HmbC, a Protein of the HMG Family, Participates in the Regulation of Carotenoid Biosynthesis in Fusarium fujikuroi
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. DNA Isolation and PCR Assays
2.3. Plasmid Construction and Transformation
2.4. Southern Blot
2.5. Expression Analyses
2.6. Nuclei Extraction
2.7. Full Protein Extraction, Streptavidin Pull-Down, and Precipitation
2.8. Mass Spectrometry
2.9. Bioinformatic and Statistical Analysis
3. Results
3.1. Identification of Proteins Binding to car Promoters by Pull-Down Assay
3.2. Proteins with HMG Domains in F. fujikuroi
3.3. Features of hmbC Gene and HmbC Protein
3.4. Deletion of hmbC
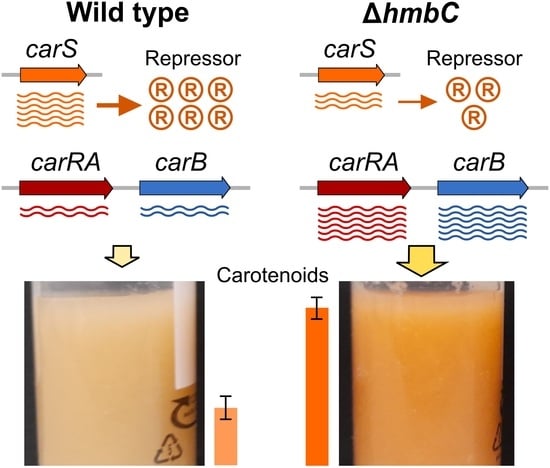
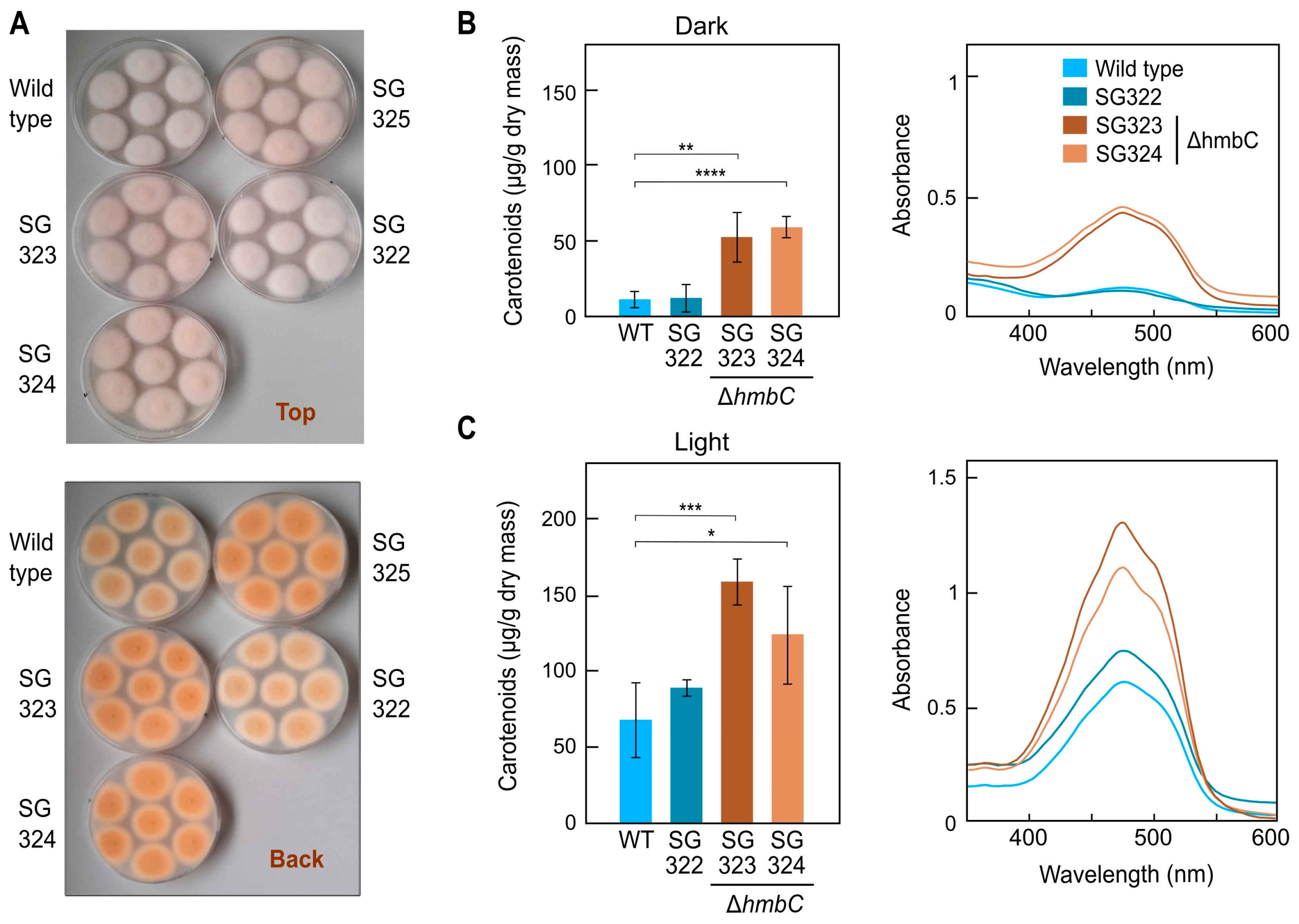
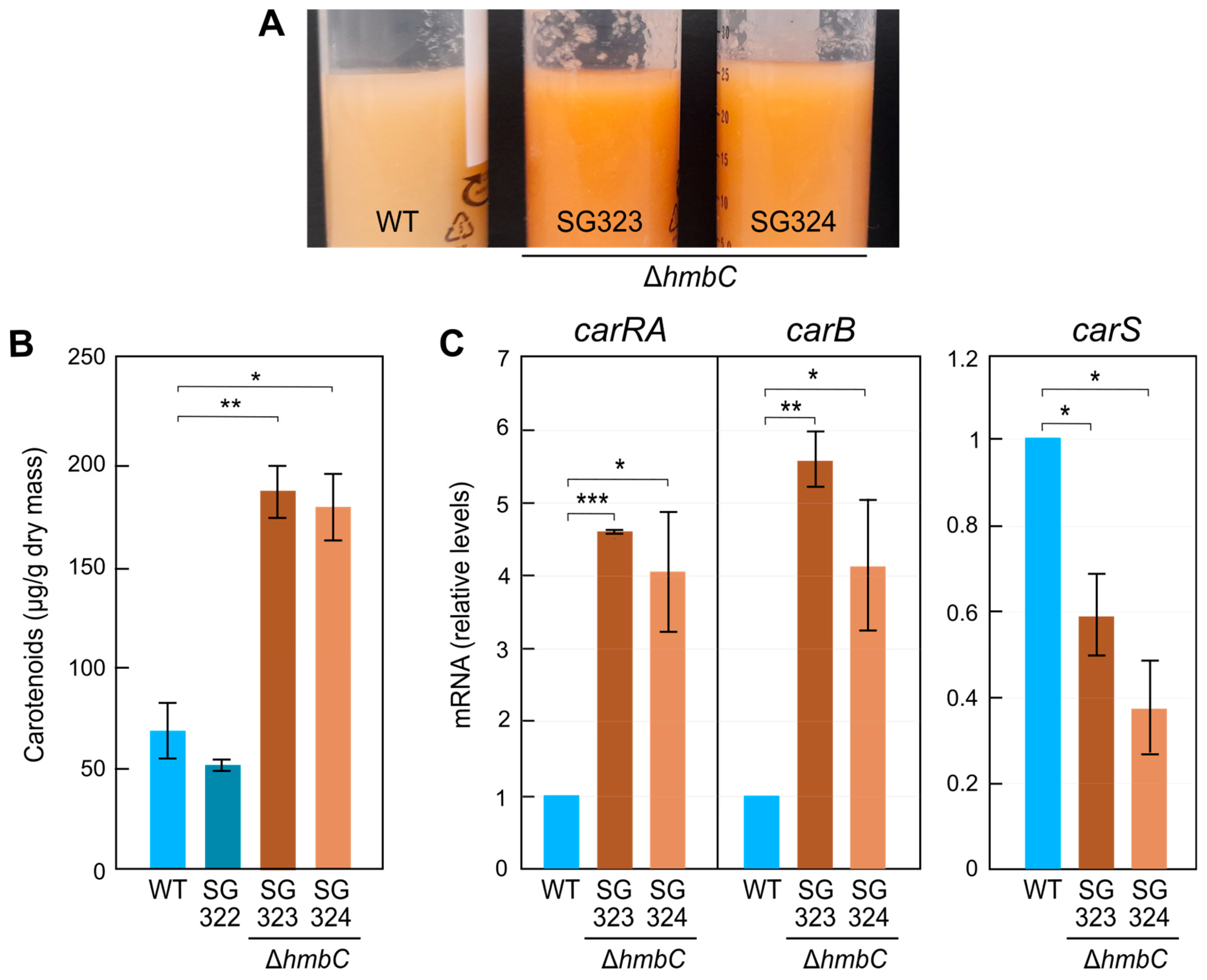
3.5. Effect of ΔhmbC Deletion on Carotenogenesis
3.6. Other ΔhmbC Phenotypic Alterations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reeves, R. Nuclear Functions of the HMG Proteins. Biochim. Biophys Acta (BBA) Gene Regul. Mech. 2010, 1799, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Mallik, R.; Kundu, A.; Chaudhuri, S. High Mobility Group Proteins: The Multifaceted Regulators of Chromatin Dynamics. Nucleus 2018, 61, 213–226. [Google Scholar] [CrossRef]
- Venters, B.J.; Pugh, B.F. How Eukaryotic Genes Are Transcribed. Crit. Rev. Biochem. Mol. Biol. 2009, 44, 117–141. [Google Scholar] [CrossRef]
- Reeves, R. High Mobility Group (HMG) Proteins: Modulators of Chromatin Structure and DNA Repair in Mammalian Cells. DNA Repair 2015, 36, 122–136. [Google Scholar] [CrossRef] [PubMed]
- Štros, M. HMGB Proteins: Interactions with DNA and Chromatin. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2010, 1799, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Kozlova, A.L.; Valieva, M.E.; Maluchenko, N.V.; Studitsky, V.M. HMGB Proteins as DNA Chaperones That Modulate Chromatin Activity. Mol. Biol. 2018, 52, 637–647. [Google Scholar] [CrossRef]
- Das, D.; Scovell, W.M. The Binding Interaction of HMG-1 with the TATA-Binding Protein/TATA Complex. J. Biol. Chem. 2001, 276, 32597–32605. [Google Scholar] [CrossRef]
