Assessment of Candidate Reference Genes for Gene Expression Studies Using RT-qPCR in Colletotrichum fructicola from Litchi
Abstract
:1. Introduction
2. Materials and Methods
2.1. Inoculation of C. fructicola
2.2. The Preparation and Collection of Samples
2.3. The Extraction of Total RNA and the Synthesis of cDNA
2.4. The Design of Primers and the Cloning of Genes
2.5. RT-qPCR
2.6. Assessment of Gene Expression Stability
2.7. Statistical Analysis
3. Results
3.1. Detection of Primer Specificity and Efficiency
3.2. Expression Levels of Candidate Reference Genes
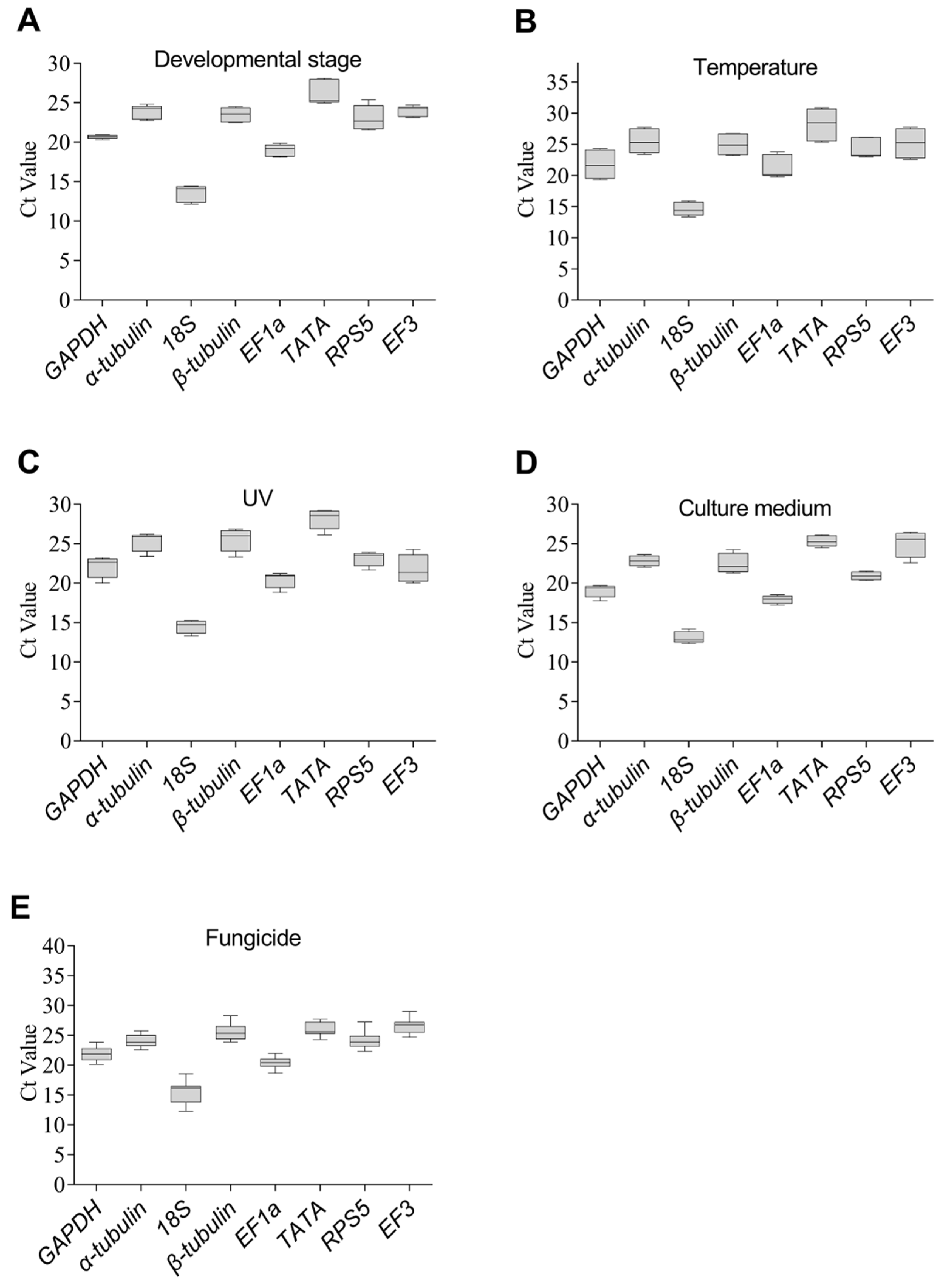
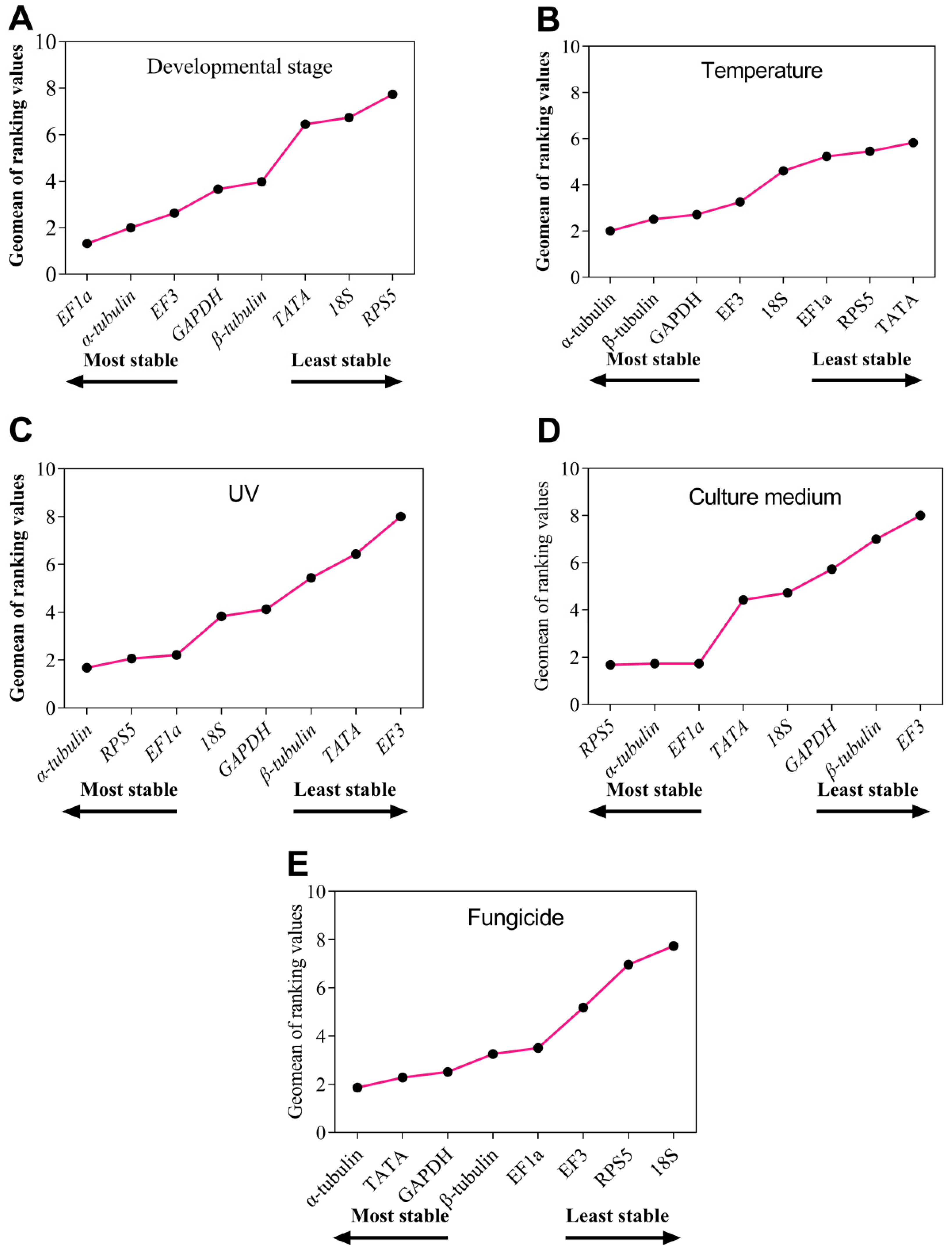
3.3. The Stability of the Eight Reference Genes under Specific Experimental Conditions
3.4. The RefFinder Ranking for Eight Reference Gene Expression Stability
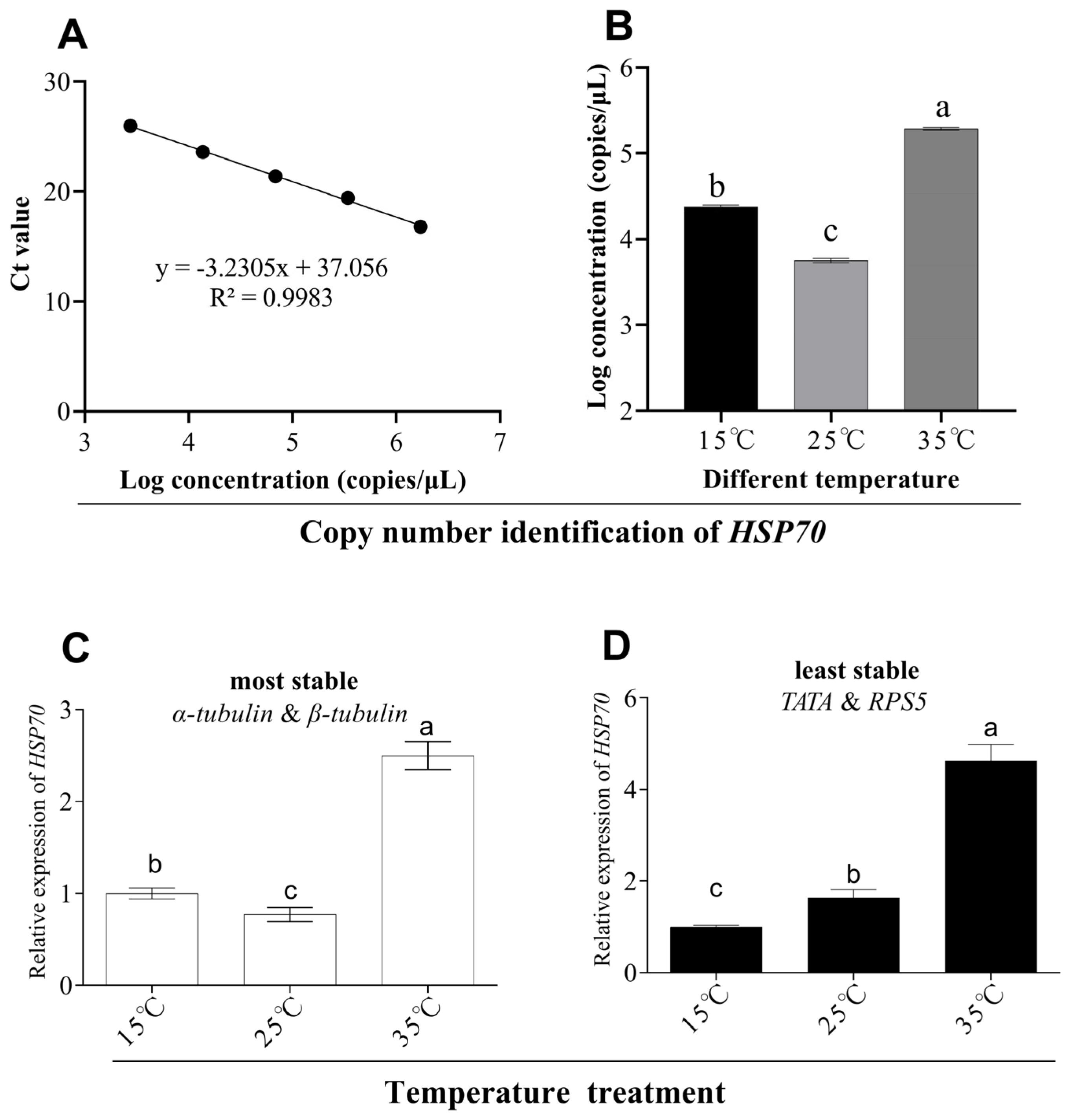
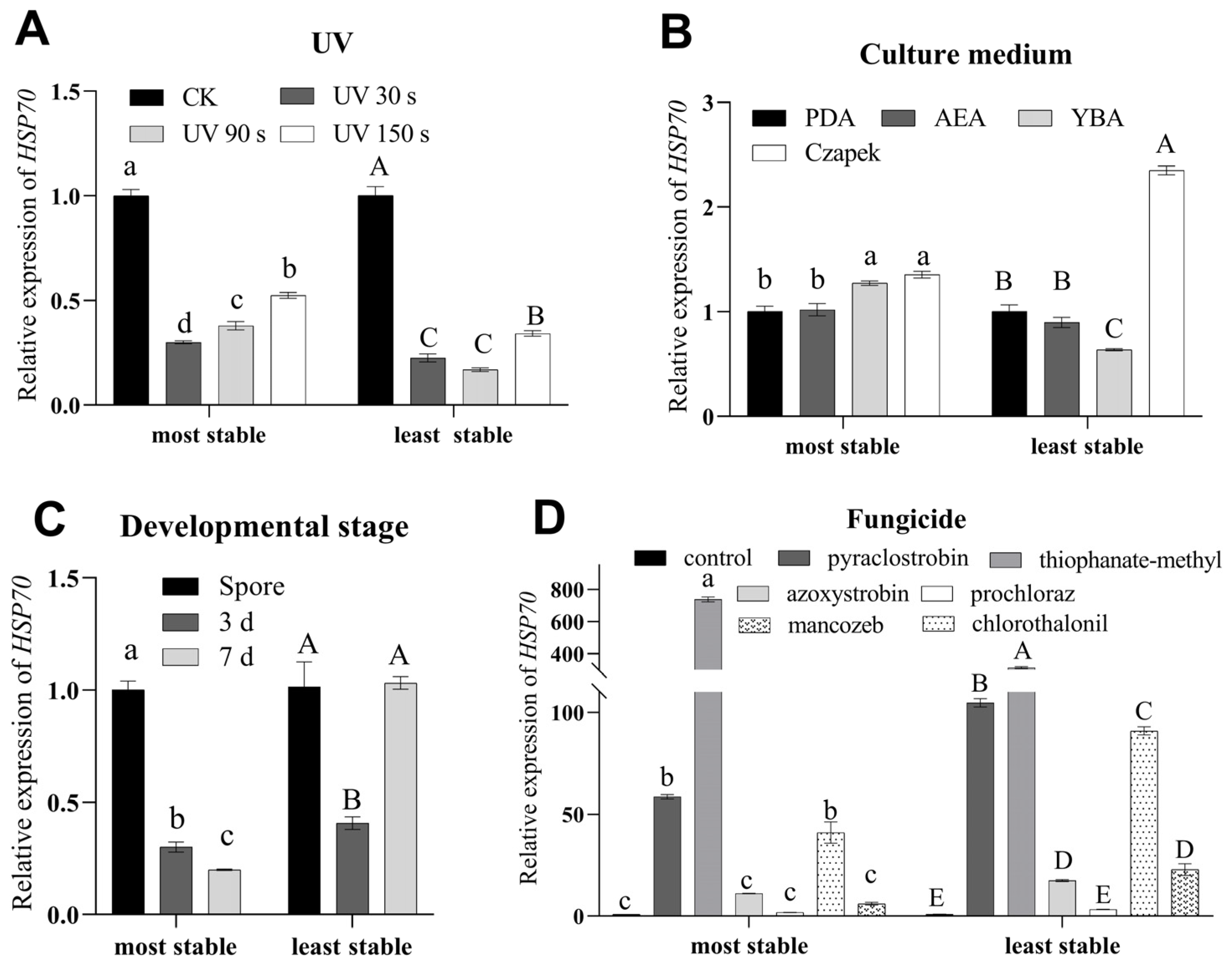
3.5. Evaluation of the Selected Reference Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anderson, J.M.; Aitken, E.A.B.; Dann, E.K.; Coates, L.M. Morphological and molecular diversity of Colletotrichum spp. causing pepper spot and anthracnose of lychee (Litchi chinensis) in Australia. Plant Pathol. 2013, 62, 279–288. [Google Scholar] [CrossRef]
- De Silva, D.D.; Crous, P.W.; Ades, P.K.; Hyde, K.D.; Taylor, P.W.J. Life styles of Colletotrichum species and implications for plant biosecurity. Fungal Biol. Rev. 2017, 31, 155–168. [Google Scholar] [CrossRef]
- Royal Botanic Gardens, Kew. State of the World’s Plants 2016. Research Report. 2019-08-12. Report No.: 978-1-84246-628-5. 2016. Available online: https://kew.iro.bl.uk/concern/reports/f931f1de-72c7-46b4-b57c-28eb417c53ec?locale=en (accessed on 10 December 2023).
