CFHTF2 Is Needed for Vegetative Growth, Conidial Morphogenesis and the Osmotic Stress Response in the Tea Plant Anthracnose (Colletotrichum fructicola)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Determination of Pathogenicity
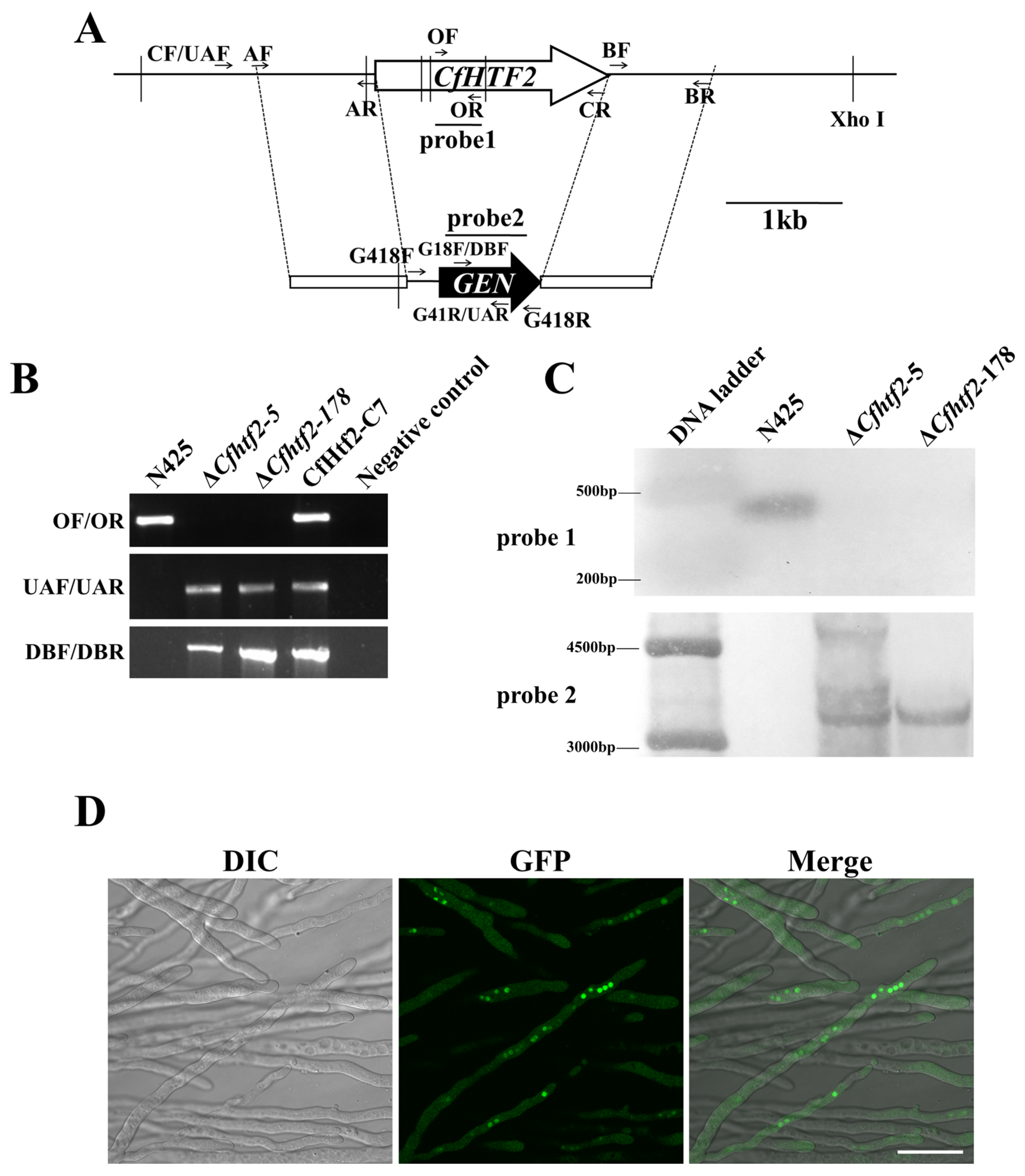
2.2. Generation of Cfhtf2 Deletion Mutants and Complementary Strains
2.3. Vegetative Growth of Colonies
2.4. Spore Yield, Conidiation and Appressorium Observation
2.5. Sensitivity to Stress Factors
2.6. RNA-Seq Analysis
2.7. qRT-PCR
3. Results
3.1. CfHTF2 Evolutionary Analysis and Acquisition of Knockout Mutants and Complementary Strains
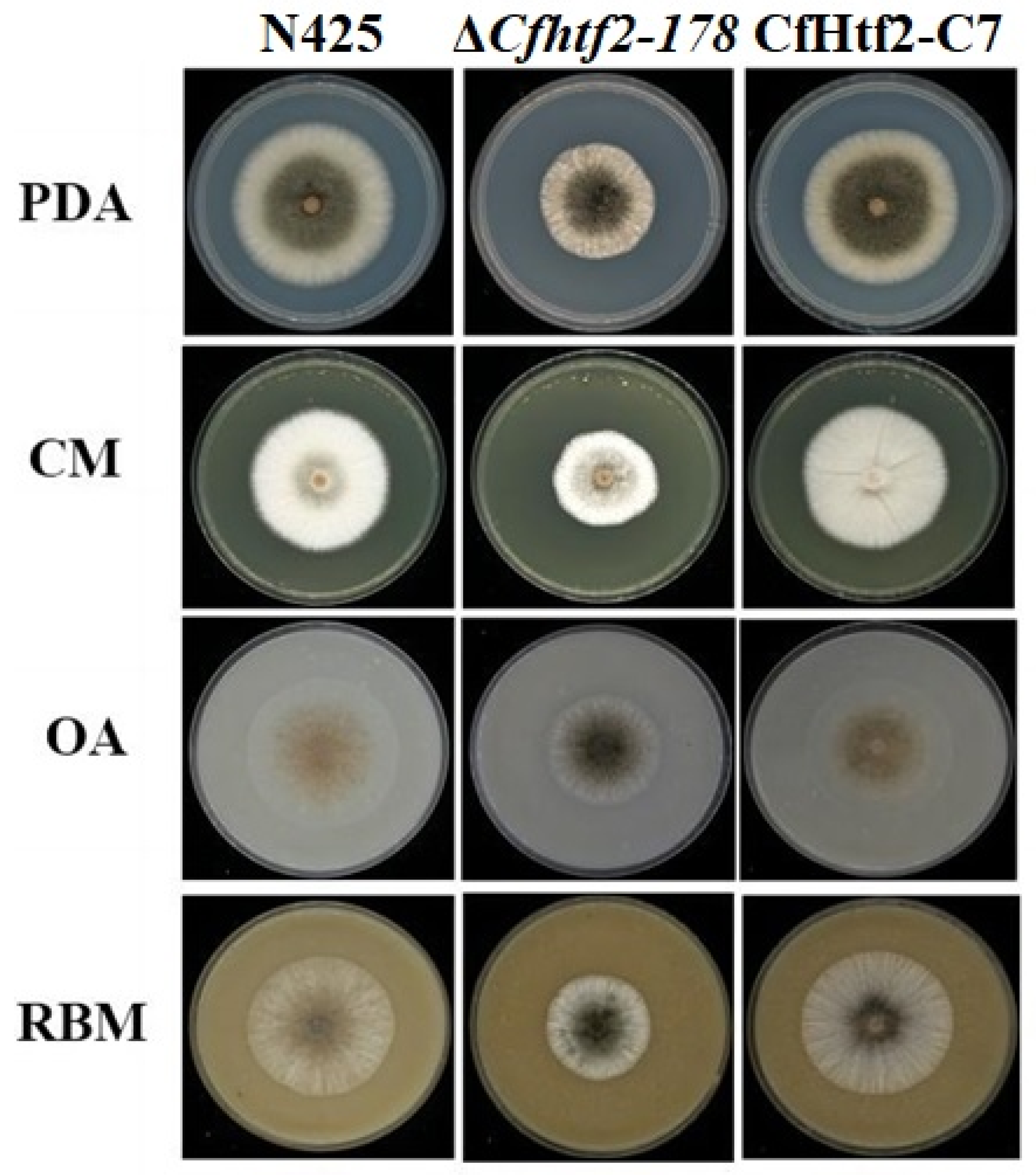
3.2. Loss of CfHTF2 Slows the Vegetative Growth of C. fructicola
3.3. Effect of The CHTF2 Gene on Asexual Reproduction in Anthracnose
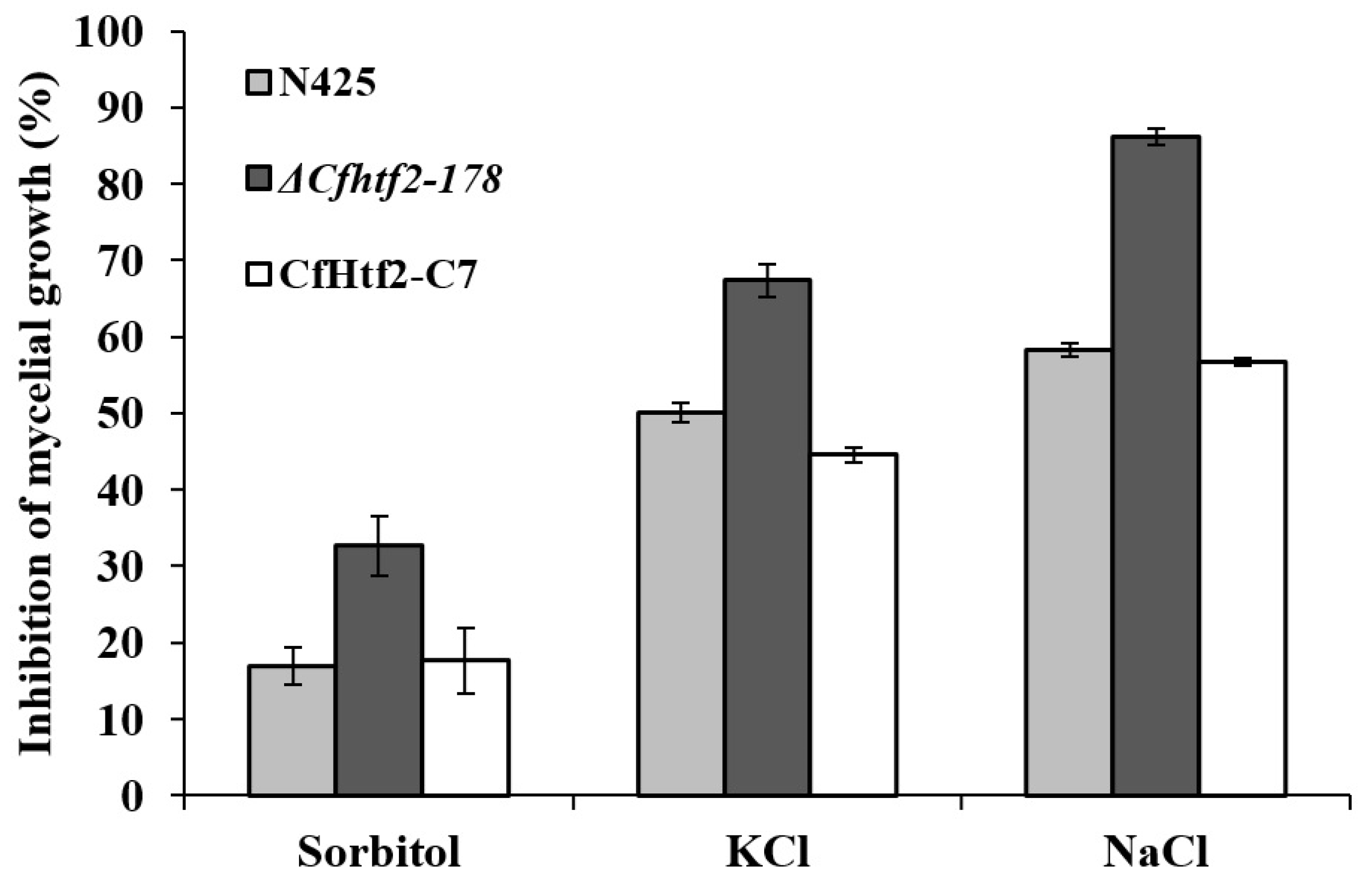
3.4. Response of CfHTF2 to Anthrax under Different Stresses
3.5. Determination of Pathogenicity
3.6. RNA-Seq Analysis
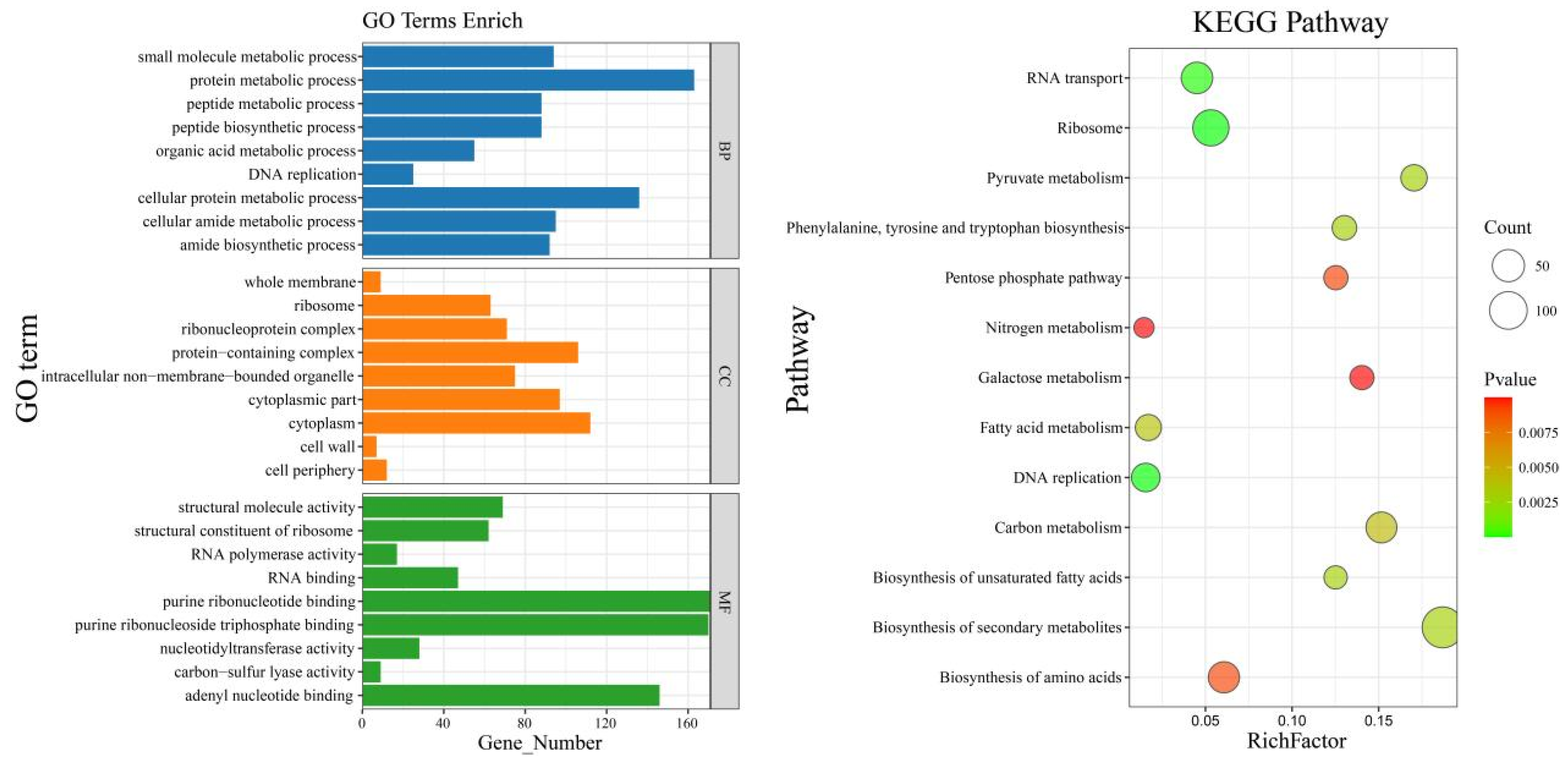
3.7. GO and KEGG Enrichment Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Diez-Ozaeta, I.; Astiazaran, O.J. Recent advances in Kombucha tea: Microbial consortium, chemical parameters, health implications and biocellulose production. Int. J. Food Microbiol. 2022, 377, 109783. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, F.; Li, J.; Yang, X.; Chen, J.; Cao, J.; Wu, X.; Lu, X.; Huang, J.; Li, Y.; et al. Tea consumption and the risk of atherosclerotic cardiovascular disease and all-cause mortality: The China-PAR project. Eur. J. Prev. Cardiol. 2020, 27, 1956–1963. [Google Scholar] [CrossRef]
- Jin, J.Q.; Chai, Y.F.; Liu, Y.F.; Zhang, J.; Yao, M.Z.; Chen, L. Hongyacha, a Naturally Caffeine-Free Tea Plant from Fujian, China. J. Agric. Food Chem. 2018, 66, 11311–11319. [Google Scholar] [CrossRef] [PubMed]
- Dou, Q.P. Tea in Health and Disease. Nutrients 2019, 11, 929. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Cannon, P.F.; Damm, U.; Johnston, P.R.; Weir, B.S. Colletotrichum—Current status and future directions. Stud. Mycol. 2012, 73, 181–213. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Lei, T.; Yuan, H.; Chen, S. First Report of Anthracnose Caused by Colletotrichum kahawae and Colletotrichum horri on Tea-oil Tree in China. Plant Dis. 2022, 107, 1944. [Google Scholar]
- Zhang, S.; Guo, Y.; Chen, S.; Li, H. The Histone Acetyltransferase CfGcn5 Regulates Growth, Development, and Pathogenicity in the Anthracnose Fungus Colletotrichum fructicola on the Tea-Oil Tree. Front. Microbiol. 2021, 12, 680415. [Google Scholar] [CrossRef]
