle a knife in a specific way for two days in their experimental model. After that, the participants shook hands with a fellow volunteer for 10 s and then stabbed a foam block repeatedly with one of their knives for a minute. | With the exception of one participant, less than 5% of the recovered profiles had non-donor DNA co-deposited. |
| Neuhuber et al., 2017, Austria [48] | The authors investigated different scenarios (indirect transfer via camera; -indirect transfer via car; -indirect transfer via desk) about a police officer’s DNA transfer on crime scene samples, generating an indirect transfer as a source of contamination. | The authors confirmed the possibility of DNA transfer of police officers’ DNA onto crime scene items through three different scenarios. |
| Pfeifer and Wiegand, 2017, Germany [49] | In the first scenario, items belonging to one person are taken in a robbery by another person. In the second scenario, items are used by one person before being handled in a less severe manner. | When the second user simulated a burglary by using a tool barehanded, the first user may not be found as a major component on their handles. When the second user broke up the burglary setup using gloves, the first user matched the DNA handle profile in 37% of the cases. |
| Szkuta et al., 2017, Australia [50] | On glass plates, both the depositor’s self-DNA and non-self-DNA from the known contributor who shook hands with them were deposited. | The experimental model’s results indicate that a considerable amount of DNA is transferred, which is linked to an individual’s ability to transfer their own DNA (shedder status). |
| Szkuta et al., 2017, Australia [51] | The objective of this study was to assess the potential risk of contamination resulting from the transfer of dried saliva and skin deposits between glass surfaces using new, unused squirrel hair and fiberglass brushes. Various scenarios were examined during the investigation. | The experimental model’s results indicate that squirrel hair and fiberglass brushes can collect and transfer varying amounts of DNA-containing material. The detectability of the transferred material on the secondary surface depends on the biological nature of the material being transferred. |
| Taylor et al., 2017, Australia [52] | Transfer from hand to object, and, subsequently, secondary transfer from object to object. | Possibility to find the secondary transfer. |
| Ruan et al., 2018, Australia [53] | The researchers conducted a laundry experiment wherein 38 individuals were provided with a cotton swatch measuring roughly 10 cm × 10 cm to be washed and dried with their laundry using their own washing machine and detergent. As negative controls, two cotton swatches were randomly selected. | The DNA profiles of most cotton swatch samples indicated either a distinct single source (21%) or a blend of DNA from multiple sources (55%). In the case of mixed profiles, the majority of them (around two to three persons) showed DNA from only one source, while a few (around one in five) had a combination of DNA from four individuals. |
| Szkuta et al., 2018, Australia [54] | After exchanging two handshakes, the participants resumed their regular activities for either 40 min, 5 h, or 8 h. Later, they held a polished wooden axe handle with their right hand and rotated it to produce friction for 10 s. | The profiles obtained from the axe handles after they were in contact with the known contributor for 40 min, 5 h, or 8 h showed a diverse range of alleles. In all the profiles from the axe handle, except for one four-person mixture generated after the depositor’s contact 40 min post-handshake, the depositor was the primary or sole contributor. |
| Voskoboinik et al., 2018, Israel [55] | Under various washing conditions, a group of eight new socks made of different cotton blends were washed together with the regular laundry of four households. | The possibility of a secondary transfer was confirmed by 7/32 samples (22%). |
| Butcher et al., 2019, United Kingdom [56] | The researchers designed an experiment where a person used knives for 4 min over two days before another individual used them for 2, 30, or 60 s to determine how shorter durations of second use affect the resulting DNA profiles. | The DNA ratios of the first user to the second user were around 4:1, 2:1, and 1:1 for durations of 2, 30, and 60 s respectively. The analysis of the DNA quantities showed that the trend occurred due to a decrease in the DNA of the first user, transferred to the second user’s hands, rather than an increase in DNA deposition from the second user. This trend was observed after the knives were used by the first user for a total of 4 min over two days before being used by the second user for the specified durations. |