- Joshi, R. A Review on Colletotrichum spp. virulence mechanism against host plant defensive factors. J. Med. Plants. 2018, 6, 64–67. [Google Scholar] [CrossRef]
- Villa-Rivera, G.M.; Conejo-Saucedo, U.; Lara-Marquez, A.; Cano-Camacho, H.; Lopez-Romero, E.; Zavala-Paramo, G.M. The role of virulence factors in the pathogenicity of Colletotrichum sp. Curr. Protein Pept. Sci. 2017, 18, 1005–1018. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.Q.; Yuan, Q.F.; Tang, J.T.; Huang, J.B.; Hsiang, T.; Wei, Y.D.; Zheng, L. Colletotrichum higginsianum as a model for understanding host–pathogen interactions: A review. Int. J. Mol. Sci. 2018, 19, 2142. [Google Scholar] [CrossRef] [PubMed]
- Alaniz, S.; Hernández, L.; Mondino, P. Colletotrichum fructicola is the dominant and one of the most aggressive species causing bitter rot of apple in Uruguay. Trop Plant Pathol. 2015, 40, 265–274. [Google Scholar] [CrossRef]
- Li, Q.L.; Bu, J.Y.; Yu, Z.H.; Tang, L.H.; Huang, S.P.; Guo, T.X.; Mo, J.Y.; Hsiang, T. Draft genome sequence of an isolate of Colletotrichum fructicola, a causal agent of mango anthracnose. Genome Announc. 2018, 6, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zheng, X.J.; Khaskheli, M.I.; Sun, X.F.; Chang, X.L.; Gong, G.S. Identification of Colletotrichum species associated with blueberry anthracnose in Sichuan, China. Pathogens 2020, 9, 718. [Google Scholar] [CrossRef] [PubMed]
- Gan, P.; Nakata, N.; Suzuki, T.; Shirasu, K. Markers to differentiate species of anthracnose fungi identify Colletotrichum fructicola as the predominant virulent species in strawberry plants in Chiba Prefecture of Japa. J. Gen. Plant Pathol. 2017, 83, 14–22. [Google Scholar] [CrossRef]
- Dowling, M.; Peres, N.; Villani, S.; Schnabel, G. Managing Colletotrichum on fruit crops: A “complex” challenge. Plant Dis. 2020, 104, 2301–2316. [Google Scholar] [CrossRef]
- Cortaga, C.Q.; Cordez, B.W.P.; Dacones, L.S.; Balendres, M.A.O.; Dela Cueva, F.M. Mutations associated with fungicide resistance in Colletotrichum species: A review. Phytoparasitica 2023, 51, 569–592. [Google Scholar] [CrossRef]
- Chen, Y.; Jin, L.H.; Zhou, M.G. Effect of azoxystrobin on oxygen consumption and cyt b gene expression of Colletotrichum capsici from chilli fruits. Agric. Sci. China 2009, 8, 628–631. [Google Scholar] [CrossRef]
- Nakaune, R.; Nakano, M. Benornyl resistance of Colletotrichum acutatum is caused by enhanced expression of β-tubulin 1 gene regulated by putative leucine zipper protein CaBEN1. Fungal Genet. Biol. 2007, 44, 1324–1335. [Google Scholar] [CrossRef]
- Wei, L.L.; Li, X.J.; Chen, B.; Chen, W.C.; Wei, L.H.; Zhou, D.M.; Chen, C.J.; Wu, C.D. Sterol 14 α-demethylase CaCYP51A and CaCYP51B are functionally redundant, but differentially regulated in Colletotrichum acutatum: Responsibility for DMI-fungicide resistance. J. Agric. Food Chem. 2022, 70, 11911–11922. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Diao, Y.Z.; Wang, W.Z.; Hao, J.J.; Imran, M.; Duan, H.X.; Liu, X.L. Assessing the risk for resistance and elucidating the genetics of Colletotrichum truncatum that is only sensitive to some DMI fungicides. Front. Microbiol. 2017, 8, 1779. [Google Scholar] [CrossRef]