- Hassan, O.; Kim, S.H.; Kim, K.M.; Chang, T. First report of leaf anthracnose caused by Colletotrichum camelliae on tea plants (Camellia sinensis) in South Korea. Plant Dis. 2023, 107, 2881. [Google Scholar] [CrossRef]
- Lin, S.R.; Yu, S.Y.; Chang, T.D.; Lin, Y.J.; Wen, C.J.; Lin, Y.H. First report of anthracnose caused by Colletotrichum fructicola on tea in Taiwan. Plant Dis. 2020, 105, 710. [Google Scholar] [CrossRef]
- Zheng, X.R.; Zhang, M.J.; Shang, X.L.; Fang, S.Z.; Chen, F.M. Etiology of Cyclocarya paliurus Anthracnose in Jiangsu Province, China. Front. Plant Sci. 2021, 11, 613499. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liu, C.; Liu, J.A.; Zhou, G.Y. First Report of Colletotrichum fructicola Causing Anthracnose on Camellia yuhsienensis Hu in China. Plant Dis. 2021, 106, 321. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Ye, C.; Zhang, D.; Bai, R.; Zheng, S.; Hu, W.; Yuan, Y. The Effects of Differentially-Expressed Homeobox Family Genes on the Prognosis and HOXC6 on Immune Microenvironment Orchestration in Colorectal Cancer. Front. Immunol. 2021, 12, 781221. [Google Scholar] [CrossRef] [PubMed]
- Garber, R.L.; Kuroiwa, A.; Gehring, W.J. Genomic and cDNA clones of the homeotic locus Antennapedia in Drosophila. EMBO J. 1983, 2, 2027–2036. [Google Scholar] [CrossRef] [PubMed]
