Genomic Landscape of Copy Number Variations and Their Associations with Climatic Variables in the World’s Sheep
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collecting, CNV Calling, and Quality Controls
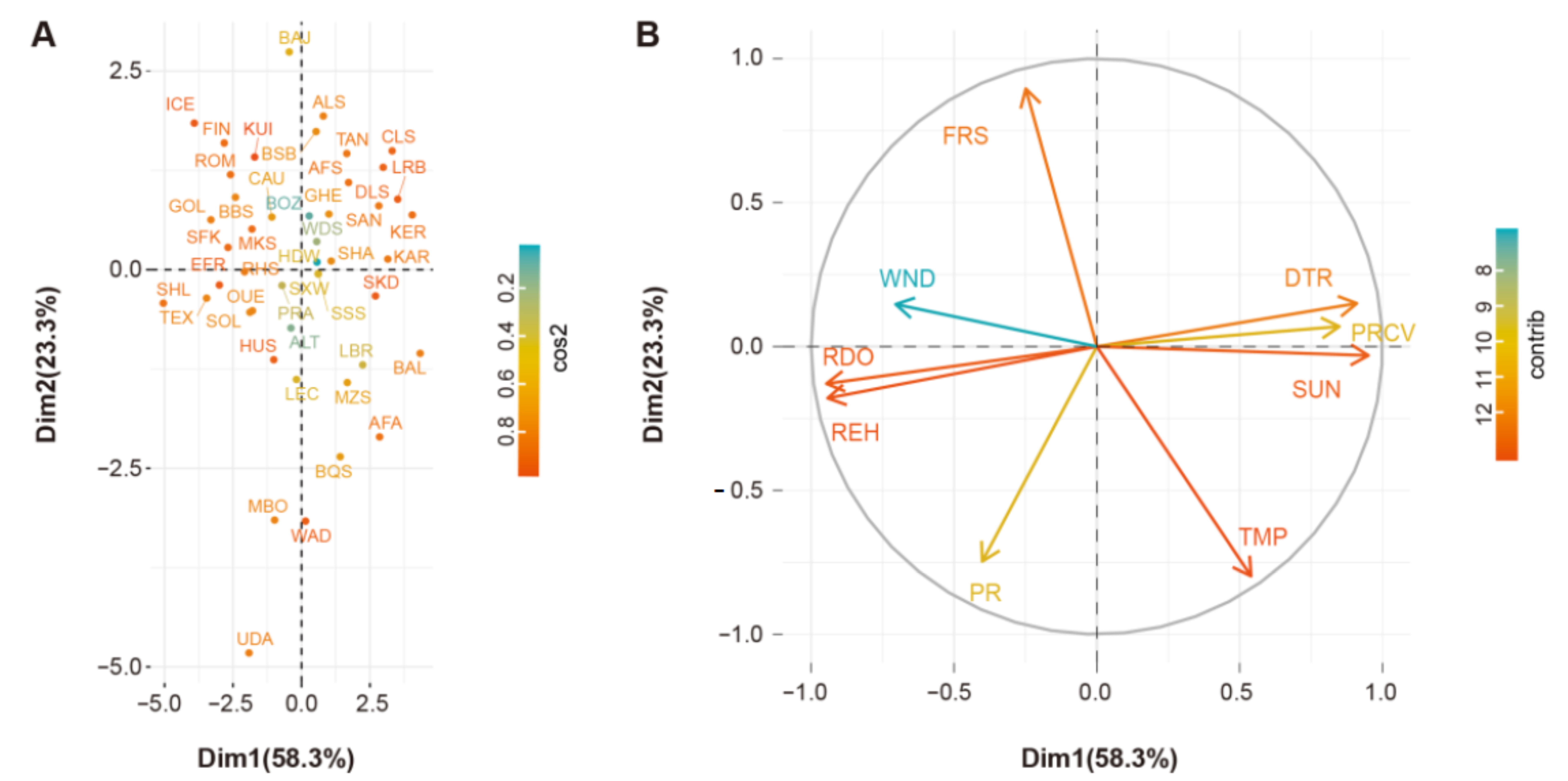
2.2. Environmental Data
2.3. Estimation of Solar Irradiation
2.4. Testing for Genomic Signatures Associated with Climate Variables
2.5. Testing for Genomic Signatures Associated with Solar Radiation-Mediated Selective Pressure
2.6. Gene Annotation and Overlapping with QTLs
3. Results
3.1. CNV Detection and Population Differential Analyses
3.2. Function Annotation of CNVRs
3.3. Climate-Driven Candidate Selective Signatures Testing
3.4. Solar Radiation-Driven Candidate Signatures Test
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zeder, M.A. Domestication and early agriculture in the Mediterranean Basin: Origins, diffusion, and impact. Proc. Natl. Acad. Sci. USA 2008, 105, 11597–11604. [Google Scholar] [CrossRef] [PubMed]
- Driscoll, C.A.; Macdonald, D.W.; O’Brien, S.J. From wild animals to domestic pets, an evolutionary view of domestication. Proc. Natl. Acad. Sci. USA 2009, 106, 9971–9978. [Google Scholar] [CrossRef] [PubMed]
- Tapio, M.; Marzanov, N.; Ozerov, M.; Ćinkulov, M.; Gonzarenko, G.; Kiselyova, T.; Murawski, M.; Viinalass, H.; Kantanen, J. Sheep Mitochondrial DNA Variation in European, Caucasian, and Central Asian Areas. Mol. Biol. Evol. 2006, 23, 1776–1783. [Google Scholar] [CrossRef] [PubMed]
- Chessa, B.; Pereira, F.; Arnaud, F.; Amorim, A.; Goyache, F.; Mainland, I.; Kao, R.R.; Pemberton, J.M.; Beraldi, D.; Stear, M.J.; et al. Revealing the History of Sheep Domestication Using Retrovirus Integrations. Science 2009, 324, 532–536. [Google Scholar] [CrossRef]
- Lv, F.H.; Agha, S.; Kantanen, J.; Colli, L.; Stucki, S.; Kijas, J.W.; Joost, S.; Li, M.-H.; Marsan, P.A. Adaptations to Climate-Mediated Selective Pressures in Sheep. Mol. Biol. Evol. 2014, 31, 3324–3343. [Google Scholar] [CrossRef]
- Lv, F.H.; Peng, W.F.; Yang, J.; Zhao, Y.X.; Li, W.R.; Liu, M.J.; Ma, Y.H.; Zhao, Q.J.; Yang, G.L.; Wang, F.; et al. Mitogenomic Meta-Analysis Identifies Two Phases of Migration in the History of Eastern Eurasian Sheep. Mol. Biol. Evol. 2015, 32, 2515–2533. [Google Scholar] [CrossRef]
- Kim, E.S.; Elbeltagy, A.R.; Aboul-Naga, A.M.; Rischkowsky, B.; Sayre, B.; Mwacharo, J.M.; Rothschild, M.F. Multiple genomic signatures of selection in goats and sheep indigenous to a hot arid environment. Heredity 2016, 116, 255–264. [Google Scholar] [CrossRef]
- Yang, J.; Li, W.R.; Lv, F.H.; He, S.G.; Tian, S.L.; Peng, W.F.; Sun, Y.W.; Zhao, Y.X.; Tu, X.L.; Zhang, M.; et al. Whole-Genome Sequencing of Native Sheep Provides Insights into Rapid Adaptations to Extreme Environments. Mol. Biol. Evol. 2016, 33, 2576–2592. [Google Scholar] [CrossRef]
- Cao, Y.H.; Xu, S.S.; Shen, M.; Chen, Z.H.; Gao, L.; Lv, F.H.; Xie, X.L.; Wang, X.H.; Yang, H.; Liu, C.B.; et al. Historical Introgression from Wild Relatives Enhanced Climatic Adaptation and Resistance to Pneumonia in Sheep. Mol. Biol. Evol. 2021, 38, 838–855. [Google Scholar] [CrossRef]
- Senczuk, G.; Criscione, A.; Mastrangelo, S.; Biscarini, F.; Marletta, D.; Pilla, F.; Laloë, D.; Ciampolini, R. How Geography and Climate Shaped the Genomic Diversity of Italian Local Cattle and Sheep Breeds. Animals 2022, 12, 2198. [Google Scholar] [CrossRef]