- Radonić, A.; Thulke, S.; Mackay, I.M.; Landt, O.; Siegert, W.; Nitsche, A. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2004, 313, 856–862. [Google Scholar] [CrossRef] [PubMed]
- Marcial-Quino, J.; Fierro, F.; De la Mora-De la Mora, I.; Enríquez-Flores, S.; Gómez-Manzo, S.; Vanoye-Carlo, A.; Garcia-Torres, I.; Sierra-Palacios, E.; Reyes-Vivas, H. Validation of housekeeping genes as an internal control for gene expression studies in Giardia lamblia using quantitative real-time PCR. Gene 2016, 581, 21–30. [Google Scholar] [CrossRef]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef]
- Provenzano, M.; Mocellin, S. Complementary Techniques. In Microarray Technology and Cancer Gene Profiling; Mocellin, S., Ed.; Springer: New York, NY, USA, 2007; pp. 66–73. [Google Scholar] [CrossRef]
- Guénin, S.; Mauriat, M.; Pelloux, J.; Van Wuytswinkel, O.; Bellini, C.; Gutierrez, L. Normalization of qRT-PCR data: The necessity of adopting a systematic, experimental conditions-specific, validation of references. J. Exp. Bot. 2009, 60, 487–493. [Google Scholar] [CrossRef]
- Pombo, M.A.; Ramos, R.N.; Zheng, Y.; Fei, Z.; Martin, G.B.; Rosli, H.G. Transcriptome-based identification and validation of reference genes for plant-bacteria interaction studies using Nicotiana benthamiana. Sci. Rep. 2019, 9, 1632. [Google Scholar] [CrossRef]
- Vieira, A.; Cabral, A.; Fino, J.; Azinheira, H.G.; Loureiro, A.; Talhinhas, P.; Pires, A.S.; Varzea, V.; Moncada, P.; Oliveira, H.; et al. Comparative validation of conventional and RNA-seq data-derived reference genes for qPCR expression studies of Colletotrichum kahawae. PLoS ONE 2016, 11, e0150651. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Z.; Chen, X.G.; Tan, Q.; He, Y.; Wang, Z.K.; Zhou, G.Y.; Liu, J.A. Selection of potential reference genes for RT-qPCR in the plant pathogenic fungus Colletotrichum fructicola. Front. Microbiol. 2022, 13, 982748. [Google Scholar] [CrossRef] [PubMed]
- Nandakumar, M.; Viswanathan, R.; Malathi, P.; Sundar, A.R. Selection of reference genes for normalization of microRNA expression in sugarcane stalks during its interaction with Colletotrichum falcatum. 3 Biotech 2021, 11, 72. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, A.; Loureiro, A.; Batista, D.; Monteiro, F.; Várzea, V.; Pais, M.S.; Gichuru, E.K.; Silva, M.C. Validation of reference genes for normalization of qPCR gene expression data from Coffea spp. hypocotyls inoculated with Colletotrichum kahawae. BMC Res. Notes 2013, 6, 388. [Google Scholar] [CrossRef] [PubMed]
- He, S.; An, T.; Liu, S. Validation of reliable reference genes for RT-qPCR studies of target gene expression in Colletotrichum camelliae during spore germination and mycelial growth and interaction with host plants. Front. Microbiol. 2019, 10, 2055. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Kulcheski, F.R.; Marcelino-Guimaraes, F.C.; Nepomuceno, A.L.; Abdelnoor, R.V.; Margis, R. The use of microRNAs as reference genes for quantitative polymerase chain reaction in soybean. Anal. Biochem. 2010, 406, 185–192. [Google Scholar] [CrossRef]