- Panday, A.; Grove, A. Yeast HMO1: Linker Histone Reinvented. Microbiol. Mol. Biol. Rev. 2017, 81, e00037-16. [Google Scholar] [CrossRef]
- Voong, C.K.; Goodrich, J.A.; Kugel, J.F. Interactions of HMGB Proteins with the Genome and the Impact on Disease. Biomolecules 2021, 11, 1451. [Google Scholar] [CrossRef]
- Atanasoff-Kardjalieff, A.K.; Studt, L. Secondary Metabolite Gene Regulation in Mycotoxigenic Fusarium Species: A Focus on Chromatin. Toxins 2022, 14, 96. [Google Scholar] [CrossRef]
- Ma, L.J.; Geiser, D.M.; Proctor, R.H.; Rooney, A.P.; O’Donnell, K.; Trail, F.; Gardiner, D.M.; Manners, J.M.; Kazan, K. Fusarium Pathogenomics. Annu. Rev. Microbiol. 2013, 67, 399–416. [Google Scholar] [CrossRef] [PubMed]
- Janevska, S.; Tudzynski, B. Secondary Metabolism in Fusarium fujikuroi: Strategies to Unravel the Function of Biosynthetic Pathways. Appl. Microbiol. Biotechnol. 2018, 102, 615–630. [Google Scholar] [CrossRef] [PubMed]
- Salazar-Cerezo, S.; Martínez-Montiel, N.; García-Sánchez, J.; Pérez-Y-Terrón, R.; Martínez-Contreras, R.D. Gibberellin Biosynthesis and Metabolism: A Convergent Route for Plants, Fungi and Bacteria. Microbiol. Res. 2018, 208, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Wiemann, P.; Sieber, C.M.; von Bargen, K.W.; Studt, L.; Niehaus, E.M.; Espino, J.J.; Huss, K.; Michielse, C.B.; Albermann, S.; Wagner, D.; et al. Deciphering the Cryptic Genome: Genome-Wide Analyses of the Rice Pathogen Fusarium fujikuroi Reveal Complex Regulation of Secondary Metabolism and Novel Metabolites. PLoS Pathog. 2013, 9, e1003475. [Google Scholar] [CrossRef]
- Niehaus, E.-M.; Kim, H.-K.; Münsterkötter, M.; Janevska, S.; Arndt, B.; Kalinina, S.A.; Houterman, P.M.; Ahn, I.-P.; Alberti, I.; Tonti, S.; et al. Comparative Genomics of Geographically Distant Fusarium fujikuroi Isolates Revealed Two Distinct Pathotypes Correlating with Secondary Metabolite Profiles. PLoS Pathog. 2017, 13, e1006670. [Google Scholar] [CrossRef]
- Avalos, J.; Pardo-Medina, J.; Parra-Rivero, O.; Ruger-Herreros, M.; Rodríguez-Ortiz, R.; Hornero-Méndez, D.; Limón, M.C. Carotenoid Biosynthesis in Fusarium. J. Fungi 2017, 3, 39. [Google Scholar] [CrossRef]
