The Euphrates Poplar Responses to Abiotic Stress and Its Unique Traits in Dry Regions of China (Xinjiang and Inner Mongolia): What Should We Know?
Abstract
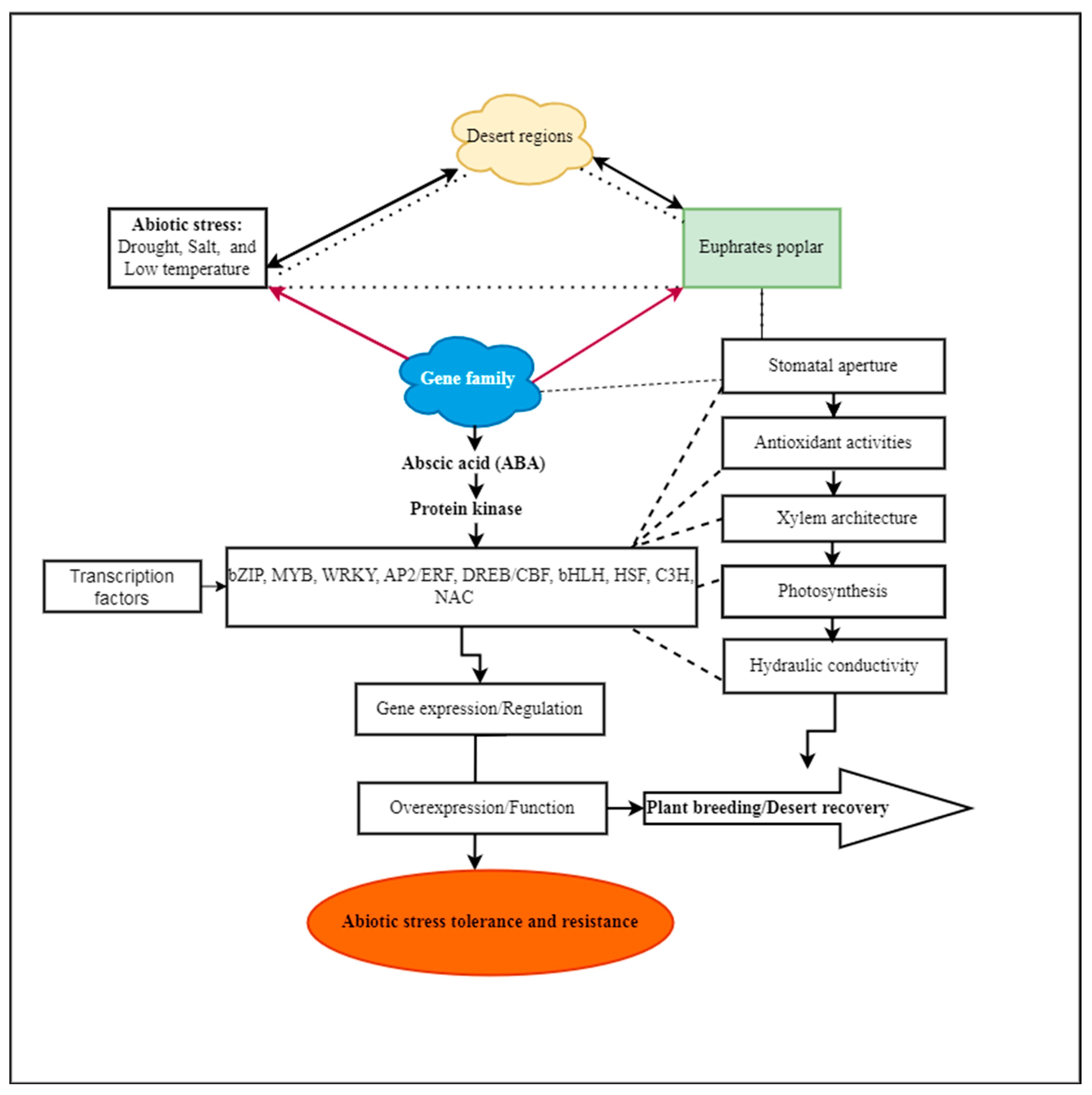
:1. Introduction
2. P. euphratica Gene Family Identification and Characterization
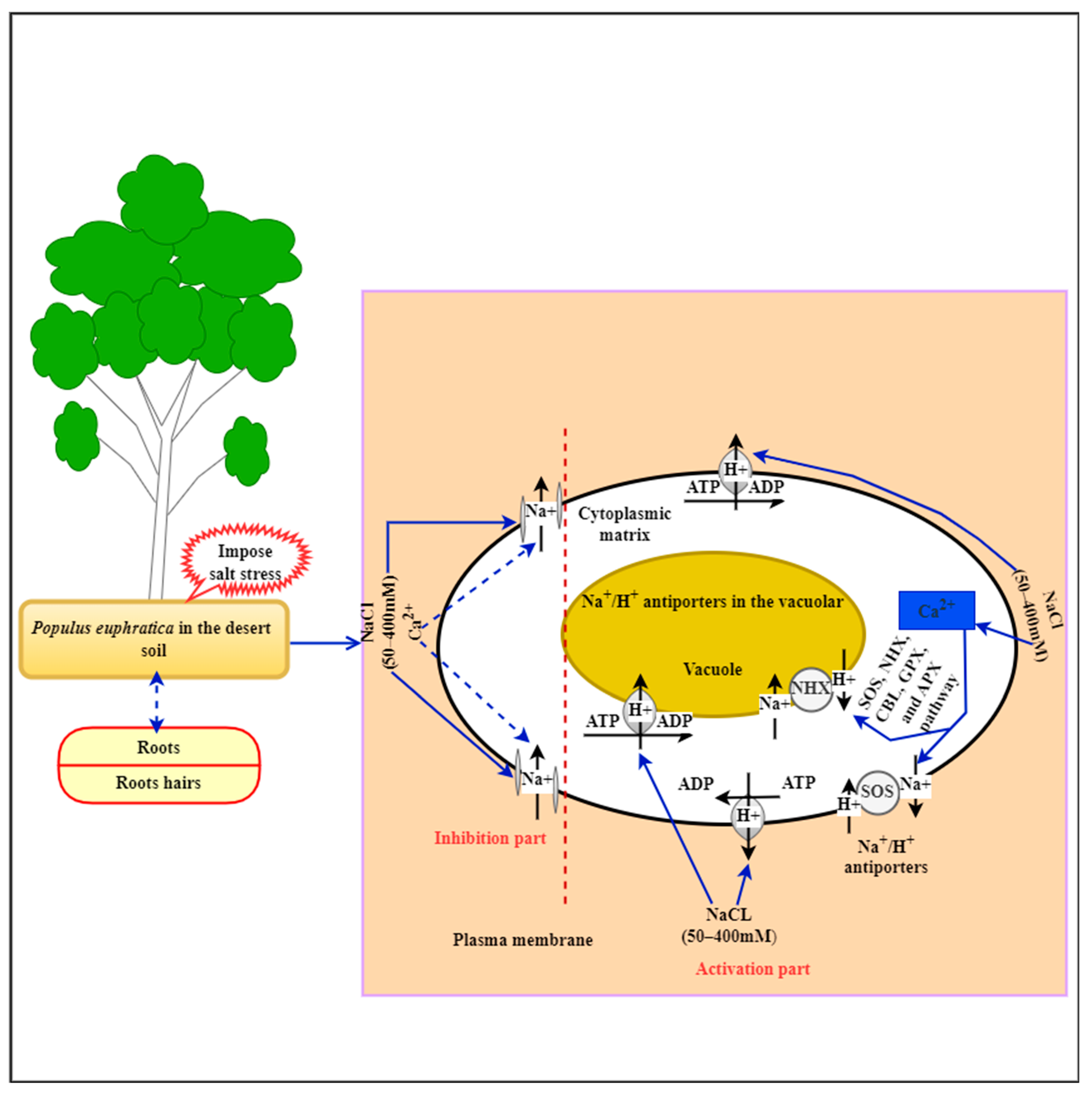
3. Effect of Salt Stress in P. euphratica
4. Effect of Drought Stress on the Physiology of P. euphratica
5. Mechanism of Abiotic Stress Tolerance in P. euphratica
6. P. euphratica, a Naturally Stress-Tolerant Tree, as a Source for Finding Stress-Adapted Genes
7. Effect of Water Stress on Hydraulic Traits in P. euphratica
8. Conclusions and Future Research Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, S.; Li, J.; Wang, S.; Hüttermann, A.; Altman, A. Salt, nutrient uptake and transport, and ABA of Populus euphratica; a hybrid in response to increasing soil NaCl. Trees 2001, 15, 186–194. [Google Scholar] [CrossRef]
- Huang, X.; Lv, R.; Zhou, Z.; Fan, M.; Bai, Y.; Ding, Y.; Yang, G. CiteSpace Software Visualization Analyses of the Last Thirty Years of Research on Populus euphratica. Forests 2023, 14, 714. [Google Scholar] [CrossRef]
- Tavakoli Neko, H.; Shirvani, A.; Assareh, M.H.; Morshedloo, M.R. Physiological Response to Salinity Stress in Various Populus euphratica Oliv. Ecotypes in Iran. Ecopersia 2019, 7, 97–103. Available online: http://ecopersia.modares.ac.ir/article-24-25997-en.html1 (accessed on 15 October 2023).