| Champion et al., 2019, Australia [57] | In this research, the contact types adopted by Goray et al. [18], namely, passive, pressure, friction, and friction with pressure, were employed to explore the transfer between aluminum and the substrates. | For the first time, researchers were able to visually detect the transfer of DNA from one substrate to another by using fluorescent Diamond™ Dye (DD) to visualize the cellular transfer. |
| Otten et al., 2019, Germany [58] | In this study, the goal was to evaluate the extent to which the DNA of an innocent person is transferred to a crime scene through work gloves, taking into account whether the suspect is a shedder or not. | The results of this study showed that the glove, especially its exterior, could act as a vector for secondary transfer in real-life scenarios. |
| Romero-García et al., 2019, Spain [59] | The researchers instructed five individuals to hold hands for five minutes and then wash their hands with soap having a neutral pH level. Next, they dried their hands using different towels each day. To serve as a control group, the researchers analyzed a portion of each towel that was not used, and they also collected saliva samples from all participants to determine their reference profiles. | The researchers were unable to obtain a comprehensive profile from either the towel or the individual who had made contact with the object, as well as their partner. |
| Szkuta et al., 2019, Australia [60] | This study examined various situations involving four participants, also known as “wearers” (P1-4). Two upper garments that belonged to the primary participant and had been worn before were chosen. Each of the selected garments was worn on both a workday (WD) and a non-workday (ND). | The research suggests that in certain situations, the presence of close associates may overshadow the wearer’s contributions, depending on the situation and the area of the garment. As a result, the wearer’s contributions may be minor or even absent. |
| Gosch et al., 2020, Germany [61] | Four firearm handling scenarios, simulating different actions of the shooter. | The amount of DNA subsequent to indirect transfer is strictly related to handling conditions and surface types. |
| Samie et al., 2020, Switzerland [62] | For all the experiments, a pair of identical knives were utilized, with one assigned to each participant. The participants, categorized as either a good or bad shedder, were instructed to shake hands before using their respective knives to stab the ballistic soap. | The secondary transfer is related to the shedder status, target surfaces, and alleged transfer mechanisms. Compared to the primary transfer, it occurs with a percentage between 1 and 3%. |
| Szkuta et al., 2020, Australia [63] | This study recruited four participants, also referred to as “wearers,” from four different laboratories. The participants wore the selected garments for an average of 5.1 h before hugging another individual, an average of 5.2 h (minimum 3.5 h, maximum 6.5 h) before going out with another individual, and an average of 1.5 h before spending a day in another individual’s office. The time spent wearing the garments includes both at home, during commuting, and at work before engaging in the respective activity. | According to this study, the DNA of a person of interest was successfully recovered from a piece of clothing after direct contact, close proximity, and physical absence. The experiment involved embracing an individual or occupying their office space, after which several DNA profiles were identified from the clothing. The transfer of DNA was more likely to occur following prolonged and/or recurring contact, as well as direct contact. |
| Tanzhaus et al., 2021, Germany [64] | This study involved the use of gloves of various materials such as cloth, leather, and rubber, which were sorted based on the material present on the exterior of the glove. The gloves were kept in separate plastic bags and handled by a perpetrator for a period of 4 weeks. Following each touch, the item was tested for DNA transfer. | Out of all the experiments conducted in this study, it was found that only one instance of secondary transfer could be detected. |
