A Comprehensive Metabolism-Related Gene Signature Predicts the Survival of Patients with Acute Myeloid Leukemia
Abstract
:1. Introduction
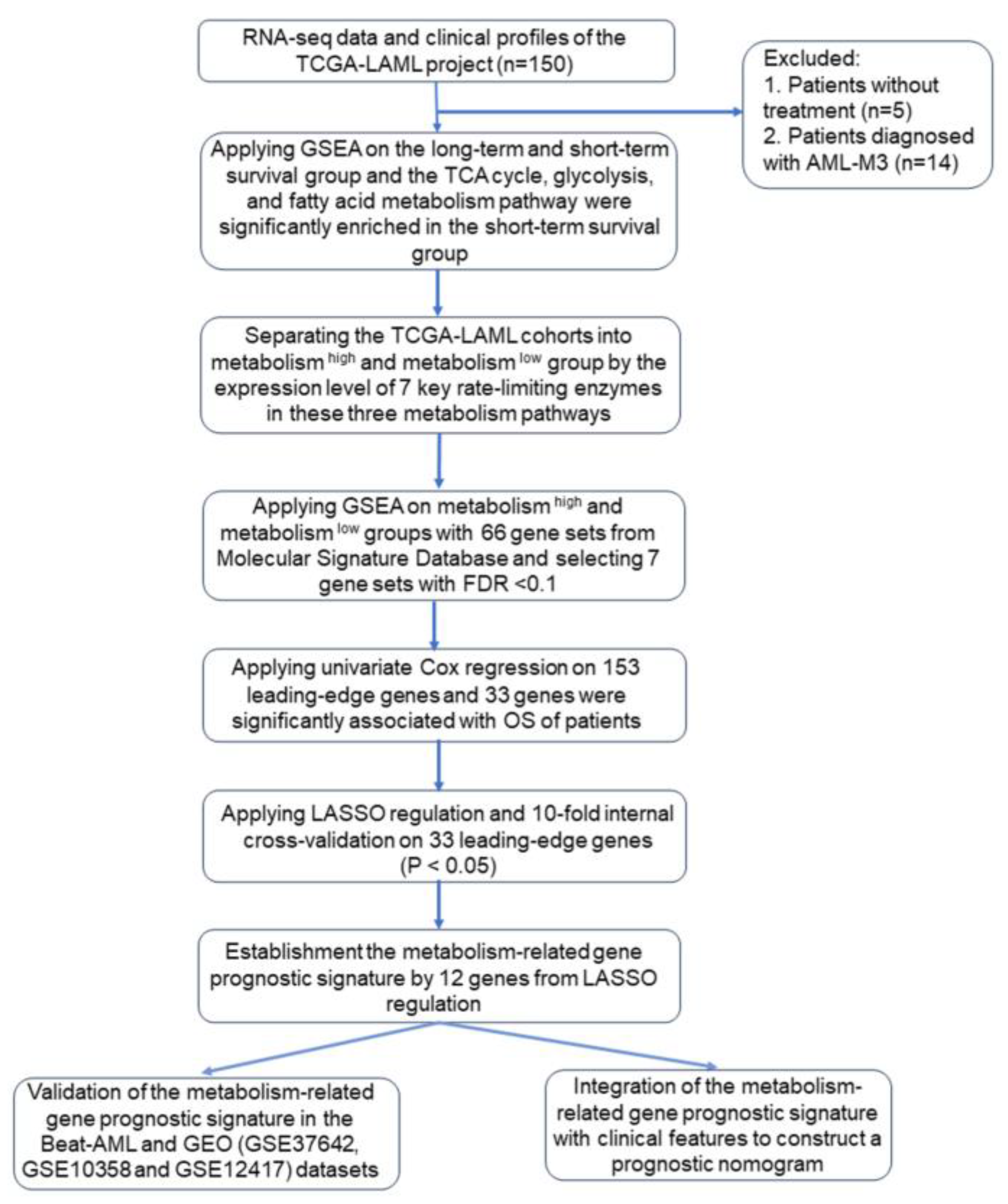
2. Materials and Methods
2.1. Data Sources and Patient Characteristics
2.2. Differentiation of Metabolic Status of the Patients in the TCGA-LAML Dataset
2.3. Gene Set Enrichment Analysis
2.4. Establishment and Validation of Prognostic Model
2.5. Statistical Analysis
2.6. Summary of the Methods
3. Results
3.1. Comparison of the Metabolic Pathways between Long-Term and Short-Term Survival Groups in AML Patients from the TCGA-LAML Dataset
3.2. Identification of Leading-Edge Gene
3.3. Establishment of Metabolism-Related Gene Prognostic Signature from Metabolism-Related Genes Associated with OS in the TCGA-LAML Dataset
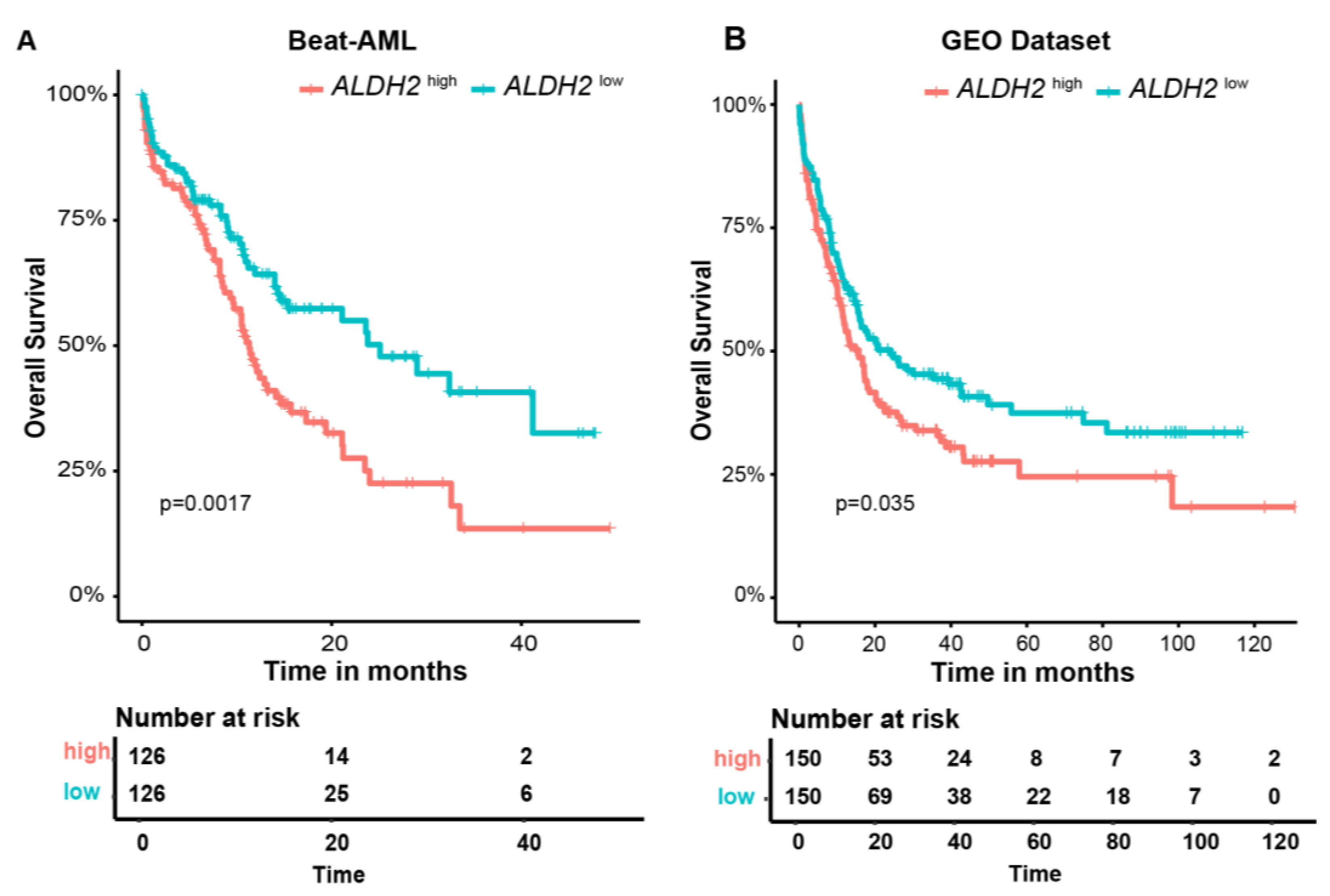
3.4. External Validation of Metabolism-Related Gene Prognostic Signature in GEO AML (GSE37642, GSE10358, and GSE12417) and the Beat-AML Datasets
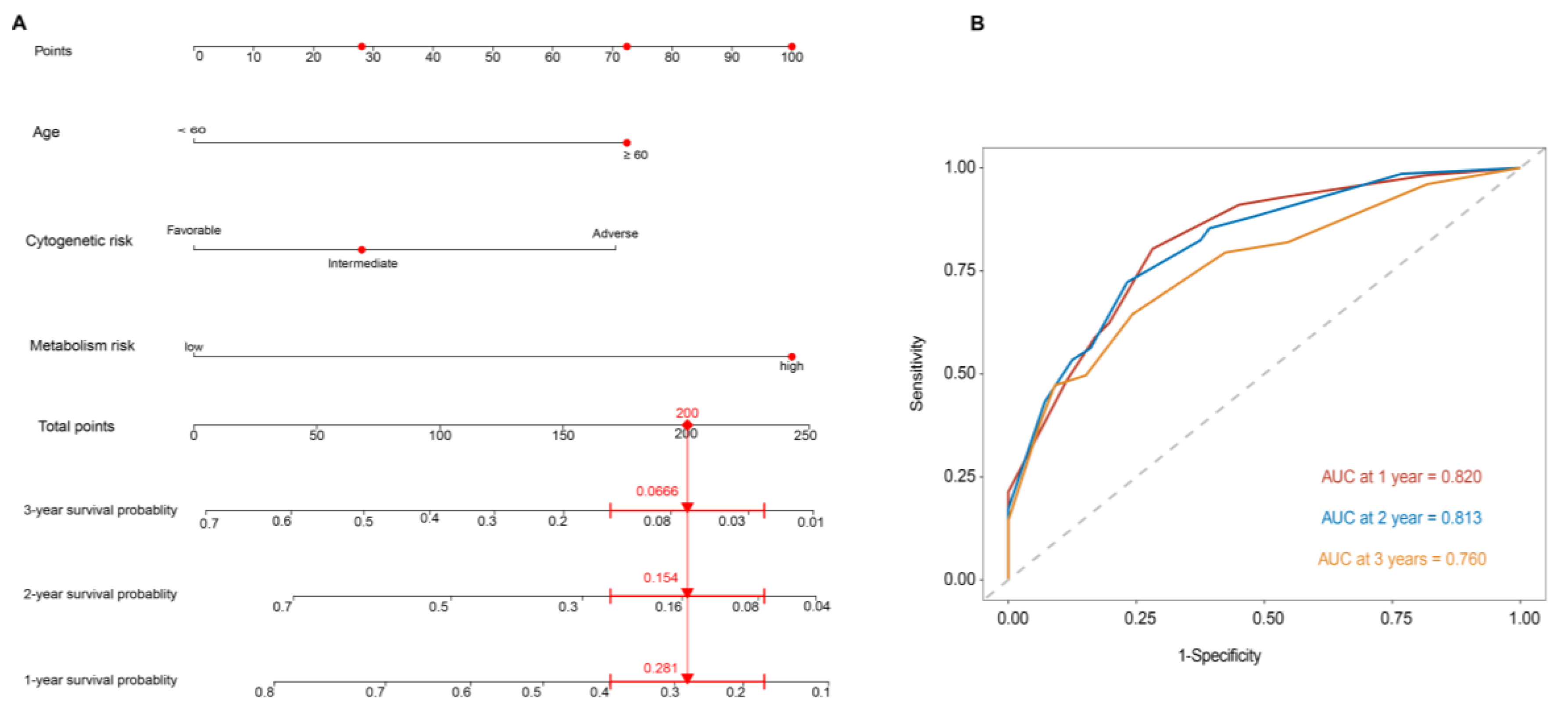
3.5. Metabolism-Related Gene Prognostic Signature Is an Independent Prognostic Factor
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Manola, K.N.; Panitsas, F.; Polychronopoulou, S.; Daraki, A.; Karakosta, M.; Stavropoulou, C.; Avgerinou, G.; Hatzipantelis, E.; Pantelias, G.; Sambani, C.; et al. Cytogenetic abnormalities and monosomal karyotypes in children and adolescents with acute myeloid leukemia: Correlations with clinical characteristics and outcome. Cancer Genet. 