- Kansu, Ç.; Kaya, Z. Genetic diversity of marginal populations of Oliv. from highly fragmented river ecosystems. Silvae Genet. 2020, 69, 139–151. [Google Scholar] [CrossRef]
- Wang, T.Y.; Wang, P.; Wang, Z.L.; Niu, G.Y.; Yu, J.J.; Ma, N.; Wu, Z.N.; Pozdniakov, S.P.; Yan, D.H. Drought adaptability of phreatophytes: Insight from vertical root distribution in drylands of China. J. Plant Ecol. 2021, 14, 1128–1142. [Google Scholar] [CrossRef]
- Qiu, Q.; Ma, T.; Hu, Q.; Liu, B.; Wu, Y.; Zhou, H.; Wang, Q.; Wang, J.; Liu, J. Genome-scale transcriptome analysis of the desert poplar, Populus euphratica. Tree Physiol. 2011, 31, 452–461. [Google Scholar] [CrossRef] [PubMed]
- Eusemann, P.; Petzold, A.; Thevs, N.; Schnittler, M. Growth patterns and genetic structure of Populus euphratica Oliv.(Salicaceae) forests in NW China–Implications for conservation and management. For. Ecol. Manag. 2013, 297, 27–36. [Google Scholar] [CrossRef]
- Monda, Y.; Miki, N.; Yoshikawa, K. Stand structure and regeneration of Populus euphratica forest in the lower reaches of the Heihe River, NW China. Landsc. Ecol. Eng. 2008, 4, 115–124. [Google Scholar] [CrossRef]
- Lam, T.Y.; Kleinn, C.; Coenradie, B. Double sampling for stratification for the monitoring of sparse tree populations: The example of Populus euphratica Oliv. forests at the lower reaches of Tarim River, Southern Xinjiang, China. Environ. Monit. Assess. 2011, 175, 45–61. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, P.; Feng, Q.; Wang, T.; Wang, T. The spatiotemporal responses of Populus euphratica to global warming in Chinese oases between 1960 and 2015. J. Geogr. Sci. 2018, 28, 579–594. [Google Scholar] [CrossRef]
- Du, F.K.; Xu, F.; Qu, H.; Feng, S.; Tang, J.; Wu, R. Exploiting the transcriptome of Euphrates Poplar, Populus euphratica (Salicaceae) to develop and characterize new EST-SSR markers and construct an EST-SSR database. PLoS ONE 2013, 8, e61337. [Google Scholar] [CrossRef]
- Gai, Z.; Zhai, J.; Chen, X.; Jiao, P.; Zhang, S.; Sun, J.; Qin, R.; Liu, H.; Wu, Z.; Li, Z. Phylogeography reveals geographic and environmental factors driving genetic differentiation of Populus sect. Turanga in Northwest China. Front. Plant Sci. 2021, 12, 5083. [Google Scholar] [CrossRef] [PubMed]
- Phan, C.T.; Jörgensen, J.; Jouve, L.; Hausman, J.F.; Polle, A.; Teichmann, T. Micropropagation of Populus euphratica Olivier. Belg. J. Bot. 2004, 137, 175–180. Available online: https://www.jstor.org/stable/20794551 (accessed on 15 October 2023).
- Ma, T.; Wang, K.; Hu, Q.; Xi, Z.; Wan, D.; Wang, Q.; Feng, J.; Jiang, D.; Ahani, H.; Abbott, R.J.; et al. Ancient polymorphisms and divergence hitchhiking contribute to genomic islands of divergence within a poplar species complex. Proc. Natl. Acad. Sci. USA 2018, 115, E236–E243. [Google Scholar] [CrossRef] [PubMed]
- Overdieck, D.; Ziche, D.; Yu, R. Gas exchange of Populus euphratica leaves in a riparian zone. J. Arid Land 2013, 5, 531–541. [Google Scholar] [CrossRef]
- Lu, K.Q.; Qin, F.; Li, Y.; Xie, G.; Li, J.F.; Cui, Y.-M.; Ferguson, D.K.; Yao, Y.F.; Wang, G.H.; Wang, Y.F. A new approach to interpret vegetation and ecosystem changes through time by establishing a correlation between surface pollen and vegetation types in the eastern central Asian desert. Palaeogeography 2020, 551, 109762. [Google Scholar] [CrossRef]
- Zhang, P.; Deng, X.; Long, A.; Xu, H.; Ye, M.; Li, J. Change in spatial distribution patterns and regeneration of Populus euphratica under different surface soil salinity conditions. Sci. Rep. 2019, 9, 9123. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Polle, A. Salinity tolerance of Populus. Plant Biol. 2010, 12, 317–333. [Google Scholar] [CrossRef]
- Junghans, U.; Polle, A.; Düchting, P.; Weiler, E.; Kuhlman, B.; Gruber, F.; Teichmann, T. Adaptation to high salinity in poplar involves changes in xylem anatomy and auxin physiology. Plant Cell Environ. 2006, 29, 1519–1531. [Google Scholar] [CrossRef] [PubMed]
- Yue, H.; Zhao, L.; Yang, D.; Zhang, M.; Wu, J.; Zhao, Z.; Xing, X.; Zhang, L.; Qin, Y.; Guo, F. Comparative analysis of the endophytic bacterial diversity of Populus euphratica oliv. in environments of different salinity intensities. Microbiol. Spectr. 2022, 10, e00500–e00522. [Google Scholar] [CrossRef] [PubMed]
- Tavakoli Neko, H.; Shirvany, A.; Assareh, M.; Naghavi, M.; Pessarakli, M.; Pourmeidani, A. Effects of NaCl on growth, yield and ion concentration of various Populus euphratica Oliv. ecotypes in Iran. Desert 2018, 23, 189–198. Available online: https://jdesert.ut.ac.ir/article_69116_072933987c764fffcebcd5fe41363399.pdf (accessed on 15 September 2023).