- Chen, L.; Geng, X.; Ma, Y.; Zhao, J.; Chen, W.; Xing, X.; Shi, Y.; Sun, B.; Li, H. The ER Lumenal Hsp70 Protein FpLhs1 Is Important for Conidiation and Plant Infection in Fusarium pseudograminearum. Front. Microbiol. 2019, 10, 1401. [Google Scholar] [CrossRef]
- Mota, T.M.; Oshiquiri, L.H.; Lopes, É.C.V.; Barbosa Filho, J.R.; Ulhoa, C.J.; Georg, R.C. Hsp genes are differentially expressed during Trichoderma asperellum self-recognition, mycoparasitism and thermal stress. Microbiol. Res. 2019, 227, 126296. [Google Scholar] [CrossRef]
- Archer, M.; Xu, J.P. Current practices for reference gene selection in RT-qPCR of Aspergillus: Outlook and recommendations for the future. Genes 2021, 12, 960. [Google Scholar] [CrossRef]
- Zhu, H.X.; Ma, Y.Q.; Guo, Q.Y. Expression stability of internal reference gene in response to Trichoderma polysporum infection in Avena fatua L. Curr. Genet. 2021, 67, 909–918. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.M.; Zhou, C.L.; Wan, J.N.; Guo, T.; Ji, G.Y.; Luo, S.Z.; Ji, K.P.; Cao, Y.; Tan, Q.; Bao, D.P.; et al. Selection and validation of internal control genes for quantitative real-time RT–qPCR normalization of Phlebopus portentosus gene expression under different conditions. PLoS ONE 2023, 18, e0288982. [Google Scholar] [CrossRef] [PubMed]
- Che, J.X.; Shi, J.L.; Lu, Y.; Liu, Y.L. Validation of reference genes for normalization of gene expression by qRT-PCR in a resveratrol-producing entophytic fungus (Alternaria sp. MG1). AMB Express 2016, 6, 106. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.F.; Pan, H.P.; Zhang, L.H.; Ou, D.; Lu, Z.T.; Khan, M.M.; Qiu, B.L. Comprehensive assessment of candidate reference genes for gene expression studies using RT-qPCR in Tamarixia radiata, a predominant parasitoid of Diaphorina citri. Genes 2020, 11, 1178. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Hellemans, J.; Vandesompele, J. Selection of reliable reference genes for RT-qPCR analysis. Methods Mol. Biol. 2014, 1160, 19–26. [Google Scholar] [CrossRef]
- Song, Y.; Wang, Y.; Guo, D.; Jing, L. Selection of reference genes for quantitative real-time PCR normalization in the plant pathogen Puccinia helianthi Schw. BMC Plant Biol. 2019, 19, 20. [Google Scholar] [CrossRef]
- Lü, J.; Yang, C.; Zhang, Y.; Pan, H. Selection of reference genes for the normalization of RT-qPCR data in gene expression studies in insects: A systematic review. Front. Physiol. 2018, 9, 1560. [Google Scholar] [CrossRef]
- Gao, F.Y.; Li, L.; Wang, J.Y.; Wang, Y.L.; Sun, G.C. The functions of PEX genes in peroxisome biogenesis and pathogenicity in phytopathogenic fungi. Yi Chuan Hered. 2017, 39, 908–917. [Google Scholar] [CrossRef]
- Li, H. Population genetic analyses of the fungal pathogen Colletotrichum on tea-oil trees in China and characterization of a MAPK gene CfPMK1 in the pathogen. J. Cent. South Univ. 2018, 11, e0156841. [Google Scholar] [CrossRef]
- Skinner, W.; Keon, J.; Hargreaves, J. Gene information for fungal plant pathogens from expressed sequences. Curr. Opin. Microbiol. 2001, 4, 381–386. [Google Scholar] [CrossRef]
