miR-425-5p Regulates Proliferation of Bovine Mammary Epithelial Cells by Targeting TOB2
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. HS Models
2.3. Transfection
2.4. Biological Analysis of Sequencing Data
2.5. Cell Counting Kit-8 (CCK-8) Detection of Cell Viability
2.6. EDU Assay to Detect Cell Proliferation Efficiency
2.7. RNA Extraction and Real-Time Fluorescence Quantitative PCR (RT-qPCR)
2.8. Western Blotting
2.9. Dual Luciferase Reporter Gene Assay
2.10. Results Statistical Analysis of Data
3. Results
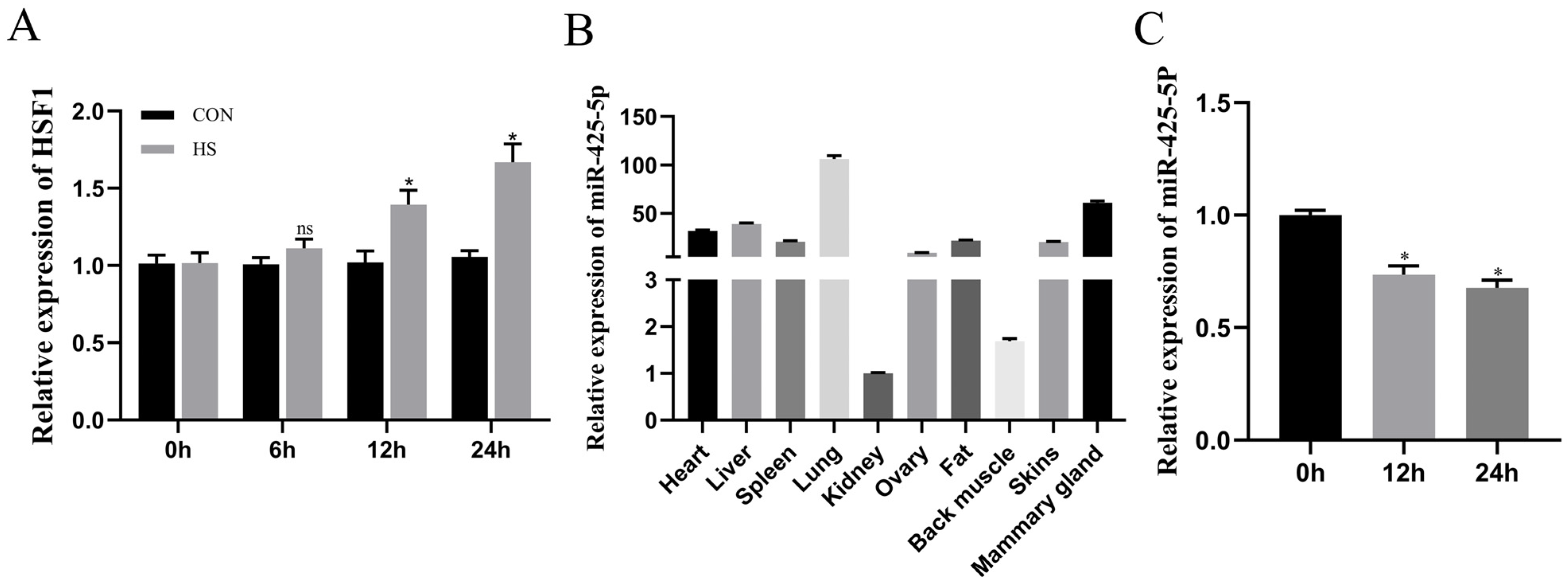
3.1. miR-425-5p Is Associated with Breast Development
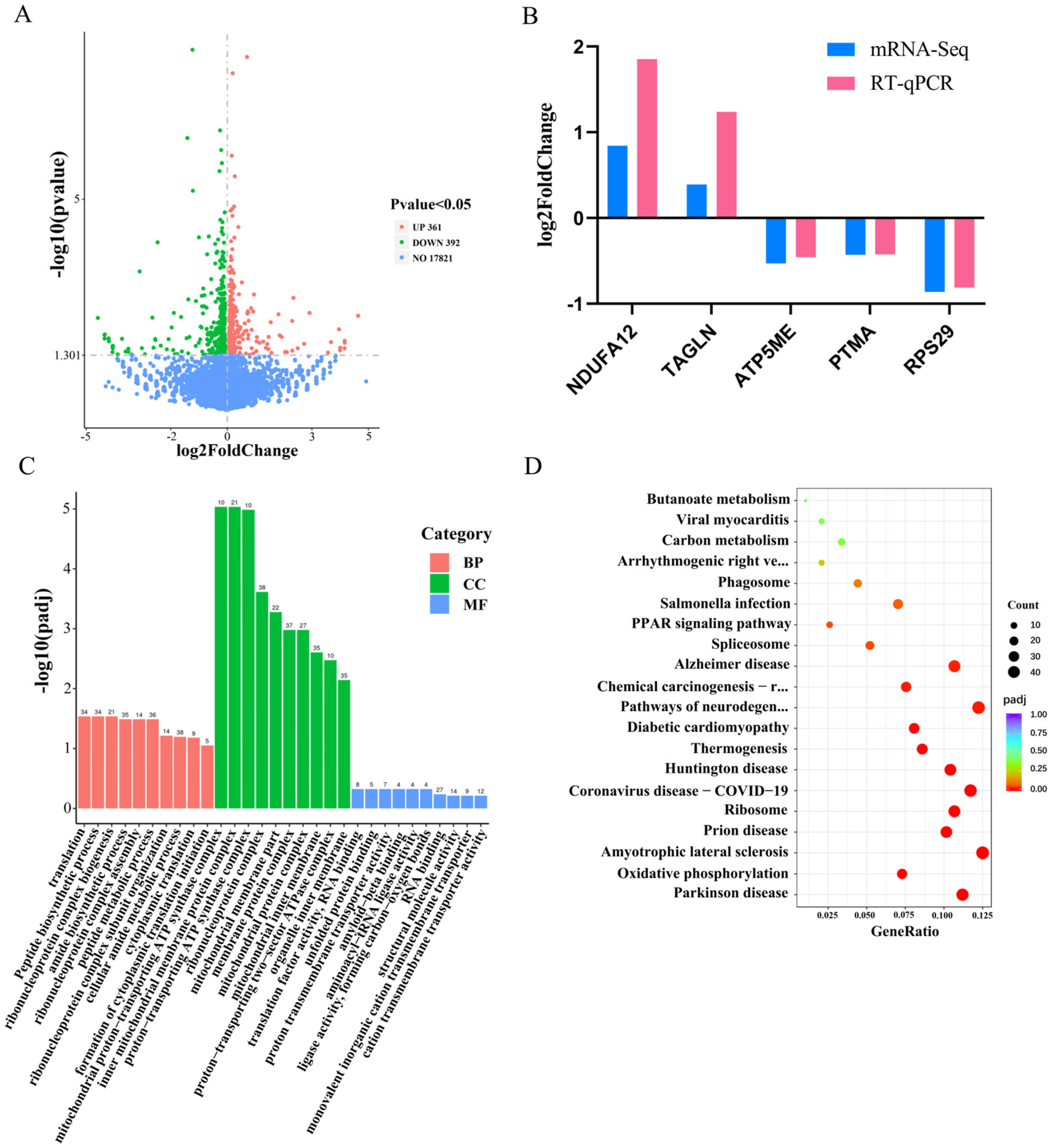
3.2. The Investigation of the Functionality by