| Thornbury et al., 2021, Australia [65] | This study aimed to investigate the potential of DNA transfer without direct contact by analyzing tapping and stretching agitation for dried blood, saliva, semen, touch, and vaginal fluid that were deposited on four substrates. | This study found that it was possible for DNA to be transferred indirectly without any physical contact, as long as dried biological materials were present on different surfaces. The success of this transfer seemed to depend on various factors, such as the type of agitation, the type of biological material, and the surface it was transferred onto. |
| Thornbury et al., 2021, Australia [66] | This study focused on exploring the possibility of DNA transfer through indirect means without physical contact. The researchers achieved this by gently shaking used clothing, pillowcases, and towels, all of which had a known usage history, of 10 volunteers to check for DNA transfer onto a secondary surface. The results indicate that DNA transfer was a common occurrence and could take place from all three items that were tested. | This study’s experimental model revealed that DNA transfer to the secondary surface was observed in all samples except for four. |
| Reither et al., 2022, Australia [67] | The authors investigated two possible scenarios of indirect transfer: from a worn garment to a floor and vice-versa. | Based on their findings, the authors demonstrated the possibility of an indirect DNA transfer from clothing to flooring and from flooring to clothing in both ‘active’ and ‘passive’ situations. Obviously, the DNA transfer was greater in the active simulation (i.e., application of pressure and friction). |
| Carrara et al., 2023, Switzerland [7] | This study investigated a secondary transfer mediated by gloves during simulated burglary simulations. | This study confirmed the possibility of an indirect transfer in the applied experimental model. |
| McCrane and Mulligan, 2023, USA [68] | Different scenarios were investigated: a male and female alternately held the pistol, and subsequently, the female’s hand was swabbed to evaluate the secondary transfer. | Possibility to indirectly transfer the DNA on the female’s hand. |
| Onofri et al., 2023, Italy [69] | The authors performed a secondary transfer scenario simulating that the owner of a credit card, after his personal use for a month, places and moves it around the surface of a co-worker’s desk, applying slight pressure, for 30 s. | The authors reported a high value of secondary transfer (about 50% of secondary transfer DNA traces), and it was demonstrated that the co-worker could be identified as the major contributor. |
| Monkman et al., 2023, Australia [70] | The authors explored the possibility of an indirect transfer mediated by a domestic dog. | Based on their findings, the authors concluded that dogs could be a vector for human DNA transfer, demonstrating a transfer from the dog to a gloved hand during patting and a bed sheet while walking. |
| Reference | Sampling Methods | DNA Extraction | Quantification | Amplification |
|---|---|---|---|---|
| Ladd et al. [13] | Moistened (dH2O) sterile swabs. | Organic extraction/microcon-100 purification, following in-house protocol. | QuantiBlot Kit (Perkin Elmer Applied Biosystems, Shelton, WA, USA). | AmpFlSTRR Profiler Plus and COfiler DNA typing kits (Perkin Elmer Applied Biosystems). |
| Lowe et al. [24] | Swabs of surfaces. | Qiagen QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). | DNA was not quantified. | AmpFlSTRR Profiler Plus and COfiler DNA typing kits (Perkin Elmer Applied Biosystems). |
| Goray et al. [25] | The 1 cm × 1 cm small squares (plus a surrounding margin of approximately 0.3 cm) were cut into smaller pieces and placed into 10 mL tubes. | DNA was extracted via 5% Chelex. | Quantifiler Human DNA Quantification (Perkin Elmer Applied Biosystems). | AmpFlSTRR Profiler Plus kit (Perkin Elmer Applied Biosystems). |
| Goray et al. [26] | The 1 cm × 1 cm small squares (plus a surrounding margin of approximately 0.3 cm) were cut into smaller pieces and placed into 10 mL tubes. | DNA was extracted via 5% Chelex. | Quantifiler Human DNA Quantification (Perkin Elmer Applied Biosystems). | AmpFlSTRR Profiler Plus kit (Perkin Elmer Applied Biosystems). |