- Pramila, T.; Miles, S.; GuhaThakurta, D.; Jemiolo, D.; Breeden, L.L. Conserved homeodomain proteins interact with MADS box protein Mcm1 to restrict ECB-dependent transcription to the M/G1 phase of the cell cycle. Genes Dev. 2002, 16, 3034–3045. [Google Scholar] [CrossRef] [PubMed]
- Ostapenko, D.; Solomon, M.J. Anaphase promoting complex-dependent degradation of transcriptional repressors Nrm1 and Yhp1 in Saccharomyces cerevisiae. Mol. Biol. Cell 2011, 22, 2175–2184. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Park, S.Y.; Kim, K.S.; Rho, H.S.; Chi, M.H.; Choi, J.; Park, J.; Kong, S.; Park, J.; Goh, J.; et al. Homeobox transcription factors are required for conidiation and appressorium development in the rice blast fungus Magnaporthe oryzae. PLoS Genet. 2009, 5, e1000757. [Google Scholar] [CrossRef]
- Liu, W.; Xie, S.; Zhao, X.; Chen, X.; Zheng, W.; Lu, G.; Xu, J.R.; Wang, Z. A homeobox gene is essential for conidiogenesis of the rice blast fungus Magnaporthe oryzae. Mol. Plant Microbe Interact. 2010, 23, 366–375. [Google Scholar] [CrossRef]
- Zheng, W.; Zhao, X.; Xie, Q.; Huang, Q.; Zhang, C.; Zhai, H.; Xu, L.; Lu, G.; Shim, W.B.; Wang, Z. A conserved homeobox transcription factor Htf1 is required for phialide development and conidiogenesis in Fusarium species. PLoS ONE 2012, 7, e45432. [Google Scholar] [CrossRef]