- Hou, Y.; Liu, G.E.; Bickhart, D.M.; Cardone, M.F.; Wang, K.; Ventura, M.; Song, J.; VanRaden, P.M.; Sonstegard, T.S.; Van Tassell, C.P.; et al. Genomic characteristics of cattle copy number variations. BMC Genom. 2011, 12, 127. [Google Scholar] [CrossRef]
- Zarrei, M.; MacDonald, J.R.; Merico, D.; Scherer, S.W. A copy number variation map of the human genome. Nat. Rev. Genet. 2015, 16, 172–183. [Google Scholar] [CrossRef]
- Hastings, P.J.; Lupski, J.R.; Rosenberg, S.M.; Ira, G. Mechanisms of change in gene copy number. Nat. Rev. Genet. 2009, 10, 551–564. [Google Scholar] [CrossRef]
- Bruder, C.E.G.; Piotrowski, A.; Gijsbers, A.A.C.J.; Andersson, R.; Erickson, S.; de Stahl, T.D.; Menzel, U.; Sandgren, J.; von Tell, D.; Poplawski, A.; et al. Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am. J. Hum. Genet. 2008, 82, 763–771. [Google Scholar] [CrossRef]
- Nguyen, D.Q.; Webber, C.; Ponting, C.P. Bias of selection on human copy-number variants. PLoS Genet. 2006, 2, e20. [Google Scholar] [CrossRef]
- Dumas, L.; Kim, Y.H.; Karimpour-Fard, A.; Cox, M.; Hopkins, J.; Pollack, J.R.; Sikela, J.M. Gene copy number variation spanning 60 million years of human and primate evolution. Genome Res. 2007, 17, 1266–1277. [Google Scholar] [CrossRef]
- Conrad, D.F.; Pinto, D.; Redon, R.; Feuk, L.; Gokcumen, O.; Zhang, Y.; Aerts, J.; Andrews, T.D.; Barnes, C.; Campbell, P.; et al. Origins and functional impact of copy number variation in the human genome. Nature 2010, 464, 704–712. [Google Scholar] [CrossRef]
- Kibriya, M.G.; Jasmine, F.; Parvez, F.; Argos, M.; Roy, S.; Paul-Brutus, R.; Islam, T.; Ahmed, A.; Rakibuz-Zaman, M.; Shinkle, J.; et al. Association between genome-wide copy number variation and arsenic-induced skin lesions: A prospective study. Environ. Health 2017, 16, 75. [Google Scholar] [CrossRef] [PubMed]
- Fontanesi, L.; Beretti, F.; Riggio, V.; Gómez González, E.; Dall’Olio, S.; Davoli, R.; Russo, V.; Portolano, B. Copy number variation and missense mutations of the agouti signaling protein (ASIP) gene in goat breeds with different coat colors. Cytogenet. Genome Res. 2009, 126, 333–347. [Google Scholar] [CrossRef] [PubMed]
- Giuffra, E.; Törnsten, A.; Marklund, S.; Bongcam-Rudloff, E.; Chardon, P.; Kijas, J.M.; Anderson, S.I.; Archibald, A.L.; Andersson, L. A large duplication associated with dominant white color in pigs originated by homologous recombination between LINE elements flanking KIT. Mamm. Genome 2002, 13, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Fan, H.; Yuan, Z.; Hu, S.; Ma, X.; Xuan, J.; Wang, H.; Zhang, L.; Wei, C.; Zhang, Q.; et al. Genome-wide detection of CNVs in Chinese indigenous sheep with different types of tails using ovine high-density 600K SNP arrays. Sci. Rep. 2016, 6, 27822. [Google Scholar] [CrossRef]
- Li, X.; Yang, J.; Shen, M.; Xie, X.L.; Xu, Y.X.; Lv, F.H.; Shen, M.; Yang, H.; Yang, Y.L.; Liu, C.B.; et al. Whole-genome resequencing of wild and domestic sheep identifies genes associated with morphological and agronomic traits. Nat. Commun. 2020, 11, 2815. [Google Scholar] [CrossRef]
- Hancock, A.M.; Witonsky, D.B.; Alkorta-Aranburu, G.; Beall, C.M.; Gebremedhin, A.; Sukernik, R.; Utermann, G.; Pritchard, J.K.; Coop, G.; Di Rienzo, A. Adaptations to Climate-Mediated Selective Pressures in Humans. PLoS Genet. 2011, 7, e1001375. [Google Scholar] [CrossRef]