2013, 206, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Herst, P.M.; Howman, R.A.; Neeson, P.J.; Berridge, M.V.; Ritchie, D.S. The level of glycolytic metabolism in acute myeloid leukemia blasts at diagnosis is prognostic for clinical outcome. J. Leukoc. Biol. 2011, 89, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.L.; Wang, J.H.; Zhao, A.H.; Xu, X.; Wang, Y.H.; Chen, T.L.; Li, J.M.; Mi, J.Q.; Zhu, Y.M.; Liu, Y.F.; et al. A distinct glucose metabolism signature of acute myeloid leukemia with prognostic value. Blood 2014, 124, 1645–1654. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Ward, P.S.; Kapoor, G.S.; Rohle, D.; Turcan, S.; Abdel-Wahab, O.; Edwards, C.R.; Khanin, R.; Figueroa, M.E.; Melnick, A.; et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 2012, 483, 474–478. [Google Scholar] [CrossRef] [PubMed]
- Losman, J.A.; Looper, R.E.; Koivunen, P.; Lee, S.; Schneider, R.K.; McMahon, C.; Cowley, G.S.; Root, D.E.; Ebert, B.L.; Kaelin, W.G., Jr. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science 2013, 339, 1621–1625. [Google Scholar] [CrossRef] [PubMed]
- Pollyea, D.A.; Stevens, B.M.; Jones, C.L.; Winters, A.; Pei, S.; Minhajuddin, M.; D’Alessandro, A.; Culp-Hill, R.; Riemondy, K.A.; Gillen, A.E.; et al. Venetoclax with azacitidine disrupts energy metabolism and targets leukemia stem cells in patients with acute myeloid leukemia. Nat. Med. 2018, 24, 1859–1866. [Google Scholar] [CrossRef] [PubMed]
- Carracedo, A.; Cantley, L.C.; Pandolfi, P.P. Cancer metabolism: Fatty acid oxidation in the limelight. Nat. Rev. Cancer 2013, 13, 227–232. [Google Scholar] [CrossRef]
- Mishra, S.K.; Millman, S.E.; Zhang, L. Metabolism in acute myeloid leukemia: Mechanistic insights and therapeutic targets. Blood 2023, 141, 1119–1135. [Google Scholar] [CrossRef]