| Wiegand et al. [27] | Cotton wool swabs moistened with sterile water. | First-DNA all-tissue DNA kit (GEN-IAL GmbH, Troisdorf, Germany) DNA IQ extraction protocol (Promega, Madison, WI, USA). | Plexor DNA Quantification Kit (Promega). | SEfilerPlus kit (Applied Biosystems, Waltham, MA, USA). |
| Warshauer et al. [28] | Swab. | Qiagen QIAamp DNA Mini (Qiagen). | Quantifiler Human DNA Quantification Kit (Life Technologies, Carlsbad, CA, USA). | AmpFlSTR Identifiler Plus PCR Amplification Kit (Life Technologies). |
| Lehmann et al. [29] | This study involved cutting-out cotton substrates and plastic backing using scalpels and then extracting them together. To collect DNA from glass slides, the researchers used a double swab technique where the first swab was moistened with 4 drops of deionized water, and the second swab was slightly dampened with one drop of water. | DNA IQ Automated DNA extraction Kit (Promega). | Quantifiler Human DNA Quantification Kit (Life Technologies). | PowerPlex1 21 Kit (Promega). |
| Zoppis et al. [30] | The slides were swabbed with sterile cotton swabs and distilled water. | DNA IQ Automated DNA extraction Kit (Promega). | Quantifiler Duo DNA Quantification Kit (Applied Biosystems). | AmpFlSTR1 NGM SElectTM PCR Amplification kit (Applied Biosystems). |
| Fonneløp et al. [31] | Samples were collected by swabbing the participants’ hands. | All samples were extracted by 5% Chelex. | Quantifiler Duo Kit (Thermo Fisher, Waltham, MA, USA). | PowerPlex ESX 17 Fast System kit (Promega). |
| Fonneløp et al. [32] | DNA was recovered from all items using DNA-free mini-lifting tapes (Scenesafe FAST). | DNA was extracted by the 5% Chelex. | Quantifiler Duo Kit (Applied Biosystems). | Powerplex ESX 17 Kit (Promega). |
| Goray and van Oorschot [33] | The wet and dry double swabbing technique was used. | DNA IQ System (Promega). | Quantifiler Human DNA Quantification Kit (Thermo Fisher). | PowerPlex1 21 System (Promega). |
| Kamphausen et al. [34] | Dried clothes were taped with self-adhesive tape, and cells were collected from the tape with a double swab technique using first a DNA-free swab, moistened with lysis buffer, and then a dry swab. | DNA extraction from artificial stains was performed using a modified phenol/chloroform method. | Quantifiler Human DNA Quantification Kit (Thermo Fisher). | Powerplex ESX 17 or Powerplex S5 Kit (Promega). |
| Montpetit and O’Donnell [35] | The cartridges or casings were swabbed using a single nano pure water moistened cotton-tipped swab. | BioRobot EZ1 (Qiagen) using the EZ1 DNA Investigator Kit. | Quantifiler Human DNA Quantification Kit (Thermo Fisher). | AmpFlSTR Identifiler Plus PCR Amplification Kit (Life Technologies). |
| Oldoni et al. [36] | Samples were collected either with the double swab technique or by direct object cutting (nurse cap). | DNA was manually extracted using the QIAshredder/QIAamp (Qiagen) kit or phenol/chloroform (nurse cap). | Investigator Quantiplex HYres (Qiagen). | AmpFlSTR NGM SElectTM PCR Amplification (Life Technologies). |
| Szkuta et al. [37] | The wet–dry swabbing technique was used to collect samples from glass slides. | DNA IQ™ (Promega, USA). | Quantifiler Human DNA Quantification Kit (Thermo Fisher). | PowerPlex 21 System (Promega). |
| Szkuta et al. [38] | The wet and dry double swabbing technique was used. | DNA IQ System (Promega). | Quantifiler Human DNA Quantification Kit (Thermo Fisher). | PowerPlex1 21 System (Promega). |
| Verdon et al. [39] | Two different tapelift types were used: Scenesafe FAST, and Scotch Magic tape. | Following pre-treatment with 500 μL of TNE buffer containing Proteinase K, DNA was extracted from tapes and substrates using the DNA IQ system (Promega). | Quantifiler Human DNA Quantification Kit (Thermo Fisher). | PowerPlex 21 (Promega). |
| Cale et al. [40] | The surface of each knife’s handle was immediately sampled using a wet swabbing technique. | The process of removing the swabs from both the smooth-handled and rough-handled knives, as well as the control swabs, was carried out using the DNA Purification from Buccal Swabs Spin Protocol by Qiagen, a company based in Hilden, Germany. (Hilden, Germany). | Quantifiler Human DNA Quantification Kit (Thermo Fisher). | AmpFlSTR Identifiler Plus PCR Amplification Kit (Life Technologies). |