- Rajput, V.D.; Minkina, T.; Yaning, C.; Sushkova, S.; Chapligin, V.A.; Mandzhieva, S. A review on salinity adaptation mechanism and characteristics of Populus euphratica, a boon for arid ecosystems. Acta Ecol. Sin. 2016, 36, 497–503. [Google Scholar] [CrossRef]
- Polle, A.; Chen, S.L.; Eckert, C.; Harfouche, A. Engineering Drought Resistance in Forest Trees. Front. Plant Sci. 2018, 9, 1875. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Loredana, F.C.; Pasqualina, W.; Amodio, F.; Giovanni, P.; Petronia, C. Plant Genes for Abiotic Stress; IntechOpen: Rijeka, Crotia, 2011; pp. 285–308. [Google Scholar] [CrossRef]
- Saibo, N.J.; Lourenço, T.; Oliveira, M.M. Transcription factors and regulation of photosynthetic and related metabolism under environmental stresses. Ann. Bot. 2009, 103, 609–623. [Google Scholar] [CrossRef] [PubMed]
- Carthew, R.W. Gene Regulation and Cellular Metabolism: An Essential Partnership. Trends Genet. 2021, 37, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Chen, X.; Gai, Z.; Zhai, J.; Guo, X.; Han, X.; Zhang, S.; Wu, Z.; Li, Z. Phenotypic Diversity and Variation in Natural Populus euphratica Populations Shaped by Environmental Factors. Contemp. Probl. Ecol. 2023, 16, 230–252. [Google Scholar] [CrossRef]
- Soleimani, A.; Etemad, V.; Calagari, M.; Namiranian, M.; Shirvany, A. Influence of climatic factors on fruit morphological traits in Populus euphratica Oliv. Ann. For. Res. 2014, 57, 31–38. [Google Scholar] [CrossRef]
- Wiehle, M.; Eusemann, P.; Thevs, N.; Schnittler, M. Root suckering patterns in Populus euphratica (Euphrates poplar, Salicaceae). Trees 2009, 23, 991–1001. [Google Scholar] [CrossRef]
- Thomas, F.M. Ecology of Phreatophytes; Springer: Berlin, Germany, 2013; pp. 335–375. [Google Scholar] [CrossRef]
- Zhai, J.; Li, Z.; Si, J.; Zhang, S.; Han, X.; Chen, X. Structural and Functional Responses of the Heteromorphic Leaves of Different Tree Heights on Populus euphratica Oliv. to Different Soil Moisture Conditions. Plants 2022, 11, 2376. [Google Scholar] [CrossRef]
- Li, D.; Si, J.; Zhang, X.; Gao, Y.; Wang, C.; Luo, H.; Qin, J.; Gao, G. Hydraulic characteristics of Populus euphratica in an arid environment. Forests 2019, 10, 407. [Google Scholar] [CrossRef]
- Yu, L.; Dong, H.; Li, Z.; Han, Z.; Korpelainen, H.; Li, C. Species-specific responses to drought, salinity and their interactions in Populus euphratica and P. pruinosa seedlings. Plant Ecol. 2020, 13, 563–573. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Zhou, H.; Hao, X.; Zhu, C.; Fu, A.; Yang, Y.; Li, W. Research advances in plant physiology and ecology of desert riparian forests under drought stress. Forests 2022, 13, 619. [Google Scholar] [CrossRef]
- Jia, H.; Liu, G.; Li, J.; Zhang, J.; Sun, P.; Zhao, S.; Zhou, X.; Lu, M.; Hu, J. Genome resequencing reveals demographic history and genetic architecture of seed salinity tolerance in Populus euphratica. J. Exp. Bot. 2020, 71, 4308–4320. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Huisingh, D. Combating desertification in China: Monitoring, control, management and revegetation. J. Clean. Prod. 2018, 182, 765–775. [Google Scholar] [CrossRef]
- Ma, J.X.; Huang, X.; Li, W.H.; Zhu, C.G. Sap flow and trunk maximum daily shrinkage (MDS) measurements for diagnosing water status of Populus euphratica in an inland river basin of Northwest China. Ecohydrology 2013, 6, 994–1000. [Google Scholar] [CrossRef]
- Zhang, Y.; Hao, X.; Sun, H.; Hua, D.; Qin, J. How Populus euphratica utilizes dew in an extremely arid region. Plant Soil 2019, 443, 493–508. [Google Scholar] [CrossRef]
- Fan, X.; Hao, X.; Zhang, S.; Zhao, Z.; Zhang, J.; Li, Y. Populus euphratica counteracts drought stress through the dew coupling and root hydraulic redistribution processes. Ann. Bot. 2023, 131, 451–461. [Google Scholar] [CrossRef]
- Shengyue, F.; Lihua, Z. Desertification control in China: Possible solutions. Ambio 2001, 30, 384–385. [Google Scholar] [CrossRef]
- Wang, L.; Zhu, G.; Lin, X.; Liu, Y.; Zhao, K.; Sang, L.; Zhang, W.; Qiu, D.; Zhang, Z.; Sun, Z. Water use patterns of dominant species of riparian wetlands in arid areas. Hydrol. Process. 2023, 37, e14835. [Google Scholar] [CrossRef]
- Li, J.; Song, Q.; Zuo, Z.-F.; Liu, L. MicroRNA398: A Master Regulator of Plant Development and Stress Responses. Int. J. Mol. Sci. 2022, 23, 10803. [Google Scholar] [CrossRef] [PubMed]
- Lotfi, A.; Pervaiz, T.; Jiu, S.; Faghihi, F.; Jahanbakhshian, Z.; Khorzoghi, E.G.; Fang, J.; Seyedi, S.M. Role of microRNAs and their target genes in salinity response in plants. Plant Growth Regul. 2017, 82, 377–390. [Google Scholar] [CrossRef]
- Chen, J.; Tian, Q.; Pang, T.; Jiang, L.; Wu, R.; Xia, X.; Yin, W. Deep-sequencing transcriptome analysis of low temperature perception in a desert tree, Populus euphratica. BMC Genom. 2014, 15, 326. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Jiang, Z.; Li, Z.; Jiao, P.; Zhai, J.; Liu, S.; Han, X.; Zhang, S.; Sun, J.; Gai, Z. Multi-omics analysis reveals spatiotemporal regulation and function of heteromorphic leaves in Populus. Plant Physiol. 2023, 192, 188–204. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Liang, H.; Yan, D.; Zhao, Y.; Han, X.; Carlson, J.E.; Xia, X.; Yin, W. Populus euphratica: The transcriptomic response to drought stress. Plant Mol. Biol. 2013, 83, 539–557. [Google Scholar] [CrossRef]
- Tahmasebi, A.; Niazi, A.; Akrami, S. Integration of meta-analysis, machine learning and systems biology approach for investigating the transcriptomic response to drought stress in Populus species. Sci. Rep. 2023, 13, 847. [Google Scholar] [CrossRef]
- Cobb, J.N.; Juma, R.U.; Biswas, P.S.; Arbelaez, J.D.; Rutkoski, J.; Atlin, G.; Hagen, T.; Quinn, M.; Ng, E.H. Enhancing the rate of genetic gain in public-sector plant breeding programs: Lessons from the breeder’s equation. Theor. Appl. Genet. 2019, 132, 627–645. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, Y.; Du, J.; Guo, X.; Wen, W.; Gu, S.; Wang, J.; Fan, J. Crop phenomics: Current status and perspectives. Front. Plant Sci. 2019, 10, 714. [Google Scholar] [CrossRef]
- Yan, D.H.; Fenning, T.; Tang, S.; Xia, X.; Yin, W. Genome-wide transcriptional response of Populus euphratica to long-term drought stress. Plant Sci. 2012, 195, 24–35. [Google Scholar] [CrossRef]
- Rosso, L.; Cantamessa, S.; Bergante, S.; Biselli, C.; Fricano, A.; Chiarabaglio, P.M.; Gennaro, M.; Nervo, G.; Secchi, F.; Carra, A. Responses to Drought Stress in Poplar: What Do We Know and What Can We Learn? Life 2023, 13, 533. [Google Scholar] [CrossRef]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [CrossRef] [PubMed]