- Lee, J.S.; Roberts, A.; Juarez, D.; Vo, T.T.; Bhatt, S.; Herzog, L.O.; Mallya, S.; Bellin, R.J.; Agarwal, S.K.; Salem, A.H.; et al. Statins enhance efficacy of venetoclax in blood cancers. Sci. Transl. Med. 2018, 10, eaaq1240. [Google Scholar] [CrossRef]
- Ley, T.J.; Miller, C.; Ding, L.; Raphael, B.J.; Mungall, A.J.; Robertson, A.; Hoadley, K.; Triche, T.J., Jr.; Laird, P.W.; Baty, J.D.; et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar] [CrossRef]
- Herold, T.; Jurinovic, V.; Batcha, A.M.N.; Bamopoulos, S.A.; Rothenberg-Thurley, M.; Ksienzyk, B.; Hartmann, L.; Greif, P.A.; Phillippou-Massier, J.; Krebs, S.; et al. A 29-gene and cytogenetic score for the prediction of resistance to induction treatment in acute myeloid leukemia. Haematologica 2018, 103, 456–465. [Google Scholar] [CrossRef] [PubMed]
- Tomasson, M.H.; Xiang, Z.; Walgren, R.; Zhao, Y.; Kasai, Y.; Miner, T.; Ries, R.E.; Lubman, O.; Fremont, D.H.; McLellan, M.D.; et al. Somatic mutations and germline sequence variants in the expressed tyrosine kinase genes of patients with de novo acute myeloid leukemia. Blood 2008, 111, 4797–4808. [Google Scholar] [CrossRef] [PubMed]
- Metzeler, K.H.; Hummel, M.; Bloomfield, C.D.; Spiekermann, K.; Braess, J.; Sauerland, M.C.; Heinecke, A.; Radmacher, M.; Marcucci, G.; Whitman, S.P.; et al. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood 2008, 112, 4193–4201. [Google Scholar] [CrossRef] [PubMed]