| Jones et al. [41] | The researchers used a wet sterile cotton swab that had been moistened with deionized water, followed by a dry sterile cotton swab to collect the DNA. They also took samples from specific areas of the underwear using mini-tape, including the inside and outside of the front waistband, as well as the inside front panel. | Not reported. | Not reported. | Not reported. |
| Oldoni et al. [42] | DNA traces were collected using the double swab method, except for the fabric nurse cap (cutting-out). | Within 24–48 h of sample collection, the DNA was extracted manually from the traces using the QIAshredder/QIAamp DNA mini protocol (Qiagen AG, Basel, Switzerland). | Investigator Quantiplex HYres (Qiagen). | AmpFlSTR NGM SElectTM PCR Amplification (Applied Biosystems). |
| Samie et al. [43] | DNA was collected using the double swab method. | DNA was extracted, using the combination of two kits, QIAshredder and QIAAmp kit. | Investigator Quantiplex HYres (Qiagen). | NGM Select (Applied Biosystem-Life Technologies). |
| Taylor et al. [44] | The sampling method used depended on the surface being sampled. Non-porous surfaces were sampled using foam-headed swabs called popule swabs that were soaked with isopropanol during sampling. Porous surfaces, on the other hand, were sampled using tapelifts. | DNA IQ system (Promega) using in-house validated protocols. | Quantifiler Human DNA Quantification Kit (Thermo Fisher). | GlobalFiler (Thermo Fisher). |
| Fonneløp et al. [45] | The mini-tape (Scenesafe FAST™) was used. | After sampling, the mini-tape was fragmented into smaller pieces and transferred to an extraction tube, from which DNA was extracted using the 5% Chelex method. | Quantifiler Trio Kit (Applied Biosystems). | PowerPlex ESX 17 Fast System kit (Promega). |
| McColl et al. [46] | DNA was collected using the double swab method. | DNA IQ™ System (Qiagen). | Quantifiler Human DNA Quantification Kit (Life Technologies). | PowerPlex 21 (Promega). |
| Meakin et al. [47] | DNA was recovered by mini taping. | QIAamp DNA Investigator Kit (QIAgen). | Quantifiler Human DNA Quantification Kit (Applied Biosystems). | AmpFlSTR NGM SElectTM PCR Amplification Kit (Applied Biosystems). |
| Neuhuber et al. [48] | Not reported. | “First-DNA” kit (Genial), M48 robot (Qiagen), or by organic extraction (phenol/chloroform). | Not reported. | Different DNA amplification kits. |
| Pfeifer and Wiegand [49] | The tool handles were cleaned using premoistened swabs (Sarstedt) soaked in lysis buffer (Promega). | The extraction of all samples was carried out using the Maxwell 16 Blood DNA Purification Kit (Promega) in a Maxwell extraction system. | Plexor HY System (Promega). | PowerPlex ESI 17 Fast (Promega). |
| Szkuta et al. [50] | Cotton swabs (150C, Copan) were utilized with a wet/moist swabbing protocol to collect the deposits on glass plates. | DNA IQ™ (Promega, USA). | Quantifiler Human DNA Quantification Kit (Life Technologies). | PowerPlex 21 (Promega). |
| Szkuta et al. [51] | DNA was obtained from swabs and cut bristles. | DNA IQ™ (Promega). | Quantifiler Trio (Life Technologies). | PowerPlex 21 (Promega). |
| Taylor et al. [52] | Double swab method and tapelifts (using Scotch Magic and Scenesafe FAST). | QIAshredder and QIAAmp kit. | Not reported. | Not reported. |
| Ruan et al. [53] | A DNA tapelift kit (Lovell Surgical Supplies) was used. | The DNA extraction process involved placing the tape inside an AutoLys tube manufactured by Hamilton Company, USA, followed by extraction using the PrepFiler Automated Forensic DNA Extraction Kit from Thermo Fisher Scientific. | Quantifiler Human DNA Quantification Kit (Applied Biosystems). | PowerPlex 21 (Promega). |
| Szkuta et al. [54] | Deposits on the axe handle were collected using a wet-moist swabbing technique. | Not reported. | Quantifiler Trio (Life Technologies). | Not reported. |
| Voskoboinik et al. [55] | Three-layer adhesive tapes were used to sample all garments. | Chelex extraction and subsequent purification with DNA IQ kit (Promega). | Quantifiler Human DNA Quantification Kit (Applied Biosystems). | AmpFlSTR SGM Plus (Applied Biosystems). |
| Butcher et al. [56] | DNA was recovered from each knife handle using a mini-tape within an hour of each stabbing event. | DNA extractions were performed using the QIAamp DNA Investigator Kit (Qiagen). | Quantifiler Human DNA Quantification Kit (Applied Biosystems). | AmpFlSTR NGM SElect™ PCR Amplification Kit (Applied Biosystems). |
| Champion et al. [57] | Not performed. | Not performed. | Not performed. | Not performed. |