- Hovhannisyan, G.; Harutyunyan, T.; Aroutiounian, R.; Liehr, T. DNA Copy Number Variations as Markers of Mutagenic Impact. Int. J. Mol. Sci. 2019, 20, 4723. [Google Scholar] [CrossRef]
- Tsilimigaki, S.I.; Messini-Nikolaki, N.; Kanariou, M.; Piperakis, S.M. A study on the effects of seasonal solar radiation on exposed populations. Mutagenesis 2003, 18, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Zitzelsberger, H.; Unger, K. DNA copy number alterations in radiation-induced thyroid cancer. Clin. Oncol. (R. Coll. Radiol.) 2011, 23, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Skinner, M.K.; Guerrero-Bosagna, C.; Haque, M.M. Environmentally induced epigenetic transgenerational inheritance of sperm epimutations promote genetic mutations. Epigenetics 2015, 10, 762–771. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, P.; Vollger, M.R.; Dang, V.; Porubsky, D.; Baker, C.; Cantsilieris, S.; Hoekzema, K.; Lewis, A.P.; Munson, K.M.; Sorensen, M.; et al. Adaptive archaic introgression of copy number variants and the discovery of previously unknown human genes. Science 2019, 366, eaax2083. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, Y.; Wang, X.; Jiang, Q.; Zhao, H.; Wang, J.; Ju, Z.; Yang, L.; Gao, Y.; Wei, X.; et al. Population Structure, and Selection Signatures Underlying High-Altitude Adaptation Inferred From Genome-Wide Copy Number Variations in Chinese Indigenous Cattle. Front. Genet. 2020, 10, 1404. [Google Scholar] [CrossRef]
- Liu, M.; Zhou, Y.; Rosen, B.D.; Van Tassell, C.P.; Stella, A.; Tosser-Klopp, G.; Rupp, R.; Palhiere, I.; Colli, L.; Sayre, B.; et al. Diversity of copy number variation in the worldwide goat population. Heredity 2019, 122, 636–646. [Google Scholar] [CrossRef] [PubMed]
- Solé, M.; Ablondi, M.; Binzer-Panchal, A.; Velie, B.D.; Hollfelder, N.; Buys, N.; Ducro, B.J.; François, L.; Janssens, S.; Schurink, A.; et al. Inter- and intra-breed genome-wide copy number diversity in a large cohort of European equine breeds. BMC Genom. 2019, 20, 759. [Google Scholar] [CrossRef] [PubMed]
- Berglund, J.; Nevalainen, E.M.; Molin, A.M.; Perloski, M.; André, C.; Zody, M.C.; Sharpe, T.; Hitte, C.; Lindblad-Toh, K.; Lohi, H.; et al. Novel origins of copy number variation in the dog genome. Genome Biol. 2012, 13, R73. [Google Scholar] [CrossRef] [PubMed]
- Perry, G.H.; Dominy, N.J.; Claw, K.G.; Lee, A.S.; Fiegler, H.; Redon, R.; Werner, J.; Villanea, F.A.; Mountain, J.L.; Misra, R.; et al. Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 2007, 39, 1256–1260. [Google Scholar] [CrossRef]
- Reiter, T.; Jagoda, E.; Capellini, T.D. Dietary Variation and Evolution of Gene Copy Number among Dog Breeds. PLoS ONE 2016, 11, e0148899. [Google Scholar] [CrossRef]
- Gautier, M.; Moazami-Goudarzi, K.; Levéziel, H.; Parinello, H.; Grohs, C.; Rialle, S.; Kowalczyk, R.; Flori, L. Deciphering the wisent demographic and adaptive histories from individual whole-genome sequences. Mol. Biol. Evol. 2016, 33, 2801–2814. [Google Scholar] [CrossRef]
- Fontanesi, L.; Martelli, P.L.; Beretti, F.; Riggio, V.; Dall’Olio, S.; Colombo, M.; Casadio, R.; Russo, V.; Portolano, B. An initial comparative map of copy number variations in the goat (Capra hircus) genome. BMC Genom. 2010, 11, 639. [Google Scholar] [CrossRef]