- Pardo-Medina, J.; Gutiérrez, G.; Limón, M.C.; Avalos, J. Impact of the White-Collar Photoreceptor WcoA on the Fusarium fujikuroi Transcriptome. Front. Microbiol. 2021, 11, 619474. [Google Scholar] [CrossRef]
- Rodríguez-Ortiz, R.; Limón, M.C.; Avalos, J. Functional Analysis of the carS Gene of Fusarium fujikuroi. Mol. Genet. Genom. 2013, 288, 157–173. [Google Scholar] [CrossRef]
- Marente, J.; Avalos, J.; Limón, M.C. Controlled Transcription of Regulator Gene carS by Tet-on or by a Strong Promoter Confirms its Role as a Repressor of Carotenoid Biosynthesis in Fusarium fujikuroi. Microorganisms 2020, 9, 71. [Google Scholar] [CrossRef]
- Marente, J.; Ortega, P.; Pardo-Medina, J.; Avalos, J.; Limón, M.C. Modulation of Activity of a Carotenoid Pathway through the Use of the TET-on Regulatory System: Application in the Fungus Fusarium fujikuroi. In Plant and Food Carotenoids: Methods and Protocols; Rodríguez-Concepción, M., Welsch, R., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2020; pp. 343–360. ISBN 978-1-4939-9952-1. [Google Scholar]
- Hilgarth, R.S.; Lanigan, T.M. Optimization of Overlap Extension PCR for Efficient Transgene Construction. MethodsX 2020, 7, 100759. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Ruger-Herreros, M.; Parra-Rivero, O.; Pardo-Medina, J.; Romero-Campero, F.J.; Limón, M.C.; Avalos, J. Comparative Transcriptomic Analysis Unveils Interactions between the Regulatory CarS Protein and Light Response in Fusarium. BMC Genom. 2019, 20, 67. [Google Scholar] [CrossRef] [PubMed]
- Bokor, E.; Ámon, J.; Keisham, K.; Karácsony, Z.; Vágvölgyi, C.; Hamari, Z. HMGB Proteins Are Required for Sexual Development in Aspergillus nidulans. PLoS ONE 2019, 14, e0216094. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Kobayashi, R.; Brill, S.J. Characterization of a High Mobility Group 1/2 Homolog in Yeast. J. Biol. Chem. 1996, 271, 33678–33685. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, K.; Ki, S.; Aoyama, K.; Takahashi, H.; Kokubo, T. Saccharomyces cerevisiae HMO1 Interacts with TFIID and Participates in Start Site Selection by RNA polymerase II. Nucleic Acids Res. 2008, 36, 1343–1357. [Google Scholar] [CrossRef]