- Tyner, J.W.; Tognon, C.E.; Bottomly, D.; Wilmot, B.; Kurtz, S.E.; Savage, S.L.; Long, N.; Schultz, A.R.; Traer, E.; Abel, M.; et al. Functional genomic landscape of acute myeloid leukaemia. Nature 2018, 562, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Leek JT, J.W.; Parker, H.S.; Fertig, E.J.; Jaffe, A.E.; Zhang, Y.; Storey, J.D.; Torres, L.C. sva: Surrogate Variable Analysis. R package Version 3.50.0. 2023. Available online: https://bioconductor.org/packages/sva (accessed on 1 October 2022).
- Döhner, H.; Wei, A.H.; Appelbaum, F.R.; Craddock, C.; DiNardo, C.D.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Godley, L.A.; Hasserjian, R.P.; et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood 2022, 140, 1345–1377. [Google Scholar] [CrossRef] [PubMed]
- Brooks, W.M.; Lynch, P.J.; Ingle, C.C.; Hatton, A.; Emson, P.C.; Faull, R.L.; Starkey, M.P. Gene expression profiles of metabolic enzyme transcripts in Alzheimer’s disease. Brain Res. 2007, 1127, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Middleton, F.A.; Mirnics, K.; Pierri, J.N.; Lewis, D.A.; Levitt, P. Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J. Neurosci. Off. J. Soc. Neurosci. 2002, 22, 2718–2729. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Therneau, T.M. A Package for Survival Analysis in R. 2020. Available online: https://CRAN.R-project.org/package=survival (accessed on 1 October 2022).
- Tay, J.K.; Narasimhan, B.; Hastie, T. Elastic Net Regularization Paths for All Generalized Linear Models. J. Stat. Softw. 2023, 106, 1–31. [Google Scholar] [CrossRef]
- FE, H.J. rms: Regression Modeling Strategies. R Package Version 6.7-1. 2023. Available online: https://CRAN.R-project.org/package=rms (accessed on 1 October 2022).