- Wang, W.; Wang, S.; Hou, C.; Xing, Y.; Cao, J.; Wu, K.; Liu, C.; Zhang, D.; Zhang, L.; Zhang, Y.; et al. Genome-wide detection of copy number variations among diverse horse breeds by array CGH. PLoS ONE 2014, 30, e86860. [Google Scholar] [CrossRef] [PubMed]
- Scheffers, B.R.; De Meester, L.; Bridge, T.C.; Hoffmann, A.A.; Pandolfi, J.M.; Corlett, R.T.; Butchart, S.H.; Pearce-Kelly, P.; Kovacs, K.M.; Dudgeon, D.; et al. The broad footprint of climate change from genes to biomes to people. Science 2016, 354, aaf7671. [Google Scholar] [CrossRef]
- Salehian-Dehkordi, H.; Xu, Y.X.; Xu, S.S.; Li, X.; Luo, L.Y.; Liu, Y.J.; Wang, D.F.; Cao, Y.H.; Shen, M.; Gao, L.; et al. Genome-Wide Detection of Copy Number Variations and Their Association With Distinct Phenotypes in the World’s Sheep. Front. Genet. 2021, 12, 670582. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hadley, D.; Liu, R.; Glessner, J.; Grant, S.F.A.; Hakonarson, H.; Bucan, M. PennCNV: An integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007, 17, 1665–1674. [Google Scholar] [CrossRef]
- Maurano, M.T.; Humbert, R.; Rynes, E.; Thurman, R.E.; Haugen, E.; Wang, H.; Reynolds, A.P.; Sandstrom, R.; Qu, H.; Brody, J.; et al. Systematic Localization of Common Disease-Associated Variation in Regulatory DNA. Science 2012, 337, 1190–1195. [Google Scholar] [CrossRef] [PubMed]
- New, M.; Lister, D.; Hulme, M.; Makin, I. A high-resolution data set of surface climate over global land areas. Clim. Res. 2002, 21, 1–25. [Google Scholar] [CrossRef]
- Suehrcke, H.; Bowden, R.S.; Hollands, K. Relationship between sunshine duration and solar radiation. Sol. Energy 2013, 92, 160–171. [Google Scholar] [CrossRef]
- Duffie, J.A.; Beckman, W.A. Solar Engineering of Thermal Processes; John Wiley & Sons: Hoboken, NJ, USA, 2013. [Google Scholar]
- Gueymard, C.A. Revised composite extraterrestrial spectrum based on recent solar irradiance observations. Sol. Energy 2018, 169, 434–440. [Google Scholar] [CrossRef]
- Duruz, S.; Sevane, N.; Selmoni, O.; Vajana, E.; Leempoel, K.; Stucki, S.; Orozco-terWengel, P.; Rochat, E.; Dunner, S.; The NEXTGEN Consortium; et al. Rapid identification and interpretation of gene–environment associations using the new R. SamBada landscape genomics pipeline. Mol. Ecol. Resour. 2019, 19, 1355–1365. [Google Scholar] [CrossRef]
- Frichot, E.; Schoville, S.D.; Bouchard, G.; François, O. Testing for Associations between Loci and Environmental Gradients Using Latent Factor Mixed Models. Mol. Biol. Evol. 2013, 30, 1687–1699. [Google Scholar] [CrossRef]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 2009, 9, 1322–1332. [Google Scholar] [CrossRef]
- Patterson, N.; Price, A.L.; Reich, D. Population Structure and Eigenanalysis. PLoS Genet. 2006, 2, e190. [Google Scholar] [CrossRef]
- Caye, K.; Jumentier, B.; Lepeule, J.; François, O. LFMM 2: Fast and Accurate Inference of Gene-Environment Associations in Genome-Wide Studies. Mol. Biol. Evol. 2019, 36, 852–860. [Google Scholar] [CrossRef]
- Glessner, J.T.; Li, J.; Hakonarson, H. ParseCNV integrative copy number variation association software with quality tracking. Nucleic Acids Res. 2013, 41, e64. [Google Scholar] [CrossRef]