- Evans, L.M.; Slavov, G.T.; Rodgers-Melnick, E.; Martin, J.; Ranjan, P.; Muchero, W.; Brunner, A.M.; Schackwitz, W.; Gunter, L.; Chen, J.-G.; et al. Population genomics of Populus trichocarpa identifies signatures of selection and adaptive trait associations. Nat. Genet. 2014, 46, 1089–1096. [Google Scholar] [CrossRef]
- Slavov, G.T.; DiFazio, S.P.; Martin, J.; Schackwitz, W.; Muchero, W.; Rodgers-Melnick, E.; Lipphardt, M.F.; Pennacchio, C.P.; Hellsten, U.; Pennacchio, L.A.; et al. Genome resequencing reveals multiscale geographic structure and extensive linkage disequilibrium in the forest tree Populus trichocarpa. New Phytol. 2012, 196, 713–725. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Wang, J.; Zhou, G.; Yue, Z.; Hu, Q.; Chen, Y.; Liu, B.; Qiu, Q.; Wang, Z.; Zhang, J. Genomic insights into salt adaptation in a desert poplar. Nat. Commun. 2013, 4, 2797. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.J.; Gao, L.Z. The complete chloroplast genome sequence of desert poplar (Populus euphratica). Mitochondrial DNA Part A 2016, 27, 721–723. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, Y.; Zhang, J.; Ma, X.; Li, Y.; Li, M.; Wang, D.; Kang, M.; Wu, H.; Yang, Y.; et al. Improved genome assembly provides new insights into genome evolution in a desert poplar (Populus euphratica). Mol. Ecol. Resour. 2020, 20, 781–794. [Google Scholar] [CrossRef]
- Zamudio, K.R.; Bell, R.C.; Mason, N.A. Phenotypes in phylogeography: Species’ traits, environmental variation, and vertebrate diversification. Proc. Natl. Acad. Sci. USA 2016, 113, 8041–8048. [Google Scholar] [CrossRef]
- Gregory, T.R. Understanding Natural Selection: Essential Concepts and Common Misconceptions. Evol. Educ. 2009, 2, 156–175. [Google Scholar] [CrossRef]
- Sarkar, P.; Bosneaga, E.; Auer, M. Plant cell walls throughout evolution: Towards a molecular understanding of their design principles. J. Exp. Bot. 2009, 60, 3615–3635. [Google Scholar] [CrossRef]
- Meyer, R.S.; Purugganan, M.D. Evolution of crop species: Genetics of domestication and diversification. Nat. Rev. Genet. 2013, 14, 840–852. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xia, X.; Yin, W. Expression profiling and functional characterization of a DREB2-type gene from Populus euphratica. Biochem. Biophys. Res. Commun. 2009, 378, 483–487. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xue, B.; Xia, X.; Yin, W. A novel calcium-dependent protein kinase gene from Populus euphratica, confers both drought and cold stress tolerance. Biochem. Biophys. Res. Commun. 2013, 441, 630–636. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Du, C.; Li, A.; Xia, X.; Yin, W.; Chen, J. Salicylic Acid Alleviated Salt Damage of Populus euphratica: A Physiological and Transcriptomic Analysis. Forests 2019, 10, 423. [Google Scholar] [CrossRef]
- He, M.; He, C.Q.; Ding, N.Z. Abiotic stresses: General defenses of land plants and chances for engineering multistress tolerance. Front. Plant Sci. 2018, 9, 1771. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jia, H.; Li, J.; Li, Y.; Lu, M.; Hu, J. Molecular evolution and expression divergence of the Populus euphratica Hsf genes provide insight into the stress acclimation of desert poplar. Sci. Rep. 2016, 6, 30050. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Wang, C.; Han, X.; Tang, S.; Liu, S.; Xia, X.; Yin, W. A novel bHLH transcription factor PebHLH35 from Populus euphratica confers drought tolerance through regulating stomatal development, photosynthesis and growth in Arabidopsis. Biochem. Biophys. Res. Commun. 2014, 450, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Xu, J.; Qu, W.; Han, X.; Qiu, C.; Gai, Z.; Zhai, J.; Qin, R.; Liu, H.; Wu, Z.; et al. Genome-wide analysis of R2R3-MYB transcription factors reveals their differential responses to drought stress and ABA treatment in desert poplar (Populus euphratica). Gene 2023, 855, 147124. [Google Scholar] [CrossRef]
- Ma, J.; Lu, J.; Xu, J.; Duan, B.; He, X.; Liu, J. Genome-wide Identification of WRKY Genes in the Desert Poplar Populus euphratica and Adaptive Evolution of the Genes in Response to Salt Stress. Evol. Bioinform. 2015, 11s1, 47–55. [Google Scholar] [CrossRef]
- Yao, J.; Shen, Z.; Zhang, Y.; Wu, X.; Wang, J.; Sa, G.; Zhang, Y.; Zhang, H.; Deng, C.; Liu, J.; et al. Populus euphratica WRKY1 binds the promoter of H+-ATPase gene to enhance gene expression and salt tolerance. J. Exp. Bot. 2019, 71, 1527–1539. [Google Scholar] [CrossRef]
- Jia, H.; Li, J.; Zhang, J.; Ren, Y.; Hu, J.; Lu, M. Genome-wide survey and expression analysis of the stress-associated protein gene family in desert poplar, Populus euphratica. Tree Genet. Genom. 2016, 12, 78. [Google Scholar] [CrossRef]
- Han, S.; Jiao, Z.; Niu, M.-X.; Yu, X.; Huang, M.; Liu, C.; Wang, H.-L.; Zhou, Y.; Mao, W.; Wang, X. Genome-wide comprehensive analysis of the GASA gene family in Populus. Int. J. Mol. Sci. 2021, 22, 12336. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Ma, J.; Fan, D.; Li, C.; Jiang, Y.; Luo, K. Genome-wide Identification of TCP Family Transcription Factors from Populus euphratica and Their Involvement in Leaf Shape Regulation. Sci. Rep. 2016, 6, 32795. [Google Scholar] [CrossRef]
- Liu, N.; Shang, W.; Li, C.; Jia, L.; Wang, X.; Xing, G.; Zheng, W. Evolution of the SPX gene family in plants and its role in the response mechanism to phosphorus stress. Open Biol. 2018, 8, 170231. [Google Scholar] [CrossRef]
- Lu, X.; Zhang, X.; Duan, H.; Lian, C.; Liu, C.; Yin, W.; Xia, X. Three stress-responsive NAC transcription factors from Populus euphratica differentially regulate salt and drought tolerance in transgenic plants. Physiol. Plant. 2018, 162, 73–97. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Dun, H.; Lian, C.; Zhang, X.; Yin, W.; Xia, X. The role of peu-miR164 and its target PeNAC genes in response to abiotic stress in Populus euphratica. Plant Physiol. Biochem. 2017, 115, 418–438. [Google Scholar] [CrossRef]
- Ma, H.S.; Liang, D.; Shuai, P.; Xia, X.L.; Yin, W.L. The salt- and drought-inducible poplar GRAS protein SCL7 confers salt and drought tolerance in Arabidopsis thaliana. J. Exp. Bot. 2010, 61, 4011–4019. [Google Scholar] [CrossRef]