- Fan, G.; Zheng, H.; Zhang, K.; Devi Ganeshan, V.; Opiyo, S.O.; Liu, D.; Li, M.; Li, G.; Mitchell, T.K.; Yun, Y.; et al. FgHtf1 Regulates Global Gene Expression towards Aerial Mycelium and Conidiophore Formation in the Cereal Fungal Pathogen Fusarium graminearum. Appl. Environ. Microbiol. 2020, 86, e03024-19. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Wangsanut, T.; Fonzi, W.A.; Rolfes, R.J. The GRF10 homeobox gene regulates filamentous growth in the human fungal pathogen Candida albicans. FEMS Yeast Res. 2015, 15, fov093. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; Xue, C.; Peng, Y.; Katan, T.; Kistler, H.C.; Xu, J.R. A mitogen-activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol. Plant Microbe Interact. 2002, 15, 1119–1127. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra–fast all–in–one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, Z.; Zhu, R.; Wang, F.; Cheng, Y.; Liu, Y. Three Differential Expression Analysis Methods for RNA Sequencing: Limma, EdgeR, DESeq2. J. Vis. Exp. 2021, 175, e62528. [Google Scholar]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Zhai, L.N.; Zhao, Y.; Song, X.L.; Qin, T.T.; Zhang, Z.J.; Wang, J.Z.; Sui, C.Y.; Zhang, L.L.; Lv, M.; Hu, L.F.; et al. Inhalable vaccine of bacterial culture supernatant extract mediates protection against fatal pulmonary anthrax. Emerg. Microbes Infect. 2023, 12, 2191741. [Google Scholar] [CrossRef]