- Kang, H.M.; Sul, J.H.; Service, S.K.; Zaitlen, N.A.; Kong, S.y.; Freimer, N.B.; Sabatti, C.; Eskin, E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010, 42, 348–354. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Fang, L.; Liu, S.; Pan, M.G.; Seroussi, E.; Cole, J.B.; Ma, L.; Chen, H.; Liu, G.E. Array CGH-based detection of CNV regions and their potential association with reproduction and other economic traits in Holsteins. BMC Genom. 2019, 20, 181. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.S.; Cheong, H.S.; Kim, L.H.; NamGung, S.; Park, T.J.; Chun, J.Y.; Kim, J.Y.; Pasaje, C.F.A.; Lee, J.S.; Shin, H.D. Identification of copy number variations and common deletion polymorphisms in cattle. BMC Genom. 2010, 11, 232. [Google Scholar] [CrossRef]
- Wang, M.D.; Dzama, K.; Hefer, C.A.; Muchadeyi, F.C. Genomic population structure and prevalence of copy number variations in South African Nguni cattle. BMC Genom. 2015, 16, 894. [Google Scholar] [CrossRef]
- Prunier, J.; Giguère, I.; Ryan, N.; Guy, R.; Soolanayakanahally, R.; Isabel, N.; MacKay, J.; Porth, I. Gene copy number variations involved in balsam poplar. Mol. Ecol. 2019, 28, 1476–1490. [Google Scholar] [CrossRef]
- Mei, C.G.; Junjvlieke, Z.; Raza, S.H.A.; Wang, H.B.; Cheng, G.; Zhao, C.; Zhu, W.; Zan, L. Copy number variation detection in Chinese indigenous cattle by whole genome sequencing. Genomics 2019, 112, 831–836. [Google Scholar] [CrossRef]
- Wang, Y.; Song, H.; Wang, W.; Zhang, Z. Generation and characterization of Megf6 null and Cre knock-in alleles. Genesis 2019, 57, e23262. [Google Scholar] [CrossRef]
- Xu, L.Y.; Yang, L.; Wang, L.; Zhu, B.; Chen, Y.; Gao, H.; Gao, X.; Zhang, L.; Liu, G.E.; Li, J. Probe-based association analysis identifies several deletions associated with average daily gain in beef cattle. BMC Genom. 2019, 20, 31. [Google Scholar] [CrossRef]
- Beckmann, J.S.; Estivill, X.; Antonarakis, S.E. Copy number variants and genetic traits: Closer to the resolution of phenotypic to genotypic variability. Nat. Rev. Genet. 2007, 8, 639–646. [Google Scholar] [CrossRef]
- O’Donovan, M.C.; Kirov, G.; Owen, M.J. Phenotypic variations on the theme of CNVs. Nat. Genet. 2008, 40, 1392–1393. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, M.; Kuroda, T.S.; Mikoshiba, K. Slac2-a/Melanophilin, the Missing Link between Rab27 and Myosin Va. J. Biol. Chem. 2002, 277, 12432–12436. [Google Scholar] [CrossRef]
- Gudbjartsson, D.F.; Sulem, P.; Stacey, S.N.; Goldstein, A.M.; Rafnar, T.; Sigurgeirsson, B.; Benediktsdottir, K.R.; Thorisdottir, K.; Ragnarsson, R.; Sveinsdottir, S.G.; et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nat. Genet. 2008, 40, 886–891. [Google Scholar] [CrossRef]
- Yamakaze, J.; Lu, Z. Deletion of the lactoperoxidase gene causes multisystem inflammation and tumors in mice. Sci. Rep. 2021, 11, 12429. [Google Scholar] [CrossRef]
- Zhu, X.; Chang, Y.P.C.; Yan, D.; Weder, A.; Cooper, R.; Luke, A.; Kan, D.; Chakravarti, A. Associations Between Hypertension and Genes in the Renin-Angiotensin System. Hypertension 2003, 41, 1027–1034. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.K.; Singh, S.; Nandhini, P.B.; Bhatia, A.K.; Dixit, S.P.; Ganguly, I. Comparative genomic diversity analysis of copy number variations (CNV) in indicine and taurine cattle thriving in Europe and Indian subcontinent. Anim. Biotechnol. 2023, 1–12. [Google Scholar] [CrossRef]