- Huang, N.; Ling, H.; Liu, F.; Su, Y.C.; Su, W.H.; Mao, H.Y.; Zhang, X.; Wang, L.; Chen, R.K.; Que, Y.X. Identification and evaluation of PCR reference genes for host and pathogen in sugarcane-Sporisorium scitamineum interaction system. BMC Genom. 2018, 19, 479. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Wang, J.; Zhang, B. RefFinder: A web-based tool for comprehensively analyzing and identifying reference genes. Funct. Integr. Genom. 2023, 23, 125. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.M.; Guo, C.F.; Peng, J.; Fan, Z.Y.; Hafeez, M.; Ali, D.; Wang, K.; Almarzoug, M.H.A.; Qiu, B.L. Screening and validation of reference genes using in RT-qPCR for gene expression studies in Paederus fuscipes, a medically and agriculturally important insect. J. King Saud Univ. Sci. 2022, 34, 101654. [Google Scholar] [CrossRef]
- Yang, C.X.; Pan, H.P.; Noland, J.E.; Zhang, D.Y.; Zhang, Z.H.; Liu, Y.; Zhou, X.G. Selection of reference genes for RT-qPCR analysis in a predatory biological control agent, Coleomegilla maculata (Coleoptera: Coccinellidae). Sci. Rep. 2015, 5, 18201. [Google Scholar] [CrossRef]
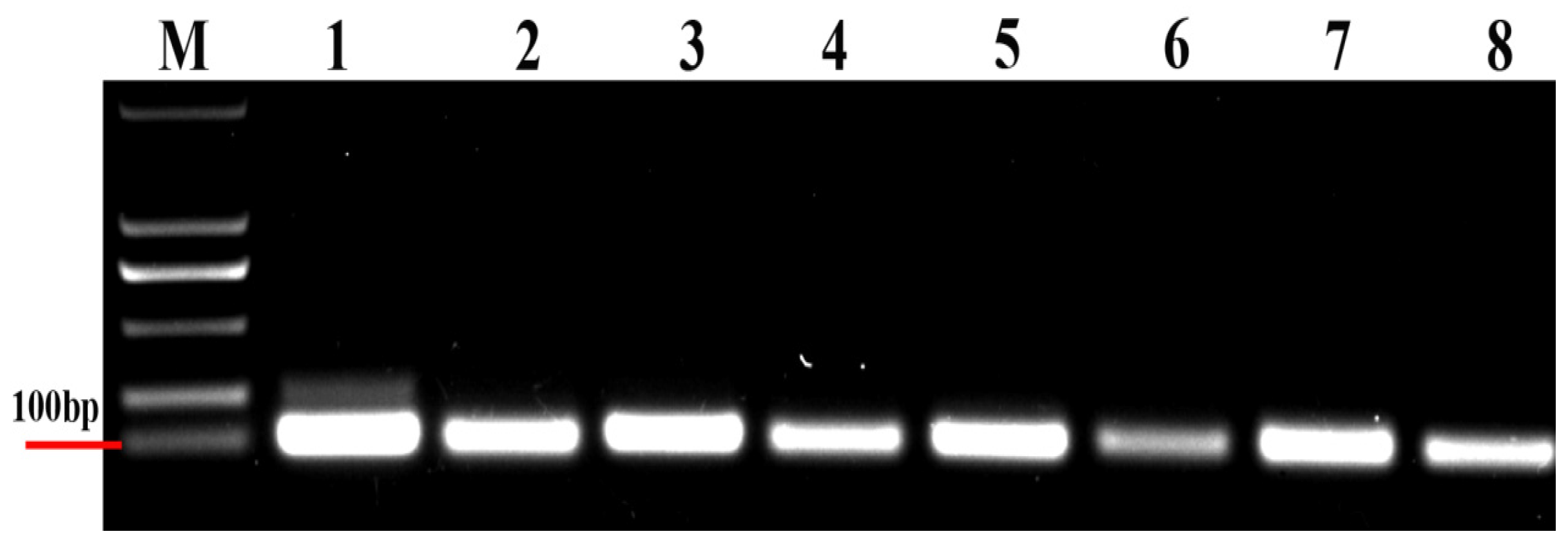
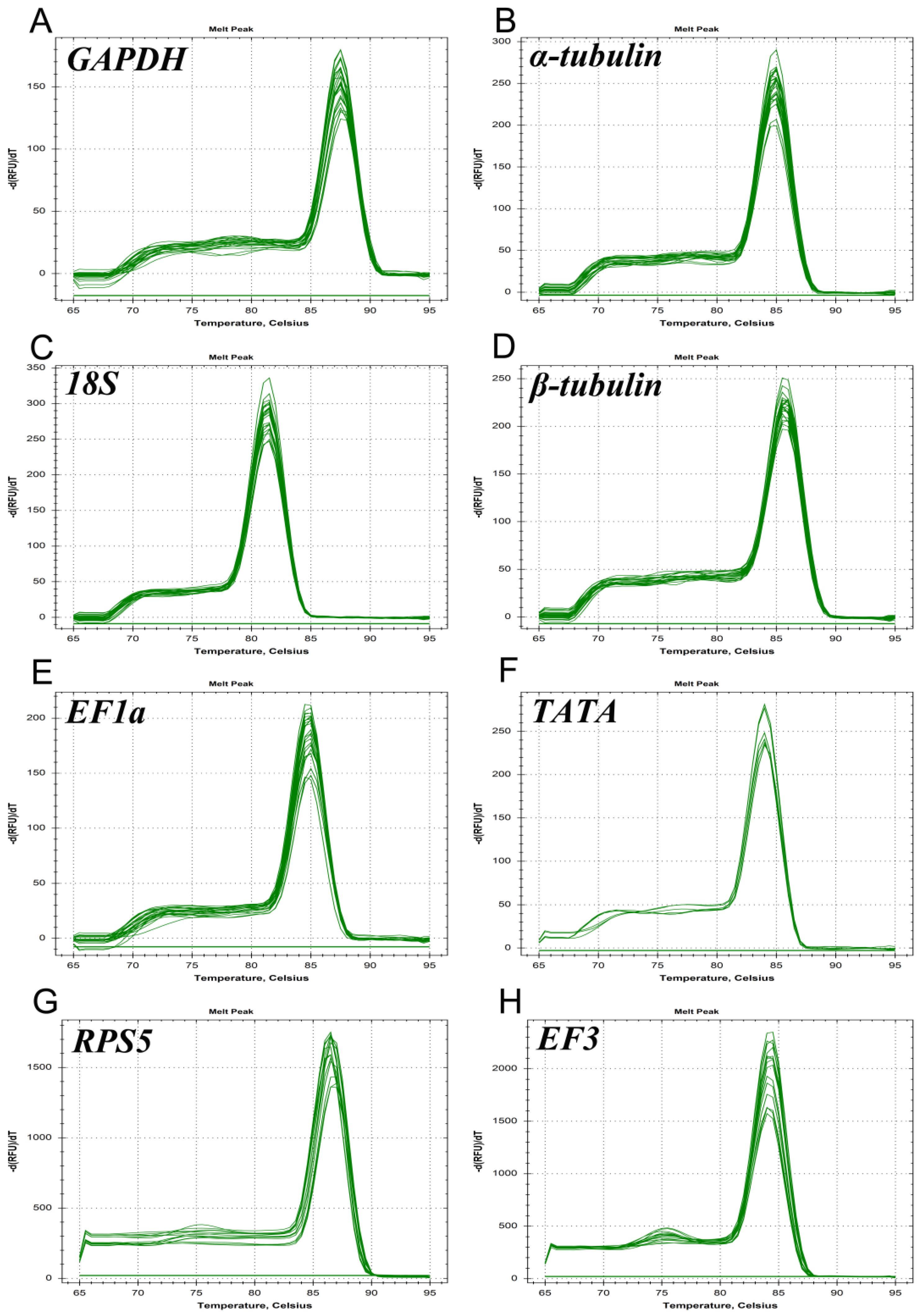
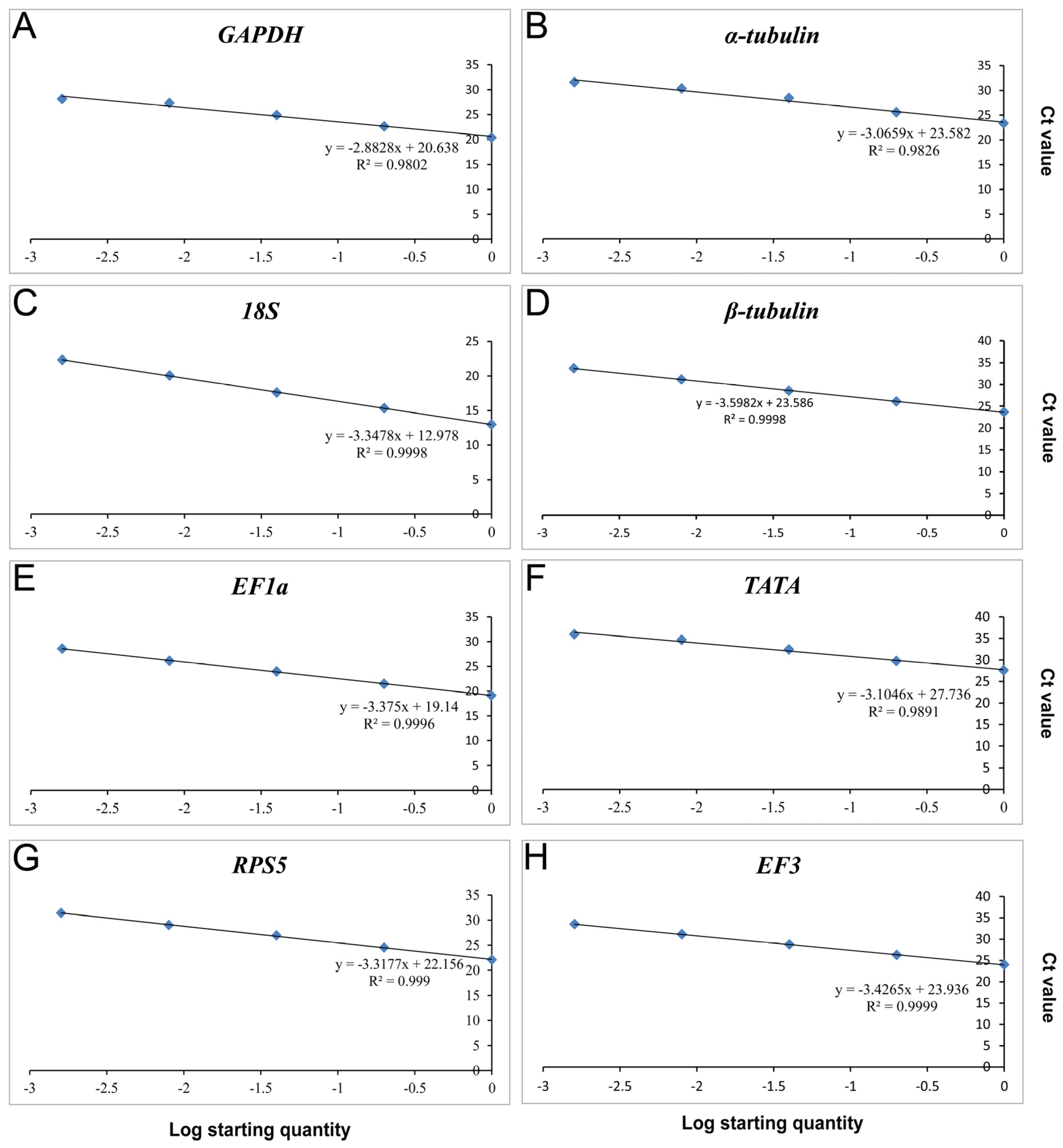

| Reference Genes | Forward/Reverse Primer Sequence (5′–3′) | Amplicon Size (bp) | Efficiency (%) | R2 | Linear Regression |
|---|---|---|---|---|---|
| GAPDH | F: GTCTCCGTCGTTGACCTGAC | 120 | 122.27 | 0.9802 | y = −2.8828x + 20.638 |
| R: GTCCTCAGTGTAGGCCAGGA | |||||
| α-tubulin | F: CTCAGGTCGTCTCCTCCATC | 110 | 111.92 | 0.9826 | y = −3.0659x + 23.582 |
| R: GGGGTAGTGGATACGAGGGT | |||||
| 18S | F: CAGGAACGAAAGTTAGGGGA | 120 | 98.93 | 0.9998 | y = −3.3478x + 12.978 |
| R: ATTTCTCGTAAGGTGCCGAA | |||||
| β-tubulin | F: CTGCCTTCTGGCAAAACATT | 108 | 89.63 | 0.9998 | y = −3.5982x + 23.586 |
| R: GCTTCGTTGAAGTAGACGCTC | |||||
| EF1a | F: CCAACGTCACCACTGAAGTC | 109 | 97.83 | 0.9996 | y = −3.375x + 19.14 |
| R: TCCTTGACGGAGACGTTCTT | |||||
| TATA | F: TTCGCCTCTGGTAAGATGGT | 120 | 109.94 | 0.9891 | y = −3.1046x + 27.736 |
| R: GAAGTCGGTGAACTTGGCAT | |||||
| RPS5 | F: CAACTGCCCCATCATTGAG | 119 | 100.18 | 0.9990 | y = −3.3177x + 22.156 |
| R: TCATCAGGTGGATGATCTCG | |||||
| EF3 | F: GGACACCAAGAAGGAGGTCA | 109 | 95.81 | 0.9990 | y = −3.4265x + 23.936 |
| R: ACACTTGATAAGCTCGGGGA |
| Conditions | Rank | geNorm | NormFinder | BestKeeper | ΔCt | ||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Stability | Gene | Stability | Gene | Stability | Gene | Stability | ||
| Developmental stage | 1 | β-tublin | 0.142 | EF1a | 0.071 | GAPDH | 0.2 | EF1a | 0.79 |
| 2 | EF1a | 0.142 | β-tubulin | 0.137 | EF3 | 0.53 | β-tubulin | 0.83 | |
| 3 | α-tubulin | 0.21 | GAPDH | 0.32 | EF1a | 0.59 | α-tubulin | 0.88 | |
| 4 | GAPDH | 0.415 | α-tubulin | 0.358 | β-tubulin | 0.66 | GAPDH | 0.93 | |
| 5 | EF3 | 0.625 | EF3 | 0.694 | α-tubulin | 0.71 | EF3 | 1.05 | |
| 6 | TATA | 0.796 | TATA | 1.13 | 18S | 0.86 | TATA | 1.34 | |
| 7 | 18S | 0.917 | 18S | 1.252 | RPS5 | 1.23 | 18S | 1.39 | |
| 8 | RPS5 | 1.113 | RPS5 | 1.625 | TATA | 1.27 | RPS5 | 1.7 | |