- Lu, Z.; Paolella, B.R.; Truex, N.L.; Loftis, A.R.; Liao, X.; Rabideau, A.E.; Brown, M.S.; Busanovich, J.; Beroukhim, R.; Pentelute, B.L. Targeting Cancer Gene Dependencies with Anthrax-Mediated Delivery of Peptide Nucleic Acids. ACS Chem. Biol. 2020, 15, 1358–1369. [Google Scholar] [CrossRef]
- Liang, X.; Zhang, E.; Zhang, H.; Wei, J.; Li, W.; Zhu, J.; Wang, B.; Dong, S. Involvement of the pagR gene of pXO2 in anthrax pathogenesis. Sci. Rep. 2016, 6, 28827. [Google Scholar] [CrossRef]
- Boufleur, T.R.; Ciampi-Guillardi, M.; Tikami, Í.; Rogério, F.; Thon, M.R.; Sukno, S.A.; Massola, N.S., Jr.; Baroncelli, R. Soybean anthracnose caused by Colletotrichum species: Current status and future prospects. Mol. Plant Pathol. 2021, 22, 393–409. [Google Scholar] [CrossRef] [PubMed]
- Peralta-Ruiz, Y.; Rossi, C.; Grande-Tovar, C.D.; Chaves-López, C. Green Management of Postharvest Anthracnose Caused by Colletotrichum gloeosporioides. J. Fungi 2023, 9, 623. [Google Scholar] [CrossRef] [PubMed]
- Song, F.Q.; Gao, X.Q.; Liang, L.L. Effects of Homeobox transcription factors on Aspergillus flavus growth and toxin synthesis. Mycosystema 2020, 39, 566–580. [Google Scholar]
- Ruiz-Roldán, C.; Pareja-Jaime, Y.; González-Reyes, J.A.; Roncero, M.I. The Transcription Factor Con7-1 Is a Master Regulator of Morphogenesis and Virulence in Fusarium oxysporum. Mol. Plant Microbe Interact. 2015, 28, 55–68. [Google Scholar] [CrossRef]