- Kalds, P.; Zhou, S.; Gao, Y.; Cai, B.; Huang, S.; Chen, Y.; Wang, X. Genetics of the phenotypic evolution in sheep: A molecular look at diversity-driving genes. Genet. Sel. Evol. 2022, 54, 61. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.X.; Wang, B.; Jing, J.N.; Ma, R.; Luo, Y.H.; Li, X.; Yan, Z.; Liu, Y.J.; Gao, L.; Ren, Y.L.; et al. Whole-body adipose tissue multi-omic analyses in sheep reveal molecular mechanisms underlying local adaptation to extreme environments. Commun. Biol. 2023, 6, 159. [Google Scholar] [CrossRef]
- Aboul-Naga, A.M.; Alsamman, A.M.; El Allali, A.; Elshafie, M.H.; Abdelal, E.S.; Abdelkhalek, T.M.; Abdelsabour, T.H.; Mohamed, L.G.; Hamwieh, A. Genome-wide analysis identified candidate variants and genes associated with heat stress adaptation in Egyptian sheep breeds. Front. Genet. 2022, 3, 898522. [Google Scholar] [CrossRef]
- Nabi Khan, R.I.; Sahu, A.R.; Malla, W.A.; Praharaj, M.R.; Hosamani, N.; Kumar, S.; Gupta, S.; Sharma, S.; Saxena, A.; Varshney, A.; et al. Systems biology under heat stress in Indian cattle. Gene 2021, 30, 145908. [Google Scholar] [CrossRef]
- Seifert, A.; Schofield, P.; Barton, G.J.; Hay, R.T. Proteotoxic stress reprograms the chromatin landscape of SUMO modification. Sci. Signal. 2015, 8, rs7. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Yang, Y.; Wang, L.; Ma, T.; Shang, H.; Ding, L.; Han, J.; Qiu, Q. Different gene expressions between cattle and yak provide insights into high-altitude adaptation. Anim. Genet. 2016, 47, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Igoshin, A.V.; Deniskova, T.E.; Yurchenko, A.A.; Yudin, N.S.; Dotsev, A.V.; Selionova, M.I.; Zinovieva, N.A.; Larkin, D.M. Copy number variants in genomes of local sheep breeds from Russia. Anim. Genet. 2022, 53, 119–132. [Google Scholar] [CrossRef]
- Maiuri, T.; Mocle, A.J.; Hung, C.L.; Xia, J.; van Roon-Mom, W.M.; Truant, R. Huntingtin is a scaffolding protein in the ATM oxidative DNA damage response complex. Hum. Mol. Genet. 2017, 26, 395–406. [Google Scholar] [CrossRef]
- Naji, M.M.; Jiang, Y.; Utsunomiya, Y.T.; Rosen, B.D.; Sölkner, J.; Wang, C.; Jiang, L.; Zhang, Q.; Zhang, Y.; Ding, X.; et al. Favored single nucleotide variants identified using whole genome Re-sequencing of Austrian and Chinese cattle breeds. Front. Genet. 2022, 13, 974787. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, S.; Dai, W.; Xie, C.; Li, J.C. A Comprehensive Prognostic and Immune Analysis of SLC41A3 in Pan-Cancer. Front. Oncol. 2021, 10, 586414. [Google Scholar] [CrossRef] [PubMed]
- Zhai, R.; Wang, W.; Zhang, D.; Li, X.; Zhang, Y.; Zhao, Y.; Zhao, L.; Wang, J.; Xu, D.; Cheng, J.; et al. Novel polymorphism at KLF15 gene and its association with growth traits in Hu sheep. Anim. Biotechnol. 2022, 1–7. [Google Scholar] [CrossRef]
- Chen, J.; Okimura, K.; Yoshimura, T. Light and Hormones in Seasonal Regulation of Reproduction and Mood. Endocrinology 2020, 161, bqaa130. [Google Scholar] [CrossRef]
- Brunet, A.G.; Santiago-Moreno, J.; Toledano-Díaz, A.; López-Sebastián, A. Reproductive seasonality and its control in Spanish sheep and goats. Biol. Trop. Subtrop. Agroecosyst. 2011, 15, S47–S70. [Google Scholar]