| Otten et al. [58] | To collect DNA from items, sterile swabs were moistened with HPLC grade water. | DNA extraction was performed using a Maxwell16 Forensic Instrument with Casework Extraction Kit and DNA IQ™ Casework Pro Kit (Promega). | PowerQuant System (Promega). | PowerPlex ESX 17 System (Promega). |
| Romero-García et al. [59] | Not reported. | DNA was extracted with Speedtools DNA extraction kit (Biotools). | Not reported. | AmpFℓSTR NGM Select Kit (Thermo Fisher). |
| Szkuta et al. [60] | Polyvinyl chloride tape stubs (in-house). Mini-tape (SceneSafe FAST). Mini-tape (SceneSafe FAST). Moistened cotton swab followed by dry cotton swab (150C, Copan). | QIAmp isolation (Qiagen). Chelex (modified method). EZ1 advanced XL (Qiagen). DNA IQ™ (Promega). | ALU assay (in-house); Quantifiler HP (Thermo Fisher Scientific); Quantifiler Trio (Thermo Fisher Scientific). | NGM (Thermo Fisher Scientific); PowerPlex ESX16 Fast (Promega); -NGM SElect (Thermo Fisher Scientific); PowerPlex 21 (Promega). |
| Gosch et al. [61] | Modified ‘double swab’ technique. | In-house method. | PowerQuant System (Promega GmbH). | PowerPlex ESX 17 Fast Kit. |
| Samie et al. [62] | After the stabbing, both knife handles were swabbed for DNA using a single moist COPAN’s FLOQSwab, covering their entire surface. | Using both the QIAshredder and QIAamp DNA mini kit from Qiagen, DNA was collected from the swabs. | Investigator Quantiplex Kit (Qiagen). | NGM SElect (Applied Biosystem). |
| Szkuta et al. [63] | Polyvinyl chloride tape stubs (in-house). Mini-tape (SceneSafe FAST™). Mini-tape (SceneSafe FAST™). Moistened cotton swab followed by dry cotton swab (150C, Copan). | QIAmp isolation (Qiagen). Chelex (modified method). EZ1 advanced XL (Qiagen). DNA IQ™ (Promega). | ALU assay (in-house); Quantifiler HP (Thermo Fisher Scientific); Quantifiler Trio (Thermo Fisher Scientific). | NGM (Thermo Fisher Scientific); PowerPlex ESX16 Fast (Promega); -NGM SElect (Thermo Fisher Scientific); PowerPlex 21 (Promega). |
| Tanzhaus et al. [64] | Secondary transfer surfaces were swabbed with DNA-free cotton swabs. | Promega Maxwell RSC 16 robot (Promega) with the Promega Maxwell RSC custom total nucleic acid kit. | PowerQuant system (Promega). | Powerplex ESX 17 fast and Powerplex ESI 17 fast (Promega). |
| Thornbury et al. [65] | Wet and dry double swabbing using cotton swabs (Copan) and wetting with a few drops of sterile distilled water was applied to collect DNA from different surfaces. | DNA IQ (Promega, USA). | Quantifiler Trio (Applied Biosystems). | PowerPlex 21 System (Promega). |
| Thornbury et al. [66] | Wet and dry double swabbing using cotton swabs (Copan). | DNA IQ System (Promega). | Quantifiler Trio DNA Quantification Kit (Applied Biosystems). | PowerPlex 21 System (Promega). |
| Reither et al. [67] | Wet and dry double swabbing. | DNA IQ system (Promega). | Quantifiler Trio DNA Quantifica-tion Kit (Applied Biosystems). | PowerPlex 21 System (Promega) |
| Carrara et al. [7] | Wet and dry double swabbing. | QIAshredder/QIAamp DNA extraction procedure (Qiagen). | Investigator Quantiplex HYres Kit (Qiagen). | AmpFLSTR NGM SElect PCR amplification kit (Life Technologies). |
| McCrane and Mulligan [68] | Single swab. | Isohelix XME-50 Xtreme DNA Isolation kit (Midwest Scientific, Fenton, Missouri). | Samples were tested using the Amelogenin qPCR assay. | Not performed. |
| Onofri et al. [69] | Adhesive tape. | Phenol–chloroform organic method. | PowerQuant System (Promega). | PowerPlex ESX17 Fast kit (Promega). |
| Monkman et al. [70] | Wet–dry swabbing technique. | DNA IQ system (Promega). | Quantifiler Trio DNA Quantifica-tion Kit (Applied Biosystems). | PowerPlex 21 System (Promega) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sessa, F.; Pomara, C.; Esposito, M.; Grassi, P.; Cocimano, G.; Salerno, M. Indirect DNA Transfer and Forensic Implications: A Literature Review. Genes 2023, 14, 2153. https://doi.org/10.3390/genes14122153
Sessa F, Pomara C, Esposito M, Grassi P, Cocimano G, Salerno M. Indirect DNA Transfer and Forensic Implications: A Literature Review. Genes. 2023; 14(12):2153. https://doi.org/10.3390/genes14122153
Chicago/Turabian StyleSessa, Francesco, Cristoforo Pomara, Massimiliano Esposito, Patrizia Grassi, Giuseppe Cocimano, and Monica Salerno. 2023. "Indirect DNA Transfer and Forensic Implications: A Literature Review" Genes 14, no. 12: 2153. https://doi.org/10.3390/genes14122153