- He, F.; Li, H.G.; Wang, J.J.; Su, Y.; Wang, H.L.; Feng, C.H.; Yang, Y.; Niu, M.X.; Liu, C.; Yin, W.; et al. PeSTZ1, a C2H2-type zinc finger transcription factor from Populus euphratica, enhances freezing tolerance through modulation of ROS scavenging by directly regulating PeAPX2. Plant Biotechnol. J. 2019, 17, 2169–2183. [Google Scholar] [CrossRef]
- Niu, Z.; Li, G.; Hu, H.; Lv, J.; Zheng, Q.; Liu, J.; Wan, D. A gene that underwent adaptive evolution, LAC2 (LACCASE), in Populus euphratica improves drought tolerance by improving water transport capacity. Hort. Res. 2021, 8, 88. [Google Scholar] [CrossRef]
- Su, M.; Zhang, M.; Liu, Y.; Han, Z. Abscisic Acid, Paclobutrazol, and Salicylic Acid Alleviate Salt Stress in Populus talassica× Populus euphratica by Modulating Plant Root Architecture, Photosynthesis, and the Antioxidant Defense System. Forests 2022, 13, 1864. [Google Scholar] [CrossRef]
- Ferreira, S.; Hjernø, K.; Larsen, M.; Wingsle, G.; Larsen, P.; Fey, S.; Roepstorff, P.; SalomÉ Pais, M. Proteome profiling of Populus euphratica Oliv. upon heat stress. Ann. Bot. 2006, 98, 361–377. [Google Scholar] [CrossRef]
- Thoday-Kennedy, E.L.; Jacobs, A.K.; Roy, S.J. The role of the CBL–CIPK calcium signalling network in regulating ion transport in response to abiotic stress. Plant Growth Regul. 2015, 76, 3–12. [Google Scholar] [CrossRef]
- Reddy, A.S.; Ali, G.S.; Celesnik, H.; Day, I.S. Coping with stresses: Roles of calcium-and calcium/calmodulin-regulated gene expression. Plant Cell 2011, 23, 2010–2032. [Google Scholar] [CrossRef]
- Mahajan, S.; Pandey, G.K.; Tuteja, N. Calcium-and salt-stress signaling in plants: Shedding light on SOS pathway. Arch. Biochem. Biophys. 2008, 471, 146–158. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, C.; Tang, R.J.; Xu, H.X.; Lan, W.Z.; Zhao, F.; Luan, S. Calcineurin B-Like proteins CBL4 and CBL10 mediate two independent salt tolerance pathways in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 2421. [Google Scholar] [CrossRef]
- Liu, J.; Ishitani, M.; Halfter, U.; Kim, C.S.; Zhu, J.K. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. USA 2000, 97, 3730–3734. [Google Scholar] [CrossRef]
- Ma, X.; Li, Q.H.; Yu, Y.N.; Qiao, Y.M.; Haq, S.u.; Gong, Z.H. The CBL–CIPK pathway in plant response to stress signals. Int. J. Mol. Sci. 2020, 21, 5668. [Google Scholar] [CrossRef]
- Zhang, H.; Lv, F.; Han, X.; Xia, X.; Yin, W. The calcium sensor PeCBL1, interacting with PeCIPK24/25 and PeCIPK26, regulates Na+/K+ homeostasis in Populus euphratica. Plant Cell Rep. 2013, 32, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.H.; Choi, M.N.; Yoon, K.H.; Kim, K.N. Ectopic expression of SjCBL1, calcineurin B-like 1 gene from Sedirea japonica, rescues the salt and osmotic stress hypersensitivity in Arabidopsis cbl1 mutant. Front. Plant Sci. 2018, 9, 1188. [Google Scholar] [CrossRef] [PubMed]
- Lv, F.; Zhang, H.; Xia, X.; Yin, W. Expression profiling and functional characterization of a CBL-interacting protein kinase gene from Populus euphratica. Plant Cell Rep. 2014, 33, 807–818. [Google Scholar] [CrossRef] [PubMed]
- Neale, D.B.; Martinez-Garcia, P.J.; De La Torre, A.R.; Montanari, S.; Wei, X.X. Novel insights into tree biology and genome evolution as revealed through genomics. Annu. Rev. Plant Biol. 2017, 68, 457–483. [Google Scholar] [CrossRef]
- Tang, S.; Dong, Y.; Liang, D.; Zhang, Z.; Ye, C.Y.; Shuai, P.; Han, X.; Zhao, Y.; Yin, W.; Xia, X. Analysis of the drought stress-responsive transcriptome of black cottonwood (Populus trichocarpa) using deep RNA sequencing. Plant Mol. Biol. Rep. 2015, 33, 424–438. [Google Scholar] [CrossRef]
- Cohen, D.; Bogeat-Triboulot, M.B.; Tisserant, E.; Balzergue, S.; Martin-Magniette, M.L.; Lelandais, G.; Ningre, N.; Renou, J.P.; Tamby, J.P.; Le Thiec, D. Comparative transcriptomics of drought responses in Populus: A meta-analysis of genome-wide expression profiling in mature leaves and root apices across two genotypes. BMC Genom. 2010, 11, 630. [Google Scholar] [CrossRef]
- Pucholt, P.; Sjödin, P.; Weih, M.; Rönnberg-Wästljung, A.C.; Berlin, S. Genome-wide transcriptional and physiological responses to drought stress in leaves and roots of two willow genotypes. BMC Plant Biol. 2015, 15, 244. [Google Scholar] [CrossRef]
- Zhang, C.H.; Ma, T.; Luo, W.C.; Xu, J.M.; Liu, J.Q.; Wan, D.S. Identification of 4CL Genes in Desert Poplars and Their Changes in Expression in Response to Salt Stress. Genes 2015, 6, 901–917. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, D.; Zhang, C.; Xia, X.; Yin, W.; Tian, Q. A putative PP2C-encoding gene negatively regulates ABA signaling in Populus euphratica. PLoS ONE 2015, 10, e0139466. [Google Scholar] [CrossRef]
- Yu, L.; Ma, J.; Niu, Z.; Bai, X.; Lei, W.; Shao, X.; Chen, N.; Zhou, F.; Wan, D. Tissue-specific transcriptome analysis reveals multiple responses to salt stress in Populus euphratica seedlings. Genes 2017, 8, 372. [Google Scholar] [CrossRef]
- Flowers, T.J.; Colmer, T.D. Salinity tolerance in halophytes. New Phytol. 2008, 179, 945–963. [Google Scholar] [CrossRef]
- Brini, F.; Masmoudi, K. Ion Transporters and Abiotic Stress Tolerance in Plants. ISRN Mol. Biol. 2012, 2012, 927436. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Ding, N.; Zhao, X.; Zhao, M.; Chang, Z.; Liu, J.; Zhang, L. Molecular characterization of PeSOS1: The putative Na+/H+ antiporter of Populus euphratica. Plant Mol. Biol. 2007, 65, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, L.; Chen, B.; Qin, Z.; Xiao, Y.; Zhang, Y.; Yao, R.; Liu, H.; Yang, H. Progress in understanding the physiological and molecular responses of Populus to salt stress. Int. J. Mol. Sci. 2019, 20, 1312. [Google Scholar] [CrossRef]
- Ye, C.Y.; Zhang, H.C.; Chen, J.H.; Xia, X.L.; Yin, W.L. Molecular characterization of putative vacuolar NHX-type Na+/H+ exchanger genes from the salt-resistant tree Populus euphratica. Physiol. Plant. 2009, 137, 166–174. [Google Scholar] [CrossRef]
- Xie, Q.; Zhou, Y.; Jiang, X. Structure, Function, and Regulation of the Plasma Membrane Na+/H+ Antiporter Salt Overly Sensitive 1 in Plants. Front. Plant Sci. 2022, 13, 866265. [Google Scholar] [CrossRef]
- Zeng, F.; Yan, H.; Arndt, S.K. Leaf and whole tree adaptations to mild salinity in field grown Populus euphratica. Tree Physiol. 2009, 29, 1237–1246. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, X.; Deng, S.; Zhang, C.; Wang, M.; Ding, M.; Zhao, R.; Shen, X.; Zhou, X.; Lu, C. Extracellular ATP signaling is mediated by H2O2 and cytosolic Ca2+ in the salt response of Populus euphratica cells. PLoS ONE 2012, 7, e53136. [Google Scholar] [CrossRef]