- Wang, M.; Li, J.; Wang, Y.; Fu, H.; Qiu, H.; Li, Y.; Li, M.; Lu, Y.; Fu, Y.V. Single-Molecule Study Reveals Hmo1, not Hho1, Promotes Chromatin Assembly in Budding Yeast. mBio 2023, e0099323. [Google Scholar] [CrossRef]
- Ait Benkhali, J.; Coppin, E.; Brun, S.; Peraza-Reyes, L.; Martin, T.; Dixelius, C.; Lazar, N.; van Tilbeurgh, H.; Debuchy, R. A Network of HMG-Box Transcription Factors Regulates Sexual Cycle in the Fungus Podospora anserina. PLoS Genet. 2013, 9, e1003642. [Google Scholar] [CrossRef]
- Cherry, J.M.; Hong, E.L.; Amundsen, C.; Balakrishnan, R.; Binkley, G.; Chan, E.T.; Christie, K.R.; Costanzo, M.C.; Dwight, S.S.; Engel, S.R.; et al. Saccharomyces Genome Database: The Genomics Resource of Budding Yeast. Nucleic Acids Res. 2012, 40, D700–D705. [Google Scholar] [CrossRef]
- Kamau, E.; Bauerle, K.T.; Grove, A. The Saccharomyces cerevisiae High Mobility Group Box Protein HMO1 Contains Two Functional DNA Binding Domains. J. Biol. Chem. 2004, 279, 55234–55240. [Google Scholar] [CrossRef]
- Albert, B.; Colleran, C.; Léger-Silvestre, I.; Berger, A.B.; Dez, C.; Normand, C.; Perez-Fernandez, J.; McStay, B.; Gadal, O. Structure-Function Analysis of Hmo1 Unveils an Ancestral Organization of HMG-Box Factors Involved in Ribosomal DNA Transcription from Yeast to Human. Nucleic Acids Res. 2013, 41, 10135–10149. [Google Scholar] [CrossRef]
- Bauerle, K.T.; Kamau, E.; Grove, A. Interactions between N- and C-terminal Domains of the Saccharomyces cerevisiae High-Mobility Group Protein HMO1 Are Required for DNA Bending. Biochemistry 2006, 45, 3635–3645. [Google Scholar] [CrossRef]
- Dunlap, J.C.; Glass, N.L.; McCluskey, K.; Plamann, M.; Galagan, J.E.; Birren, B.W.; Weiss, R.L.; Townsend, J.P.; Loros, J.J.; Nelson, M.A.; et al. Enabling a Community to Dissect an Organism: Overview of the Neurospora Functional Genomics Project. Adv. Genet. 2007, 57, 49–96. [Google Scholar] [CrossRef] [PubMed]
- Greenstein, R.A.; Ng, H.; Barrales, R.R.; Tan, C.; Braun, S.; Al-Sady, B. Local Chromatin Context Regulates the Genetic Requirements of the Heterochromatin Spreading Reaction. PLoS Genet. 2022, 18, e1010201. [Google Scholar] [CrossRef]