- Gomez-Brunet, A.; Santiago-Moreno, J.; del Campo, A.; Malpaux, B.; Chemineau, P.; Tortonese, D.J. Endogenous Circannual Cycles of Ovarian Activity and Changes in Prolactin and Melatonin Secretion in Wild and Domestic Female Sheep Maintained under a Long-Day Photoperiod. Biol. Reprod. 2008, 78, 552–562. [Google Scholar] [CrossRef]
- Chemineau, P.; Bodin, L.; Migaud, M.; Thiéry, J.C.; Malpaux, B. Neuroendocrine and Genetic Control of Seasonal Reproduction in Sheep and Goats. Reprod. Domest. Anim. 2010, 45, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Bertolini, F.; Servin, B.; Talenti, A.; Rochat, E.; Kim, E.S.; Oget, C.; Palhière, I.; Crisà, A.; Catillo, G.; Steri, R.; et al. Signatures of selection and environmental adaptation across the goat genome post-domestication. Genet. Sel. Evol. 2018, 50, 57. [Google Scholar] [CrossRef] [PubMed]
- Flori, L.; Moazami-Goudarzi, K.; Alary, V.; Araba, A.; Boujenane, I.; Boushaba, N.; Casabianca, F.; Casu, S.; Ciampolini, R.; D’Acier, A.C.; et al. A genomic map of climate adaptation in Mediterranean cattle breeds. Mol. Ecol. 2019, 28, 1009–1029. [Google Scholar] [CrossRef]
- Yuan, C.; Lu, Z.; Guo, T.; Yue, Y.; Wang, X.; Wang, T.; Zhang, Y.; Hou, F.; Niu, C.; Sun, X.; et al. A global analysis of CNVs in Chinese indigenous fine-wool sheep populations using whole-genome resequencing. BMC Genom. 2021, 22, 78. [Google Scholar] [CrossRef] [PubMed]
- Di Gerlando, R.; Mastrangelo, S.; Tolone, M.; Rizzuto, I.; Sutera, A.M.; Moscarelli, A.; Portolano, B.; Sardina, M.T. Identification of Copy Number Variations and Genetic Diversity in Italian Insular Sheep Breeds. Animals 2022, 12, 217. [Google Scholar] [CrossRef]
- Norris, B.J.; Whan, V.A. A gene duplication affecting expression of the ovine ASIP gene is responsible for white and black sheep. Genome Res. 2008, 18, 1282–1293. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Tao, H.; Li, P.; Li, L.; Zhong, T.; Wang, L.; Ma, J.; Chen, X.; Song, T.; Zhang, H. Whole-genome sequencing reveals selection signatures associated with important traits in six goat breeds. Sci. Rep. 2018, 8, 10405. [Google Scholar] [CrossRef] [PubMed]
- Braz, C.U.; Rowan, T.N.; Schnabel, R.D.; Decker, J.E. Genome-wide association analyses identify genotype-by-environment interactions of growth traits in Simmental cattle. Sci. Rep. 2021, 11, 13335. [Google Scholar] [CrossRef]
- Smith, J.L.; Wilson, M.L.; Nilson, S.M.; Rowan, T.N.; Schnabel, R.D.; Decker, J.E.; Seabury, C.M. Genome-wide association and genotype by environment interactions for growth traits in U.S. Red Angus cattle. BMC Genom. 2022, 23, 517. [Google Scholar] [CrossRef]
- Corvo, M.D.; Lazzari, B.; Capra, E.; Zavarez, L.; Milanesi, M.; Utsunomiya, Y.T.; Utsunomiya, A.T.H.; Stella, A.; de Paula Nogueira, G.; Garcia, J.F.; et al. Methylome Patterns of Cattle Adaptation to Heat Stress. Front. Genet. 2021, 12, 633132. [Google Scholar] [CrossRef]



| Type | CNVs | CNVRs | Unique CNVs | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Homozygous Deletion | Hemizygous Deletion | Duplication | Biallelic Triplication | Total | Loss | Gain | Mixed | Total | Deletion | Duplication | Mixed | Total | |
| Count | 4726 | 23,298 | 11,018 | 103 | 39,145 | 3468 | 861 | 440 | 4769 | 11,119 | 4118 | 510 | 15,747 |
| Length (Mb) | 94.4 | 801.2 | 460.9 | 3.2 | 1359.7 | 150 | 23.4 | 80.9 | 254 | 492.6 | 213.6 | 14.2 | 720.4 |