- Hoang, X.L.T.; Nhi, D.N.H.; Thu, N.B.A.; Thao, N.P.; Tran, L.P. Transcription Factors and Their Roles in Signal Transduction in Plants under Abiotic Stresses. Curr. Genom. 2017, 18, 483–497. [Google Scholar] [CrossRef]
- Khoso, M.A.; Hussain, A.; Ritonga, F.N.; Ali, Q.; Channa, M.M.; Alshegaihi, R.M.; Meng, Q.; Ali, M.; Zaman, W.; Brohi, R.D.; et al. WRKY transcription factors (TFs): Molecular switches to regulate drought, temperature, and salinity stresses in plants. Front. Plant Sci. 2022, 13, 39329. [Google Scholar] [CrossRef]
- Jiang, Y.; Tong, S.; Chen, N.; Liu, B.; Bai, Q.; Chen, Y.; Bi, H.; Zhang, Z.; Lou, S.; Tang, H.; et al. The PalWRKY77 transcription factor negatively regulates salt tolerance and abscisic acid signaling in Populus. Plant Cell Biol. 2021, 105, 1258–1273. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2006, 58, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Ullah, A.; Manghwar, H.; Shaban, M.; Khan, A.H.; Akbar, A.; Ali, U.; Ali, E.; Fahad, S. Phytohormones enhanced drought tolerance in plants: A coping strategy. Environ. Sci. Pollut. Res. 2018, 25, 33103–33118. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, S.; Wang, X.; Mubeen, I.; Kamran, M.; Kanwal, I.; Díaz, G.A.; Abbas, A.; Parveen, A.; Atiq, M.N.; Alshaya, H. Phytohormones trigger drought tolerance in crop plants: Outlook and future perspectives. Front. Plant Sci. 2022, 12, 799318. [Google Scholar] [CrossRef] [PubMed]
- Egamberdieva, D.; Wirth, S.J.; Alqarawi, A.A.; Abd Allah, E.F.; Hashem, A. Phytohormones and Beneficial Microbes: Essential Components for Plants to Balance Stress and Fitness. Front. Microbiol. 2017, 8, 2104. [Google Scholar] [CrossRef]
- Wang, S.; Ouyang, L.; Ju, X.; Zhang, L.; Zhang, Q.; Li, Y. Survey of plant drought-resistance promoting bacteria from Populus euphratica tree living in arid area. Indian J. Microbiol. 2014, 54, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Mawar, R.; Ranawat, M.; Sharma, S.K.; Sayyed, R. Plant Growth Promoting Microorganisms of Arid Region; Springer: Singapore, 2023; pp. 1–25. [Google Scholar] [CrossRef]
- Dimkpa, C.; Weinand, T.; Asch, F. Plant–rhizobacteria interactions alleviate abiotic stress conditions. Plant Cell Environ. 2009, 32, 1682–1694. [Google Scholar] [CrossRef] [PubMed]
- Gyaneshwar, P.; Naresh Kumar, G.; Parekh, L.; Poole, P. Role of soil microorganisms in improving P nutrition of plants. Plant Soil 2002, 245, 83–93. [Google Scholar] [CrossRef]
- Rodriguez, P.A.; Rothballer, M.; Chowdhury, S.P.; Nussbaumer, T.; Gutjahr, C.; Falter-Braun, P. Systems biology of plant-microbiome interactions. Mol. Plant 2019, 12, 804–821. [Google Scholar] [CrossRef]
- Boro, M.; Sannyasi, S.; Chettri, D.; Verma, A.K. Microorganisms in biological control strategies to manage microbial plant pathogens: A review. Arch. Microbiol. 2022, 204, 666. [Google Scholar] [CrossRef]
- Xia, Z.; He, Y.; Yu, L.; Li, Z.; Korpelainen, H.; Li, C. Revealing interactions between root phenolic metabolomes and rhizosphere bacterial communities in Populus euphratica plantations. Biol. Fertil. Soils 2021, 57, 421–434. [Google Scholar] [CrossRef]
- Hultine, K.R.; Grady, K.C.; Wood, T.E.; Shuster, S.M.; Stella, J.C.; Whitham, T.G. Climate change perils for dioecious plant species. Nat. Plants 2016, 2, 16109. [Google Scholar] [CrossRef]
- Bellabarba, A.; Fagorzi, C.; diCenzo, G.C.; Pini, F.; Viti, C.; Checcucci, A. Deciphering the symbiotic plant microbiome: Translating the most recent discoveries on rhizobia for the improvement of agricultural practices in metal-contaminated and high saline lands. Agronomy 2019, 9, 529. [Google Scholar] [CrossRef]
- Gazolla Volpiano, C.; Lisboa, B.; Granada, C.; São José, J.; Oliveira, A.; Beneduzi, A.; Perevalova, Y.; Passaglia, L.; Vargas, L. Rhizobia for Biological Control of Plant Diseases; Springer: Singapore, 2019; pp. 315–336. [Google Scholar] [CrossRef]
- Rozahon, M.; Ismayil, N.; Hamood, B.; Erkin, R.; Abdurahman, M.; Mamtimin, H.; Abdukerim, M.; Lal, R.; Rahman, E. Rhizobium populi sp. nov., an endophytic bacterium isolated from Populus euphratica. Int. J. Syst. Evol. Microbiol. 2014, 64, 3215–3221. [Google Scholar] [CrossRef] [PubMed]
- Tuo, Y.; Dong, Z.; Wang, X.; Gao, B.; Zhu, C.; Tuo, F. Metagenomics reveal correlations between microbial organisms in soils and the health of Populus euphratica. Front. Microbiol. 2020, 11, 2095. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.; He, Y.; Xu, J.; Zhu, Z.; Korpelainen, H.; Li, C. Rhizosphere microbe populations but not root traits induced by drought in Populus euphratica males. Soil Ecol. Lett. 2023, 5, 220152. [Google Scholar] [CrossRef]
- Juvany, M.; Munné-Bosch, S. Sex-related differences in stress tolerance in dioecious plants: A critical appraisal in a physiological context. J. Exp. Bot. 2015, 66, 6083–6092. [Google Scholar] [CrossRef]
- Hukin, D.; Cochard, H.; Dreyer, E.; Thiec, D.L.; Bogeat-Triboulot, M.B. Cavitation vulnerability in roots and shoots: Does Populus euphratica Oliv., a poplar from arid areas of Central Asia, differ from other poplar species? J. Exp. Bot. 2005, 56, 2003–2010. [Google Scholar] [CrossRef]
- Bogeat-Triboulot, M.B.A.; Brosché, M.; Renaut, J.; Jouve, L.; Le Thiec, D.; Fayyaz, P.; Vinocur, B.; Witters, E.; Laukens, K.; Teichmann, T.; et al. Gradual Soil Water Depletion Results in Reversible Changes of Gene Expression, Protein Profiles, Ecophysiology, and Growth Performance in Populus euphratica, a Poplar Growing in Arid Regions. Plant Physiol. 2006, 143, 876–892. [Google Scholar] [CrossRef]
- He, Y.; Lin, X.; Wang, L.; Ma, X.; Fang, L.; Xia, Z. Effects of long-term irrigation on soil phosphorus fractions and microbial communities in Populus euphratica plantations. For. Res. 2023, 3, 17. [Google Scholar] [CrossRef]
- Fan, Y.; Lv, G.; Chen, Y.; Chang, Y.; Li, Z. Differential effects of cow dung and its biochar on Populus euphratica soil phosphorus effectiveness, bacterial community diversity and functional genes for phosphorus conversion. Front. Plant Sci. 2023, 14, 1242469. [Google Scholar] [CrossRef]
- Bahadur, A.; Batool, A.; Nasir, F.; Jiang, S.; Mingsen, Q.; Zhang, Q.; Pan, J.; Liu, Y.; Feng, H. Mechanistic Insights into Arbuscular Mycorrhizal Fungi-Mediated Drought Stress Tolerance in Plants. Int. J. Mol. Sci. 2019, 20, 4199. [Google Scholar] [CrossRef]