- Dolinski, K.J.; Heitman, J. Hmo1p, a High Mobility Group 1/2 homolog, Genetically and Physically Interacts with the Yeast FKBP12 Prolyl Isomerase. Genetics 1999, 151, 935–944. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Santiago, F.; Navarro, F. Transcription by the Three RNA Polymerases under the Control of the TOR Signaling Pathway in Saccharomyces cerevisiae. Biomolecules 2023, 13, 642. [Google Scholar] [CrossRef] [PubMed]
- Vizoso-Vázquez, A.; Barreiro-Alonso, A.; González-Siso, M.I.; Cerdán, M.E. HMGB Proteins Involved in TOR Signaling as General Regulators of Cell Growth by Controlling Ribosome Biogenesis. Curr. Genet. 2018, 64, 1205–1213. [Google Scholar] [CrossRef]
- Karácsony, Z.; Gácser, A.; Vágvölgyi, C.; Scazzocchio, C.; Hamari, Z. A Dually Located Multi-HMG-Box Protein of Aspergillus nidulans Has a Crucial Role in Conidial and Ascospore Germination. Mol. Microbiol. 2014, 94, 383–402. [Google Scholar] [CrossRef]
- Karácsony, Z.; Gácser, A.; Vágvölgyi, C.; Hamari, Z. Further Characterization of the Role of the Mitochondrial High-Mobility Group Box Protein in the Intracellular Redox Environment of Aspergillus nidulans. Microbiology 2015, 161, 1897–1908. [Google Scholar] [CrossRef]
- Murugesapillai, D.; Murugesapillai, D.; McCauley, M.J.; Huo, R.; Holte, M.H.N.; Stepanyants, A.; Maher, L.J.; Israeloff, N.E.; William, M.C. DNA Bridging and Looping by HMO1 Provides a Mechanism for Stabilizing Nucleosome-Free Chromatin. Nucleic Acids Res. 2014, 42, 8996–9004. [Google Scholar] [CrossRef]



| PcarB | PcarRA/X | |||||
|---|---|---|---|---|---|---|
| Protein | Yeast Ortholog | Features | WT | S | WT | S |
| CCT62089 | uncharacterized protein | 62 | 108 | 41 | 12 | |
| CCT66893 | related to NAD+ ADP-ribosyltransferase | 40 | 86 | |||
| CCT62893 | Hmo1p | related to nonhistone chromosomal protein | 34 | 58 | ||
| CCT71516 | Hho1p | related to HHO1-histone H1 protein | 20 | 39 | 21 | 3 |
| CCT72522 | related to Pas7p | 14 | 64 | 3 | ||
| CCT62055 | Rpl36ap | probable 60S ribosomal protein L36 | 12 | 13 | ||
| CCT70243 | Yku80p | related to transcriptional regulator ATRX homolog | 10 | 36 | 2 | |
| CCT61955 | Apn1p | probable AP endonuclease | 8 | 57 | ||
| CCT67445 | Htb1p | probable HTB1-histone H2B | 8 | 12 | ||