- Rattigan, K.M.; Zarou, M.M.; Helgason, G.V. Metabolism in stem cell-driven leukemia: Parallels between hematopoiesis and immunity. Blood 2023, 141, 2553–2565. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, Y.; Liu, J.; Zeng, Y.; Guo, Q.; Guo, J.; Guo, L.; Lu, H.; Liu, W. Establishment and validation of a carbohydrate metabolism-related gene signature for prognostic model and immune response in acute myeloid leukemia. Front. Immunol. 2022, 13, 1038570. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liang, J.; Yang, W.; Guo, W.; Song, W.; Zhang, W.; Wu, X.; He, B. A distinct lipid metabolism signature of acute myeloid leukemia with prognostic value. Front. Oncol. 2022, 12, 876981. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Ding, L.; Luo, Q.; Li, X.; Zeng, X.; Kong, D.; Yu, X.; Feng, J.; Ye, Y.; Wang, L.; et al. Development and Validation of an Individualized Metabolism-Related Prognostic Model for Adult Acute Myeloid Leukemia Patients. Front. Oncol. 2022, 12, 829007. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, Y.; Xu, Y.; Xing, H.; Tian, Z.; Tang, K.; Rao, Q.; Wang, M.; Wang, J. AML1-ETO-Related Fusion Circular RNAs Contribute to the Proliferation of Leukemia Cells. Int. J. Mol. Sci. 2022, 24, 71. [Google Scholar] [CrossRef] [PubMed]
- Handschuh, L.; Kaźmierczak, M.; Milewski, M.C.; Góralski, M.; Łuczak, M.; Wojtaszewska, M.; Uszczyńska-Ratajczak, B.; Lewandowski, K.; Komarnicki, M.; Figlerowicz, M. Gene expression profiling of acute myeloid leukemia samples from adult patients with AML-M1 and -M2 through boutique microarrays, real-time PCR and droplet digital PCR. Int. J. Oncol. 2018, 52, 656–678. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Zhou, Z.; Wen, Y.; Li, Z.; Huang, Z.; Li, Y. The immunometabolic landscape of the bone marrow microenvironment in acute myeloid leukemia. Exp. Hematol. Oncol. 2022, 11, 81. [Google Scholar] [CrossRef]
- Kiesel, V.A.; Sheeley, M.P.; Coleman, M.F.; Cotul, E.K.; Donkin, S.S.; Hursting, S.D.; Wendt, M.K.; Teegarden, D. Pyruvate carboxylase and cancer progression. Cancer Metab. 2021, 9, 20. [Google Scholar] [CrossRef]
- Reed, M.A.C.; Ludwig, C.; Bunce, C.M.; Khanim, F.L.; Günther, U.L. Malonate as a ROS product is associated with pyruvate carboxylase activity in acute myeloid leukaemia cells. Cancer Metab. 2016, 4, 15. [Google Scholar] [CrossRef]
- D’Angelo, M.A.; Gomez-Cavazos, J.S.; Mei, A.; Lackner, D.H.; Hetzer, M.W. A change in nuclear pore complex composition regulates cell differentiation. Dev. Cell 2012, 22, 446–458. [Google Scholar] [CrossRef]
- Petersen, M.A.; Rosenberg, C.A.; Bill, M.; Enemark, M.B.; Rahbek, O.; Roug, A.S.; Hasle, H.; Honoré, B.; Ludvigsen, M. Proteomic Profiling Identifies Specific Leukemic Stem Cell-Associated Protein Expression Patterns in Pediatric AML Patients. Cancers 2022, 14, 3567. [Google Scholar] [CrossRef]
- Li, M.; Zhao, H. Bioinformatics analysis of the expression and clinical significance of the NUP210 Gene in acute myeloid leukaemia. Hematology 2022, 27, 456–462. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liao, X.; Hu, Y.; Li, M.; Tang, M.; Zhang, S.; Mo, S.; Li, X.; Chen, S.; Qian, W.; et al. SLC27A4-mediated selective uptake of mono-unsaturated fatty acids promotes ferroptosis defense in hepatocellular carcinoma. Free. Radic. Biol. Med. 2023, 201, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Strong, A.; Rader, D.J. Sortilin as a regulator of lipoprotein metabolism. Curr. Atheroscler. Rep. 2012, 14, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Garza-Veloz, I.; Martinez-Fierro, M.L.; Jaime-Perez, J.C.; Carrillo-Sanchez, K.; Ramos-Del Hoyo, M.G.; Lugo-Trampe, A.; Rojas-Martinez, A.; Gutierrez-Aguirre, C.H.; Gonzalez-Llano, O.; Salazar-Riojas, R.; et al. Identification of differentially expressed genes associated with prognosis of B acute lymphoblastic leukemia. Dis. Markers 2015, 2015, 828145. [Google Scholar] [CrossRef] [PubMed]
- Modarres, P.; Mohamadi Farsani, F.; Nekouie, A.A.; Vallian, S. Meta-analysis of gene signatures and key pathways indicates suppression of JNK pathway as a regulator of chemo-resistance in AML. Sci. Rep. 2021, 11, 12485. [Google Scholar] [CrossRef] [PubMed]