- Zou, Y.N.; Qin, Q.Y.; Ma, W.Y.; Zhou, L.J.; Wu, Q.S.; Xu, Y.J.; Kuča, K.; Hashem, A.; Al-Arjani, A.B.F.; Almutairi, K.F.; et al. Metabolomics reveals arbuscular mycorrhizal fungi-mediated tolerance of walnut to soil drought. BMC Plant Biol. 2023, 23, 118. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Zou, Y.N.; Kuča, K.; Hashem, A.; Abd Allah, E.F.; Wu, Q.S. Elucidating the Mechanisms Underlying Enhanced Drought Tolerance in Plants Mediated by Arbuscular Mycorrhizal Fungi. Front. Microbiol. 2021, 12, 809473. [Google Scholar] [CrossRef] [PubMed]
- Madouh, T.A.; Quoreshi, A.M. The Function of Arbuscular Mycorrhizal Fungi Associated with Drought Stress Resistance in Native Plants of Arid Desert Ecosystems: A Review. Diversity 2023, 15, 391. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, Y.; Cai, B.; Jie, W.; Lv, D. The arbuscular mycorrhizal symbiotic status of Populus euphratica, a drought resistant tree species from arid lands. Ecohydrology 2013, 6, 1001–1008. [Google Scholar] [CrossRef]
- Chaves, M.M.; Flexas, J.; Pinheiro, C. Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Ann. Bot. 2008, 103, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Meng, H.; Zhao, J.; Yang, Y.; Diao, K.; Zheng, G.; Li, T.; Dai, X.; Li, J. PeGSTU58, a Glutathione S-Transferase from Populus euphratica, Enhances Salt and Drought Stress Tolerance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2023, 24, 9354. [Google Scholar] [CrossRef]
- Hull, J.C.; Neufeld, H.S.; Gilliam, F.S. Plant Ecology, 2nd ed.; Elsevier: Oxford, UK, 2019; pp. 528–548. [Google Scholar] [CrossRef]
- Hatfield, J.L.; Dold, C. Water-use efficiency: Advances and challenges in a changing climate. Front. Plant Sci. 2019, 10, 103. [Google Scholar] [CrossRef]
- Pineda-García, F.; Paz, H.; Meinzer, F.C. Drought resistance in early and late secondary successional species from a tropical dry forest: The interplay between xylem resistance to embolism, sapwood water storage and leaf shedding. Plant Cell Environ. 2013, 36, 405–418. [Google Scholar] [CrossRef]
- Verslues, P.E.; Agarwal, M.; Katiyar-Agarwal, S.; Zhu, J.; Zhu, J.K. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 2006, 45, 523–539. [Google Scholar] [CrossRef]
- Akıncı, Ş.; Lösel, D.M. Plant Water-Stress Response Mechanisms; IntechOpen: Rijeka, Crotia, 2012; pp. 16–42. [Google Scholar] [CrossRef]
- Iqbal, A.; Wang, T.; Wu, G.; Tang, W.; Zhu, C.; Wang, D.; Li, Y.; Wang, H. Physiological and transcriptome analysis of heteromorphic leaves and hydrophilic roots in response to soil drying in desert Populus euphratica. Sci. Rep. 2017, 7, 12188. [Google Scholar] [CrossRef]
- Maherali, H.; Pockman, W.T.; Jackson, R.B. Adaptive variation in the vulnerability of woody plants to xylem cavitation. Ecology 2004, 85, 2184–2199. [Google Scholar] [CrossRef]
- Klein, T.; Zeppel, M.J.; Anderegg, W.R.; Bloemen, J.; De Kauwe, M.G.; Hudson, P.; Ruehr, N.K.; Powell, T.L.; von Arx, G.; Nardini, A. Xylem embolism refilling and resilience against drought-induced mortality in woody plants: Processes and trade-offs. Ecol. Res. 2018, 33, 839–855. [Google Scholar] [CrossRef]
- Skelton, R.P.; Anderegg, L.D.; Diaz, J.; Kling, M.M.; Papper, P.; Lamarque, L.J.; Delzon, S.; Dawson, T.E.; Ackerly, D.D. Evolutionary relationships between drought-related traits and climate shape large hydraulic safety margins in western North American oaks. Proc. Nat. Acad. Sci. USA 2021, 118, e2008987118. [Google Scholar] [CrossRef] [PubMed]
- Pratt, R.B.; Jacobsen, A.L. Conflicting demands on angiosperm xylem: Tradeoffs among storage, transport and biomechanics. Plant Cell Environ. 2017, 40, 897–913. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Xu, J.; Shao, X.; Luo, W.; Niu, Z.; Gao, C.; Wan, D. A novel gene coding γ-aminobutyric acid transporter may improve the tolerance of Populus euphratica to adverse environments. Front. Plant Sci. 2019, 10, 1083. [Google Scholar] [CrossRef]
- Cesarino, I.; Dello Ioio, R.; Kirschner, G.K.; Ogden, M.S.; Picard, K.L.; Rast-Somssich, M.I.; Somssich, M. Plant science’s next top models. Ann. Bot. 2020, 126, 1–23. [Google Scholar] [CrossRef]
- Jansson, S.; Douglas, C.J. Populus: A model system for plant biology. Annu. Rev. Plant Biol. 2007, 58, 435–458. [Google Scholar] [CrossRef]
- Sorce, C.; Giovannelli, A.; Sebastiani, L.; Anfodillo, T. Hormonal signals involved in the regulation of cambial activity, xylogenesis and vessel patterning in trees. Plant Cell Rep. 2013, 32, 885–898. [Google Scholar] [CrossRef]
- Balfagón, D.; Zandalinas, S.I.; Mittler, R.; Gómez-Cadenas, A. High temperatures modify plant responses to abiotic stress conditions. Physiol. Plant. 2020, 170, 335–344. [Google Scholar] [CrossRef]
- Zhu, X.; Liu, S.; Sun, L.; Song, F.; Liu, F.; Li, X. Cold tolerance of photosynthetic electron transport system is enhanced in wheat plants grown under elevated CO2. Front. Plant Sci. 2018, 9, 933. [Google Scholar] [CrossRef] [PubMed]
- Munir, N.; Hanif, M.; Abideen, Z.; Sohail, M.; El-Keblawy, A.; Radicetti, E.; Mancinelli, R.; Haider, G. Mechanisms and strategies of plant microbiome interactions to mitigate abiotic stresses. Agronomy 2022, 12, 2069. [Google Scholar] [CrossRef]
- Ling, H.; Zhang, P.; Xu, H.; Zhao, X. How to regenerate and protect desert riparian Populus euphratica forest in arid areas. Sci. Rep. 2015, 5, 15418. [Google Scholar] [CrossRef] [PubMed]
- Joniak-Lüthi, A. A road, a disappearing river and fragile connectivity in Sino-Inner Asian borderlands. Polit. Geogr. 2020, 78, 102122. [Google Scholar] [CrossRef]
- Wang, T.; Nyamtseren, M. Combating Aeolian Desertification in Northeast Asia; Springer: Singapore, 2022; pp. 17–58. [Google Scholar] [CrossRef]
- Zhou, W.-L.; Yang, X.-Q.; Hao, P.; Liu, Q.-W.; Cao, D.-C.; Baribault, T.; Li, J.-W. Plant diversity and its maintenance in Populus euphratica riparian forests in the Ejina Oasis, China. For. Stud. China 2010, 12, 55–61. [Google Scholar] [CrossRef]
- Edmondson, J.R. The flora and fauna of the Euphrates Expedition of 1836. Isr. J. Plant Sci. 2017, 64, 224–238. [Google Scholar]
- Handley, C. Sacred Species and Sites: Advances in Biocultural Conservation. Arboric. J. 2013, 35, 55–57. [Google Scholar]
- Safriel, U. Deserts and desertification: Challenges but also opportunities. Land Degrad. Dev. 2009, 20, 353–366. [Google Scholar] [CrossRef]
- Han, Y.; Wang, W.; Sun, J.; Ding, M.; Zhao, R.; Deng, S.; Wang, F.; Hu, Y.; Wang, Y.; Lu, Y. Populus euphratica XTH overexpression enhances salinity tolerance by the development of leaf succulence in transgenic tobacco plants. J. Exp. Bot. 2013, 64, 4225–4238. [Google Scholar] [CrossRef]