| Significant CNVRs | Count SNPs | Gene ID |
|---|---|---|
| chr16:60477356-60501528 | 9 | LOC101104428, LOC101120862 |
| chr16:69228726-70108247 | 26 | IRX1, LOC105602651, LOC105602652, LOC105608090 |
| chr26:399178-402542 | 4 | DLGAP2 |
| chr12:77533856-78344379 | 14 | LOC105610048, LOC105616624, LOC105610045, LOC105610042, LOC105610043, LOC106991502, RABIF, LOC106991503, LOC106991504, UBE2T, C12H1orf106, LOC105610049, CACNA1S, TMEM9, IGFN1, PKP1, LOC101109820, KLHL12, LOC105616599, SYT2, LOC105616625, LOC101110351, LOC101110859, KIF21B |
| chr19:58987471-60169432 | 29 | LOC105603604, GATA2, DNAJB8, LOC105603605, LOC105603606, SEC61A1, LOC106991760, LOC105603608, LOC105603609, LOC105603631, LOC106991788, RPN1, EEFSEC, RUVBL1, MGLL, ZXDC, SLC41A3, ALDH1L1, MCM2, KBTBD12, PLXNA1, KLF15, RAB7A |
| chr21:41984078-42773808 | 4 | SLC22A12, MEN1, PPP2R5B, CDC42EP2, GPHA2, ZFPL1, TMEM262, TM7SF2, ZNHIT2, FAU, MRPL49, SYVN1, SPDYC, LOC101107147, TIGD3, SLC25A45, SLC22A11, LOC105604109, RASGRP2, LOC105604110, MAP4K2, CDC42BPG, EHD1, C21H11orf85, BATF2, NAALADL1, CDCA5, VPS51, LOC101105958, LOC105604111, SLC22A20, POLA2, DPF2, NRXN2, SF1, ATG2A, SNX15, SAC3D1, ARL2, CAPN1, MIR194, PYGM |
| chr21:46919932-47922324 | 17 | LOC105604138, LOC101118574, PHLDA2, LOC105604139, DHCR7, NADSYN1, OSBPL5, CARS, NAP1L4, SLC22A18, LOC101118066, SHANK2, CDKN1C |
| chr6:114803668-114941025 | 22 | MSANTD1, RGS12, HTT |
| chr14:55268932-55284965 | 7 | IZUMO2, LOC101115729 |
| chr17:70557208-70575800 | 7 | LOC106991688, GSTT2B, LOC101111397, LOC101118990 |
| chr13:53886521-53904177 | 9 | SLCO4A1 |
| chr16:31746166-31765064 | 6 | CCDC152 |
| chr1:262818152-263558472 | 31 | LOC105604794, POFUT2, SLC19A1, PCBP3, ADARB1, COL18A1 |
| chr14:48269630-48282740 | 4 | EID2 |
| chr22:49712254-50377777 | 25 | LOC105604373, PWWP2B, LOC105606161, LOC105604355, JAKMIP3, DPYSL4, LOC101109500, STK32C, INPP5A, LOC101110287 |
| chr3:137339004-137374322 | 8 | LOC101123028, LOC101123287 |
| chr18:64006157-64006658 | 3 | BEGAIN |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salehian-Dehkordi, H.; Huang, J.-H.; Pirany, N.; Mehrban, H.; Lv, X.-Y.; Sun, W.; Esmailizadeh, A.; Lv, F.-H. Genomic Landscape of Copy Number Variations and Their Associations with Climatic Variables in the World’s Sheep. Genes 2023, 14, 1256. https://doi.org/10.3390/genes14061256
Salehian-Dehkordi H, Huang J-H, Pirany N, Mehrban H, Lv X-Y, Sun W, Esmailizadeh A, Lv F-H. Genomic Landscape of Copy Number Variations and Their Associations with Climatic Variables in the World’s Sheep. Genes. 2023; 14(6):1256. https://doi.org/10.3390/genes14061256
Chicago/Turabian StyleSalehian-Dehkordi, Hosein, Jia-Hui Huang, Nasrollah Pirany, Hossein Mehrban, Xiao-Yang Lv, Wei Sun, Ali Esmailizadeh, and Feng-Hua Lv. 2023. "Genomic Landscape of Copy Number Variations and Their Associations with Climatic Variables in the World’s Sheep" Genes 14, no. 6: 1256. https://doi.org/10.3390/genes14061256
APA StyleSalehian-Dehkordi, H., Huang, J.-H., Pirany, N., Mehrban, H., Lv, X.-Y., Sun, W., Esmailizadeh, A., & Lv, F.-H. (2023). Genomic Landscape of Copy Number Variations and Their Associations with Climatic Variables in the World’s Sheep. Genes, 14(6), 1256. https://doi.org/10.3390/genes14061256