- Stratmann, S.; Yones, S.A.; Garbulowski, M.; Sun, J.; Skaftason, A.; Mayrhofer, M.; Norgren, N.; Herlin, M.K.; Sundström, C.; Eriksson, A.; et al. Transcriptomic analysis reveals proinflammatory signatures associated with acute myeloid leukemia progression. Blood Adv. 2022, 6, 152–164. [Google Scholar] [CrossRef] [PubMed]
- Synowiec, E.; Hoser, G.; Wojcik, K.; Pawlowska, E.; Skorski, T.; Błasiak, J. UV Differentially Induces Oxidative Stress, DNA Damage and Apoptosis in BCR-ABL1-Positive Cells Sensitive and Resistant to Imatinib. Int. J. Mol. Sci. 2015, 16, 18111–18128. [Google Scholar] [CrossRef]
- Song, X.; Wu, X.; Zhang, Z.; Cui, Z.; Zheng, Y.; Sun, J. Subcellular Proteome Analysis Reveals Apoptotic Vulnerability of T-Cell Acute Lymphoblastic Leukemia. BioMed Res. Int. 2022, 2022, 5504475. [Google Scholar] [CrossRef]
- Li, L.T.; Li, Z.D.; Yang, Y.; Lu, Y.; Xie, X.B.; Chen, L.; Feng, J.Y.; Knisely, A.S.; Wang, J.S. ABCB11 deficiency presenting as transient neonatal cholestasis: Correlation with genotypes and BSEP expression. Liver Int. Off. J. Int. Assoc. Study Liver 2020, 40, 2788–2796. [Google Scholar] [CrossRef]
- Pan, S.T.; Li, Z.L.; He, Z.X.; Qiu, J.X.; Zhou, S.F. Molecular mechanisms for tumour resistance to chemotherapy. Clin. Exp. Pharmacol. Physiol. 2016, 43, 723–737. [Google Scholar] [CrossRef]
- Estrada, N.; Zamora, L.; Ferrer-Marín, F.; Palomo, L.; García, O.; Vélez, P.; De la Fuente, I.; Sagüés, M.; Cabezón, M.; Cortés, M.; et al. Association between Germline Single-Nucleotide Variants in ADME Genes and Major Molecular Response to Imatinib in Chronic Myeloid Leukemia Patients. J. Clin. Med. 2022, 11, 6217. [Google Scholar] [CrossRef] [PubMed]
- Thomas, G.E.; Egan, G.; García-Prat, L.; Botham, A.; Voisin, V.; Patel, P.S.; Hoff, F.W.; Chin, J.; Nachmias, B.; Kaufmann, K.B.; et al. The metabolic enzyme hexokinase 2 localizes to the nucleus in AML and normal haematopoietic stem and progenitor cells to maintain stemness. Nat. Cell Biol. 2022, 24, 872–884. [Google Scholar] [CrossRef] [PubMed]
- Garaycoechea, J.I.; Crossan, G.P.; Langevin, F.; Daly, M.; Arends, M.J.; Patel, K.J. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature 2012, 489, 571–575. [Google Scholar] [CrossRef] [PubMed]
- Moreb, J.S.; Ucar, D.; Han, S.; Amory, J.K.; Goldstein, A.S.; Ostmark, B.; Chang, L.J. The enzymatic activity of human aldehyde dehydrogenases 1A2 and 2 (ALDH1A2 and ALDH2) is detected by Aldefluor, inhibited by diethylaminobenzaldehyde and has significant effects on cell proliferation and drug resistance. Chem.-Biol. Interact. 2012, 195, 52–60. [Google Scholar] [CrossRef]
- Yuan, B.; El Dana, F.; Ly, S.; Yan, Y.; Ruvolo, V.; Shpall, E.J.; Konopleva, M.; Andreeff, M.; Battula, V.L. Bone marrow stromal cells induce an ALDH+ stem cell-like phenotype and enhance therapy resistance in AML through a TGF-β-p38-ALDH2 pathway. PLoS ONE 2020, 15, e0242809. [Google Scholar] [CrossRef]
- Wang, B.J.; Wang, S.; Xiao, M.; Zhang, J.; Wang, A.J.; Guo, Y.; Tang, Y.; Gu, J. Regulatory mechanisms of Sesn2 and its role in multi-organ diseases. Pharmacol. Res. 2021, 164, 105331. [Google Scholar] [CrossRef]
- Hernandez-Valladares, M.; Aasebø, E.; Berven, F.; Selheim, F.; Bruserud, Ø. Biological characteristics of aging in human acute myeloid leukemia cells: The possible importance of aldehyde dehydrogenase, the cytoskeleton and altered transcriptional regulation. Aging 2020, 12, 24734–24777. [Google Scholar] [CrossRef]


| TCGA-LAML | Beat-AML | GEO (GSE37642, GSE10358 and GSE12417) | p-Value | |
|---|---|---|---|---|
| Number | 131 | 252 | 300 | |
| Age (years), median (IQR) | 57.0 (43.0, 66.5) | 61.0 (44.0, 71.0) | 59.0 (45.0,67.0) | 0.197 |
| Gender (%) | 0.880 | |||
| Male | 72 (55.0) | 135 (53.6) | NA | |
| Female | 59 (45.0) | 117 (46.4) | NA | |
| WBC count at diagnosis, ×109/L, median (IQR) | 19.6 (5.5, 47.8) | 23.0 (8.1, 56.8) | 19.9 (5.1,62.7) | 0.316 |
| BM blast, %, median (IQR) | 71.0 (52.0, 83.0) | 72.0 (43.5, 89.0) | 70.0 (53.5, 86.0) | 0.959 |
| FAB, n (%) | <0.001 | |||
| M0 | 15 (11.5) | 4 (5.2) | 17 (5.7) | |
| M1 | 36 (27.5) | 7 (9.1) | 73 (24.4) | |
| M2 | 35 (26.7) | 6 (7.8) | 103 (34.4) | |
| M4 | 28 (21.4) | 23 (29.9) | 44 (14.7) | |
| M5 | 14 (10.7) | 27 (35.1) | 37 (12.4) | |
| M6 | 2 (1.5) | 0 (0.0) | 10 (3.3) | |
| M7 | 1 (0.8) | 2 (2.6) | 2 (0.7) | |
| Cytogenetics, n (%) | 0.213 | |||
| Normal | 63 (48.5) | 121 (50.4) | NA | |
| +8 | 6 (4.6) | 10 (4.2) | NA | |
| del(5) | 1 (0.8) | 4 (1.7) | NA | |