- Pan, J.; Xue, X.; Huang, C.; Peng, F.; Liao, J.; Ma, S.; You, Q.; Wang, T. Salt Tolerance Strategies of Nitraria tangutorum Bobr. and Elaeagnus angustifolia Linn. Determine the Inoculation Effects of Microorganisms in Saline Soil Conditions. Agronomy 2022, 12, 913. [Google Scholar] [CrossRef]
- Clifton-Brown, J.; Harfouche, A.; Casler, M.D.; Dylan Jones, H.; Macalpine, W.J.; Murphy-Bokern, D.; Smart, L.B.; Adler, A.; Ashman, C.; Awty-Carroll, D. Breeding progress and preparedness for mass-scale deployment of perennial lignocellulosic biomass crops switchgrass, miscanthus, willow and poplar. GCB Bioenergy 2019, 11, 118–151. [Google Scholar] [CrossRef]
- Llanes, A.; Palchetti, M.V.; Vilo, C.; Ibañez, C. Molecular control to salt tolerance mechanisms of woody plants: Recent achievements and perspectives. Ann. For. Sci. 2021, 78, 96. [Google Scholar] [CrossRef]
- Sixto, H.; González-González, B.D.; Molina-Rueda, J.J.; Garrido-Aranda, A.; Sanchez, M.M.; López, G.; Gallardo, F.; Cañellas, I.; Mounet, F.; Grima-Pettenati, J. Eucalyptus spp. and Populus spp. coping with salinity stress: An approach on growth, physiological and molecular features in the context of short rotation coppice (SRC). Trees 2016, 30, 1873–1891. [Google Scholar] [CrossRef]
- Harfouche, A.; Meilan, R.; Altman, A. Molecular and physiological responses to abiotic stress in forest trees and their relevance to tree improvement. Tree Physiol. 2014, 34, 1181–1198. [Google Scholar] [CrossRef] [PubMed]
- Hundleby, P.A.C.; Harwood, W.A. Impacts of the EU GMO regulatory framework for plant genome editing. Food Energy Secur. 2019, 8, e00161. [Google Scholar] [CrossRef] [PubMed]
- Mbaya, H.; Lillico, S.; Kemp, S.; Simm, G.; Raybould, A. Regulatory frameworks can facilitate or hinder the potential for genome editing to contribute to sustainable agricultural development. Front. Bioeng. Biotechnol. 2022, 10, 959236. [Google Scholar] [CrossRef]
- Poorter, H.; Niinemets, Ü.; Ntagkas, N.; Siebenkäs, A.; Mäenpää, M.; Matsubara, S.; Pons, T. A meta-analysis of plant responses to light intensity for 70 traits ranging from molecules to whole plant performance. New Phytol. 2019, 223, 1073–1105. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, Y.; Zhu, J.K. Thriving under Stress: How Plants Balance Growth and the Stress Response. Dev. Cell 2020, 55, 529–543. [Google Scholar] [CrossRef]
- Abobatta, W.F. Plant Stress Mitigators; Academic Press: Cambridge, MA, USA, 2023; pp. 425–434. [Google Scholar] [CrossRef]
- Lüttge, U.; Scarano, F.R. Ecophysiology. Braz. J. Bot. 2004, 27, 1–10. [Google Scholar] [CrossRef]
- Liu, H.; Gleason, S.M.; Hao, G.; Hua, L.; He, P.; Goldstein, G.; Ye, Q. Hydraulic traits are coordinated with maximum plant height at the global scale. Sci. Adv. 2019, 5, eaav1332. [Google Scholar] [CrossRef]
- Yang, X.D.; Anwar, E.; Xu, Y.L.; Zhou, J.; Sha, L.B.; Gong, X.W.; Ali, A.; Gao, Y.C.; Liu, Y.; Ge, P. Hydraulic constraints determine the distribution of heteromorphic leaves along plant vertical height. Front. Plant Sci. 2022, 13, 941764. [Google Scholar] [CrossRef]
- Yang, Q.; Han, X.M.; Gu, J.K.; Liu, Y.J.; Yang, M.J.; Zeng, Q.Y. Functional and structural profiles of GST gene family from three Populus species reveal the sequence–function decoupling of orthologous genes. New Phytol. 2019, 221, 1060–1073. [Google Scholar] [CrossRef] [PubMed]
- Zhuge, X.L.; Xie, T.; Du, X.; Zhang, X.X.; Hu, J.P.; Yang, H.L. Non-synonymous substitution of evolutionarily conserved residue in Tau class glutathione transferases alters structural and catalytic features. Int. J. Biol. Macromol. 2022, 197, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Brosché, M.; Vinocur, B.; Alatalo, E.R.; Lamminmäki, A.; Teichmann, T.; Ottow, E.A.; Djilianov, D.; Afif, D.; Bogeat-Triboulot, M.B.; Altman, A. Gene expression and metabolite profiling of Populus euphratica growing in the Negev desert. Genome Biol. 2005, 6, R101. [Google Scholar] [CrossRef] [PubMed]
| Species | Identified Genes No. | Gene Family | Functions | Reference |
|---|---|---|---|---|
| P. euphratica | 19 | HSF | Drought stress | [68] |
| 22 | HSF | Salt stress | [68] | |
| 2 (PeWRKY45) | WRKY | Salt stress | [71] | |
| (PeWRKY102) | ||||
| 1 (PebHLH35) | bHLH | Drought and abscisic acid | [69] | |
| 1 (PeCPK10) | CPK | Drought and cold stress | [65] | |
| 1 (PeSTZ1) | STZ | Freezing tolerance | [80] | |
| 1 (PeuGASA15) | GASA | Drought stress | [74] | |
| 1 (PeuLAC2) | LACCASE | Drought stress | [81] | |
| 3 | 4CL | Salt stress | [97] | |
| 1 (PeCBL1; CIPK24, | CBL1 | Salt stress | [90] | |
| CIPK25, and cipk26) | ||||
| AUX-IAA/ARF | Not identified yet | |||
| 1 (PeHAB1) | PP2C | Induced by drought and ABA as a negative regulator | [98] | |
| 1 (PeDREB2) | DREB2-type | Salt stress | [64] | |
| AB13-VP1 | Not identified yet | |||
| C2C2-Dof | Not identified yet | |||
| SBP | Not identified yet | |||
| MADS | Not identified yet | |||
| C2C2-GATA | Not identified yet | |||
| FHA | Not identified yet | |||
| GARP-G2 like | Not identified yet | |||
| PeG | Not identified yet | |||
| C2H2 | Not identified yet | |||
| bZIP | Not identified yet | |||
| HB | Not identified yet | |||
| C3H | Not identified yet | |||
| PHD | Not identified yet | |||
| PeSCL7 | GRAS | Salt and drought tolerance | [79] | |
| AP2-EREBP | Not identified yet | |||
| MYB-related | Not identified yet | |||
| SPL | Not identified yet |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ndayambaza, B.; Si, J.; Deng, Y.; Jia, B.; He, X.; Zhou, D.; Wang, C.; Zhu, X.; Liu, Z.; Qin, J.; et al. The Euphrates Poplar Responses to Abiotic Stress and Its Unique Traits in Dry Regions of China (Xinjiang and Inner Mongolia): What Should We Know? Genes 2023, 14, 2213. https://doi.org/10.3390/genes14122213
Ndayambaza B, Si J, Deng Y, Jia B, He X, Zhou D, Wang C, Zhu X, Liu Z, Qin J, et al. The Euphrates Poplar Responses to Abiotic Stress and Its Unique Traits in Dry Regions of China (Xinjiang and Inner Mongolia): What Should We Know? Genes. 2023; 14(12):2213. https://doi.org/10.3390/genes14122213
Chicago/Turabian StyleNdayambaza, Boniface, Jianhua Si, Yanfang Deng, Bing Jia, Xiaohui He, Dongmeng Zhou, Chunlin Wang, Xinglin Zhu, Zijin Liu, Jie Qin, and et al. 2023. "The Euphrates Poplar Responses to Abiotic Stress and Its Unique Traits in Dry Regions of China (Xinjiang and Inner Mongolia): What Should We Know?" Genes 14, no. 12: 2213. https://doi.org/10.3390/genes14122213