| CCT68541 | Rfa1p | probable single-stranded DNA-binding protein | 7 | 54 | ||
| CCT66187 | Rim1p | related to single-stranded DNA-binding protein | 6 | 14 | 3 | |
| CCT67540 | Gbp2p | related to Gbp2p | 6 | 14 | ||
| CCT69891 | Mgm101p | Probable mitochondrial genome maintenance protein | 5 | 30 | 2 | |
| CCT73695 | uncharacterized protein | 5 | 18 | 6 | 6 | |
| CCT63952 | related to excision repair protein RAD4 | 2 | 28 | |||
| CCT66006 | Top1p | probable TOP1-DNA topoisomerase I | 2 | 18 | ||
| CCT73736 | related to Glu/Asp-tRNA amidotransferase subunit | 20 | ||||
| Gene | Protein | Residues | Features |
|---|---|---|---|
| FFUJ_01566 | CCT61934 | 512 | related to mating type 1–2 protein |
| FFUJ_14410 | CCT62089 | 310 | uncharacterized protein |
| FFUJ_00897 | CCT62544 | 102 | probable NHP6B-nonhistone chromosomal protein |
| FFUJ_00508 | CCT62893 | 354 | related to nonhistone chromosomal protein |
| FFUJ_02729 | CCT65758 | 540 | related to nonhistone chromosomal protein |
| FFUJ_02997 | CCT66004 | 1085 | related to VPS33-vacuolar sorting protein |
| FFUJ_03111 | CCT66114 | 269 | uncharacterized protein |
| FFUJ_13386 | CCT67192 | 672 | uncharacterized protein |
| FFUJ_13684 | CCT67469 | 698 | uncharacterized protein |
| FFUJ_05685 | CCT69772 | 223 | mating type protein (Mat1-2-1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Franco-Losilla, M.; Nordzieke, S.; Feldmann, I.; Limón, M.C.; Avalos, J. HmbC, a Protein of the HMG Family, Participates in the Regulation of Carotenoid Biosynthesis in Fusarium fujikuroi. Genes 2023, 14, 1661. https://doi.org/10.3390/genes14081661
Franco-Losilla M, Nordzieke S, Feldmann I, Limón MC, Avalos J. HmbC, a Protein of the HMG Family, Participates in the Regulation of Carotenoid Biosynthesis in Fusarium fujikuroi. Genes. 2023; 14(8):1661. https://doi.org/10.3390/genes14081661
Chicago/Turabian StyleFranco-Losilla, Marta, Steffen Nordzieke, Ingo Feldmann, M. Carmen Limón, and Javier Avalos. 2023. "HmbC, a Protein of the HMG Family, Participates in the Regulation of Carotenoid Biosynthesis in Fusarium fujikuroi" Genes 14, no. 8: 1661. https://doi.org/10.3390/genes14081661