| del(7) | 4 (3.1) | 8 (3.3) | NA | |
| MLL rearrangement | 8 (6.2) | 18 (7.5) | NA | |
| inv(16) | 10 (7.7) | 17 (7.1) | NA | |
| t(8;21) | 7 (5.4) | 9 (3.8) | NA | |
| complex | 21 (16.2) | 22 (9.2) | NA | |
| Cytogenetic risk, n (%) | <0.001 | |||
| Adverse | 34 (26.4) | 88 (34.9) | NA | |
| Favorable | 17 (13.2) | 85 (33.7) | NA | |
| Intermediate | 78 (60.5) | 79 (31.3) | ||
| Follow-up duration, months, median (IQR) | 18.1 (6.8, 35.7) | 8.6 (4.3, 15.3) | 14.4 (5.7, 37.4) | <0.001 |
| OS events, n (%) | 0.001 | |||
| Dead | 88 (67.2) | 122 (48.4) | 185 (61.7) | |
| Alive | 43 (32.8) | 130 (51.6) | 115 (38.3) | |
| Metabolic Pathways | Key Rate-Limiting Enzymes | Gene Symbols |
|---|---|---|
| Glycolysis | Hexokinase | HK1, HK2, HK3 |
| Phosphofructokinase-1 | PFKL, PFKM, PFKP | |
| Pyruvate kinase | PKLR, PKM | |
| TCA cycle | Citrate synthase | CS |
| Isocitrate dehydrogenase | IDH1, IDH2, IDH3A, IDH3B, IDH3G | |
| Oxoglutarate dehydrogenase complex | OGDH, DLST, DLD | |
| Fatty acid metabolism | Carnitine palmitoyltransferase I | CPT1A, CPT1B, CPT1C |
| Univariable | Multivariable | |||
|---|---|---|---|---|
| HR (95% CI) | p-Value | HR (95% CI) | p-Value | |
| TCGA-LAML (Training cohort) | ||||
| Gender (female vs. male) | 1.03 (0.68–1.58) | 0.874 | \ | \ |
| Age (≥60 vs. <60) | 2.38 (1.56–3.64) | <0.001 | 2.23 (1.42–3.52) | 0.001 |
| WBC count (≥100 vs. <100 × 109/L) | 2.08 (1.04–4.16) | 0.039 | 1.75 (0.82–3.75) | 0.149 |
| Bone marrow blast (≥70% vs. <70%) | 1.36 (0.89–2.07) | 0.157 | \ | \ |
| Cytogenetic risk * | 2.88 (1.24–6.69) | 0.014 | 1.28 (0.51–3.18) | 0.600 |
| Metabolism risk score (high vs. low) | 3.39 (2.18–5.29) | <0.001 | 2.69 (1.66–4.35) | <0.001 |
| Beat-AML (Validation cohort 1) | ||||
| Gender (female vs. male) | 1.41 (0.98–2.03) | 0.065 | \ | \ |
| Age (≥60 vs. <60) | 2.47 (1.36–3.62) | <0.001 | 2.13 (1.45–3.14) | 0.001 |
| WBC count (≥100 vs. <100 ×109/L) | 1.76 (0.94–3.32) | 0.080 | \ | \ |
| Bone marrow blast (≥70% vs. <70%) | 1.15 (0.76–1.76) | 0.508 | \ | \ |
| Cytogenetic risk | 2.71 (1.63–4.53) | <0.001 | 1.78 (1.02–3.10) | 0.041 |
| Metabolism risk score (high vs. low) | 1.72 (1.19–2.49) | 0.004 | 1.74 (1.13–2.68) | 0.032 |
| Metabolism Signature | Enrolled Markers | Training Dataset | Validation Datasets | What Was Studied | Methods | Significance | Results Related to the Signature | Reference |
|---|---|---|---|---|---|---|---|---|
| A panel of serum glucose metabolism markers | Lactate, 2-oxoglutarate, pyruvate, 2-HG, glycerol-3-phosphate, and citrate | 229 de novo AML patients enrolled in 2007 to 2010 from Rui Jin Hospital in Shanghai | 171 newly diagnosed AML patients enrolled in 2011 to 2012 from 6 hematology centers | Metabolomic profiles of all serum samples, focused on the glucose metabolism | A predictive principal component analysis model | Provided strong evidence for the use of serum metabolites and metabolic pathways as novel prognostic markers and potential therapeutic targets for AML | -- | Chen et al., 2014 [3] |
| A carbohydrate-metabolism-related gene signature | PFKL, IDH3G, G6PD, DCXR, CYB5R3, CYB5R4, ACADS, MLYCD, PIK3CA, and CDIPT | 117 AML samples from the TCGA cohort | GSE37642 (n = 140), GSE37642 (n = 422), and 106 de novo AML patients in Affiliated Hospital of Southwest Medical University from January 2019 to June 2022 | 355 carbohydrate-metabolism-related genes were derived from the Kyoto Encyclopedia of Genes and Genomes pathway database | LASSO analysis and a multivariate Cox regression | The carbohydrate metabolism related signature was reliable and may provide theoretical support for AML prognostic judgment and treatment | RUNX1, IDH2, WT1, and KRAS mutations were more frequently in the low-risk group, and TP53, KIT, and TTN mutations were more common in the high-risk group | Yang et al., 2022 [24] |
| A lipid-metabolism-related gene signature | LDLRAP1, PNPLA6, DGKA, PLA2G4A, CBR1, and EBP | 144 AML patients are extracted from UCSC Xena Browser | GSE71014, GSE12417, and GSE37642 | 26 lipid-metabolism-related pathways including 1045 genes extracted from the MSigDB | Survival analysis and then LASSO analysis | Contributed to better understanding of the use of metabolites and metabolic pathways as the potential prognostic biomarkers and therapeutic targets for AML | The common immune checkpoints were significantly upregulated in the high-risk group, indicating an immunosuppressive TME of bone marrow in the high-risk group | Li et al., 2022 [25] |