- Pasquali, M.; Spanu, F.; Scherm, B.; Balmas, V.; Hoffmann, L.; Hammond-Kosack, K.E.; Beyer, M.; Migheli, Q. FcStuA from Fusarium culmorum controls wheat foot and root rot in a toxin dispensable manner. PLoS ONE 2013, 8, e57429. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.; Zhou, Z.; Guo, T.; Huang, X.; Peng, C.; Lin, Z.; Chen, M.; Liu, W. CFHTF2 Is Needed for Vegetative Growth, Conidial Morphogenesis and the Osmotic Stress Response in the Tea Plant Anthracnose (Colletotrichum fructicola). Genes 2023, 14, 2235. https://doi.org/10.3390/genes14122235
Zhang C, Zhou Z, Guo T, Huang X, Peng C, Lin Z, Chen M, Liu W. CFHTF2 Is Needed for Vegetative Growth, Conidial Morphogenesis and the Osmotic Stress Response in the Tea Plant Anthracnose (Colletotrichum fructicola). Genes. 2023; 14(12):2235. https://doi.org/10.3390/genes14122235
Chicago/Turabian StyleZhang, Chengkang, Ziwen Zhou, Tianlong Guo, Xin Huang, Chengbin Peng, Zhideng Lin, Meixia Chen, and Wei Liu. 2023. "CFHTF2 Is Needed for Vegetative Growth, Conidial Morphogenesis and the Osmotic Stress Response in the Tea Plant Anthracnose (Colletotrichum fructicola)" Genes 14, no. 12: 2235. https://doi.org/10.3390/genes14122235