| Temperature | 1 | GAPDH | 0.176 | α-tubulin | 0.137 | 18S | 0.83 | α-tubulin | 0.58 |
| 2 | EF3 | 0.176 | β-tubulin | 0.174 | β-tubulin | 1.19 | β-tubulin | 0.62 | |
| 3 | TATA | 0.276 | GAPDH | 0.376 | RPS5 | 1.32 | GAPDH | 0.64 | |
| 4 | α-tubulin | 0.381 | EF3 | 0.541 | α-tubulin | 1.41 | EF3 | 0.7 | |
| 5 | β-tubulin | 0.461 | EF1a | 0.672 | EF1a | 1.58 | EF1a | 0.84 | |
| 6 | EF1a | 0.617 | RPS5 | 0.753 | GAPDH | 1.64 | TATA | 0.88 | |
| 7 | RPS5 | 0.701 | 18S | 0.797 | EF3 | 1.67 | RPS5 | 0.88 | |
| 8 | 18S | 0.758 | TATA | 0.81 | TATA | 1.84 | 18S | 0.93 | |
| UV treatment | 1 | α-tubulin | 0.184 | RPS5 | 0.117 | 18S | 0.66 | α-tubulin | 0.51 |
| 2 | EF1a | 0.184 | α-tubulin | 0.135 | RPS5 | 0.69 | EF1a | 0.53 | |
| 3 | RPS5 | 0.243 | GAPDH | 0.221 | EF1a | 0.8 | RPS5 | 0.53 | |
| 4 | GAPDH | 0.29 | EF1a | 0.225 | α-tubulin | 0.92 | GAPDH | 0.56 | |
| 5 | β-tubulin | 0.332 | β-tubulin | 0.277 | TATA | 0.93 | β-tubulin | 0.61 | |
| 6 | 18S | 0.387 | 18S | 0.323 | GAPDH | 1.04 | 18S | 0.65 | |
| 7 | TATA | 0.456 | TATA | 0.746 | β-tubulin | 1.08 | TATA | 0.82 | |
| 8 | EF3 | 0.708 | EF3 | 1.439 | EF3 | 1.39 | EF3 | 1.46 | |
| Culture medium | 1 | EF1a | 0.113 | α-tubulin | 0.072 | EF1a | 0.46 | α-tubulin | 0.42 |
| 2 | RPS5 | 0.113 | RPS5 | 0.264 | RPS5 | 0.48 | RPS5 | 0.46 | |
| 3 | α-tubulin | 0.145 | EF1a | 0.268 | α-tubulin | 0.57 | EF1a | 0.48 | |
| 4 | TATA | 0.206 | TATA | 0.289 | 18S | 0.59 | TATA | 0.49 | |
| 5 | 18S | 0.267 | 18S | 0.384 | GAPDH | 0.61 | 18S | 0.55 | |
| 6 | GAPDH | 0.353 | GAPDH | 0.4 | TATA | 0.65 | GAPDH | 0.61 | |
| 7 | β-tubulin | 0.448 | β-tubulin | 0.574 | β-tubulin | 1 | β-tubulin | 0.73 | |
| 8 | EF3 | 0.599 | EF3 | 0.999 | EF3 | 1.22 | EF3 | 1.05 | |
| Fungicide | 1 | α-tubulin | 0.546 | GAPDH | 0.293 | EF1a | 0.8 | TATA | 0.89 |
| 2 | β-tubulin | 0.546 | α-tubulin | 0.302 | α-tubulin | 0.82 | GAPDH | 0.89 | |
| 3 | TATA | 0.577 | TATA | 0.32 | TATA | 0.96 | α-tubulin | 0.9 | |
| 4 | EF3 | 0.609 | β-tubulin | 0.392 | GAPDH | 1 | β-tubulin | 0.9 | |
| 5 | GAPDH | 0.639 | EF1a | 0.404 | EF3 | 1.08 | EF1a | 0.92 | |
| 6 | EF1a | 0.657 | EF3 | 0.436 | RPS5 | 1.13 | EF3 | 0.94 | |
| 7 | RPS5 | 0.905 | 18S | 1.642 | β-tubulin | 1.14 | RPS5 | 1.71 | |
| 8 | 18S | 1.108 | RPS5 | 1.648 | 18S | 1.47 | 18S | 1.72 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, D.; Huang, R.; Hu, Y.; Yang, X.; Xu, D.; Jiang, Z. Assessment of Candidate Reference Genes for Gene Expression Studies Using RT-qPCR in Colletotrichum fructicola from Litchi. Genes 2023, 14, 2216. https://doi.org/10.3390/genes14122216
Dong D, Huang R, Hu Y, Yang X, Xu D, Jiang Z. Assessment of Candidate Reference Genes for Gene Expression Studies Using RT-qPCR in Colletotrichum fructicola from Litchi. Genes. 2023; 14(12):2216. https://doi.org/10.3390/genes14122216
Chicago/Turabian StyleDong, Dingming, Rong Huang, Yuzhuan Hu, Xinyan Yang, Dagao Xu, and Zide Jiang. 2023. "Assessment of Candidate Reference Genes for Gene Expression Studies Using RT-qPCR in Colletotrichum fructicola from Litchi" Genes 14, no. 12: 2216. https://doi.org/10.3390/genes14122216