| A metabolism-related prognostic signature index consisting of gene pairs | FADS1| NEU1, SLC2A5| TBXAS1, FADS1| PDE4B | 151 AML patients from the TCGA cohort | 162 AML patients from the GSE12417 cohort and 417 AML patients from the GSE37642 cohort | Metabolism-related genes that are differentially expressed between TCGA cohort and normal bone marrow | A pairwise comparison, a univariate Cox regression, and LASSO analysis | Provided a composite metabolism and clinical model as a novel prognostic stratification method and identified several potential therapeutic drugs for AML | -- | Wei et al., 2022 [26] |
| A metabolism-related prognostic model | 12 genes shown in Table 5 | 131 AML patients from the TCGA-LAML cohort | 252 patients from the Beat-AML cohort and 300 from GEO cohort (GSE37642, GSE10358, and GSE12417) | Genes related to glycolysis, fatty acid metabolism, and the TCA cycle pathways | A univariate Cox regression, and then LASSO analysis | Contributed to better use of metabolism reprogramming factors as prognostic marker and provide novel insights into potential metabolism target for AML treatment | -- | Our study |
| Genes | Full Name | Function | Effects | References |
|---|---|---|---|---|
| ENO1 | α-Enolase 1 | Glycolytic enzyme | Overexpressed in several types of AML | [27,28,29] |
| PC | Pyruvate carboxylase | Catalyzing the ATP-dependent carboxylation of pyruvate to oxaloacetate | Highly expressed in AML K562 cell line | [30,31] |
| NUP210 | Nucleoporin 210 | Involved in nucleocytoplasmic transport | Overexpressed in LSCs of pediatric AML | [32,33,34] |
| SLC27A4 | Solute carrier family 27 member 4 | Fatty acid transporter protein | High expression associated with poorer clinical outcomes in several cancer types | [35] |
| SORT1 | Sortilin 1 | Regulating lipoprotein metabolism | Significantly associated with relapse and/or B-ALL-related death and upregulated in chemo-resistant AML samples | [36,37,38] |
| INSR | Insulin receptor | Binding of insulin to activate the insulin signaling pathway | Downregulated as a predictive gene for relapse among AML | [39] |
| SDHB | Succinate dehydrogenase complex iron sulfur subunit B | Encoding the iron–sulfur protein subunit of the succinate dehydrogenase enzyme complex, a complex of the mitochondrial respiratory chain | Decreased in imatinib-resistant BCR-ABL1 cells. SDHB mutations in leukemic T cells involved in cellular pre-adaptation to hypoxia. | [40] |
| SDHA | Succinate dehydrogenase complex flavoprotein subunit A | Encoding a major catalytic subunit of dehydrogenase enzyme complex, a complex of the mitochondrial respiratory chain | Significantly associated with poor survival of leukemia patients. Inhibition of SDHA with venetoclax and azacitidine led to LSC death | [41] |
| ABCB11 | ATP binding cassette subfamily B member 11 | The major canalicular bile salt export pump | Significantly associated with the achievement of major molecular response with first-line imatinib treatment | [42,43,44] |
| HK2 | Hexokinase 2 | Phosphorylating glucose to produce glucose-6-phosphate | Overexpression resulted in chemoresistance of LSCs to DNA-damaging agents | [45] |
| ALDH2 | Aldehyde dehydrogenase 2 | Oxidizing aldehydes | Overexpression in leukemia cell lines resulted in increased resistance to doxorubicin | [46,47,48] |
| SESN2 | Sestrin 2 | Catalyzing the reduction of hyperoxidized peroxiredoxins | Knockdown in T-ALL cells decreased the rate of mitochondrial respiration | [49] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhai, Y.; Shen, H.; Wei, H. A Comprehensive Metabolism-Related Gene Signature Predicts the Survival of Patients with Acute Myeloid Leukemia. Genes 2024, 15, 63. https://doi.org/10.3390/genes15010063
Zhai Y, Shen H, Wei H. A Comprehensive Metabolism-Related Gene Signature Predicts the Survival of Patients with Acute Myeloid Leukemia. Genes. 2024; 15(1):63. https://doi.org/10.3390/genes15010063
Chicago/Turabian StyleZhai, Yujia, Heng Shen, and Hui Wei. 2024. "A Comprehensive Metabolism-Related Gene Signature Predicts the Survival of Patients with Acute Myeloid Leukemia" Genes 15, no. 1: 63. https://doi.org/10.3390/genes15010063