Differentially Expressed Genes in Response to miR-425-5p
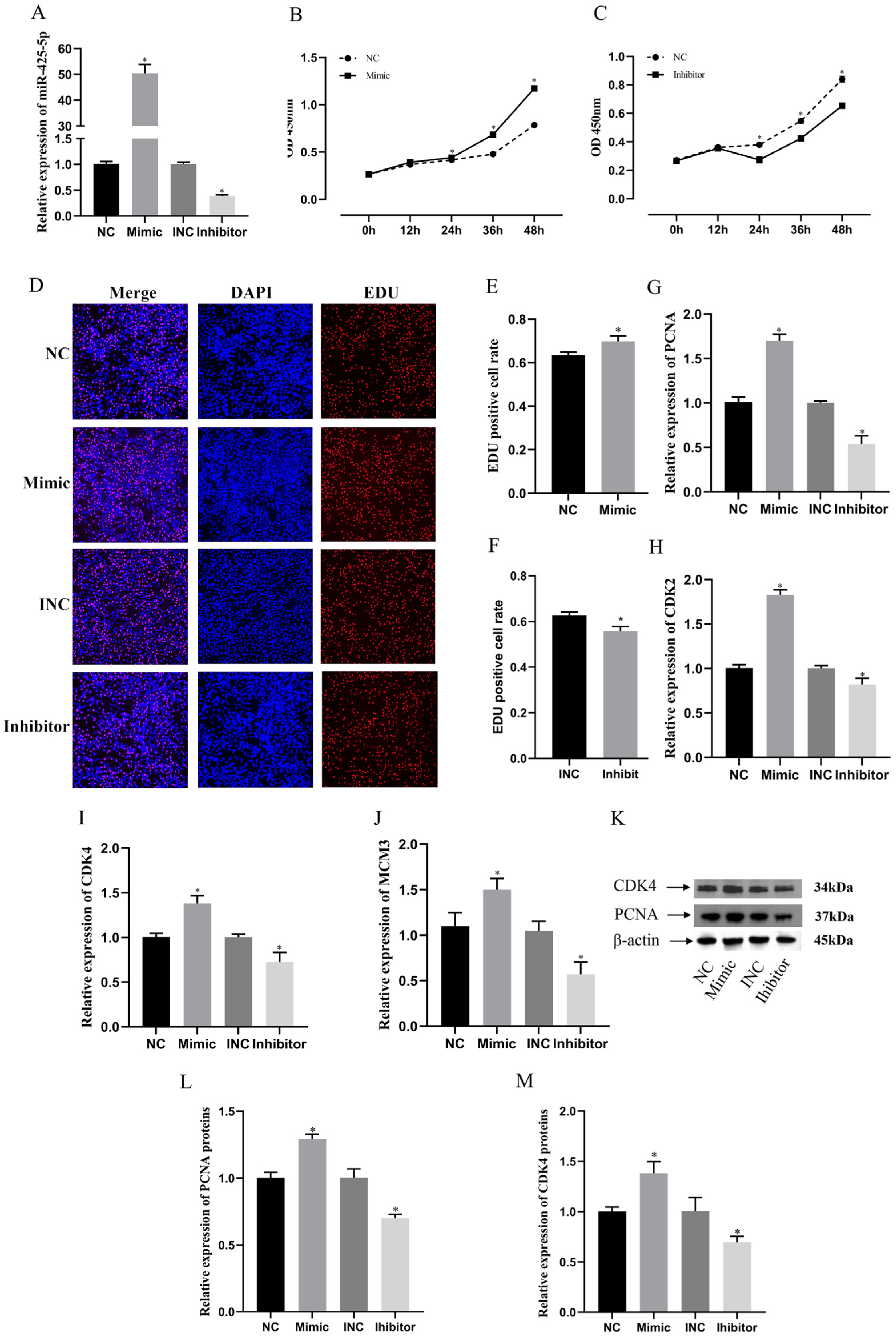
3.3. miR-425-5p Promotes Proliferation of BMECs
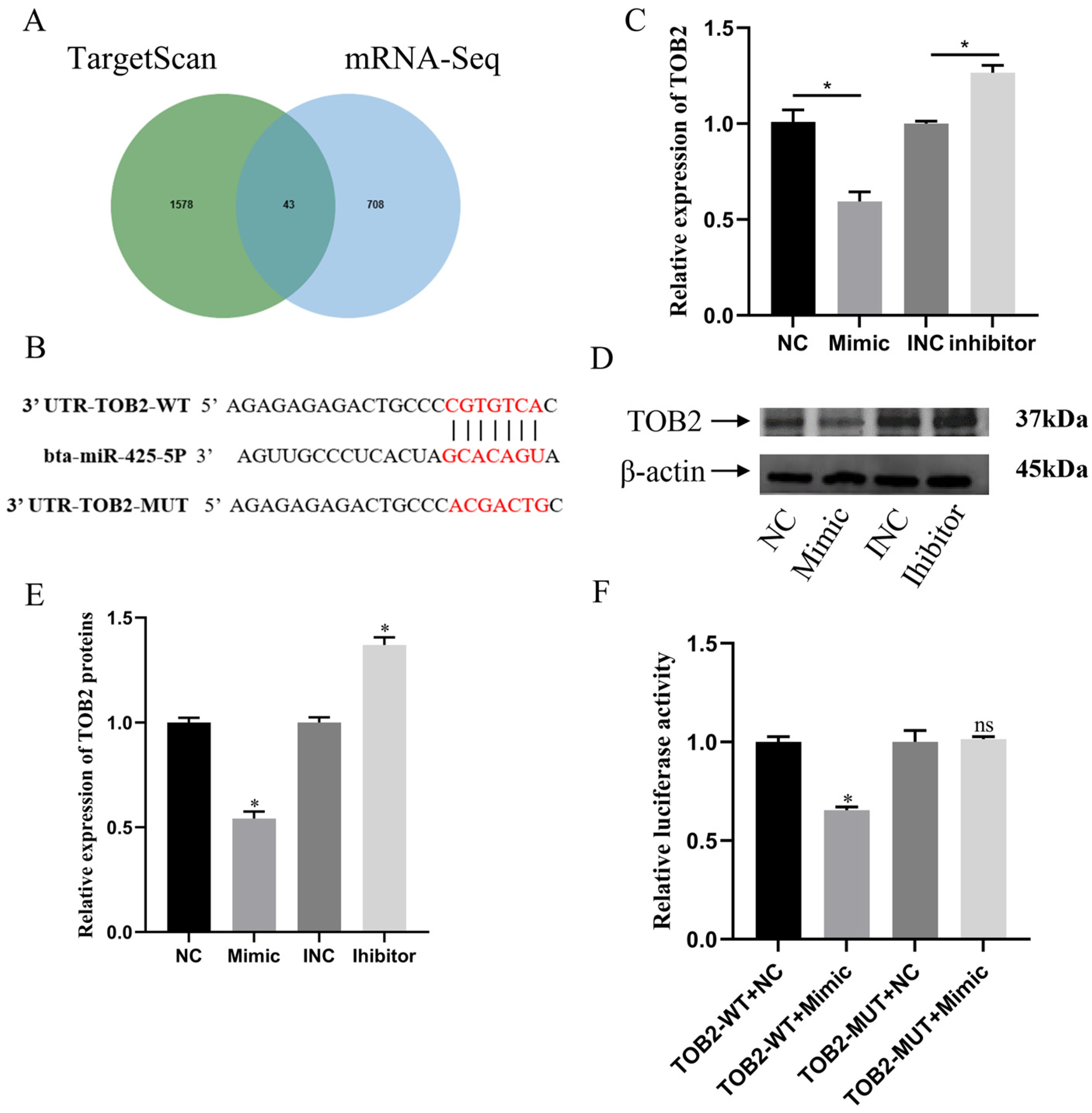
3.4. miR-425-5p Regulates BMECs by Targeting TOB2
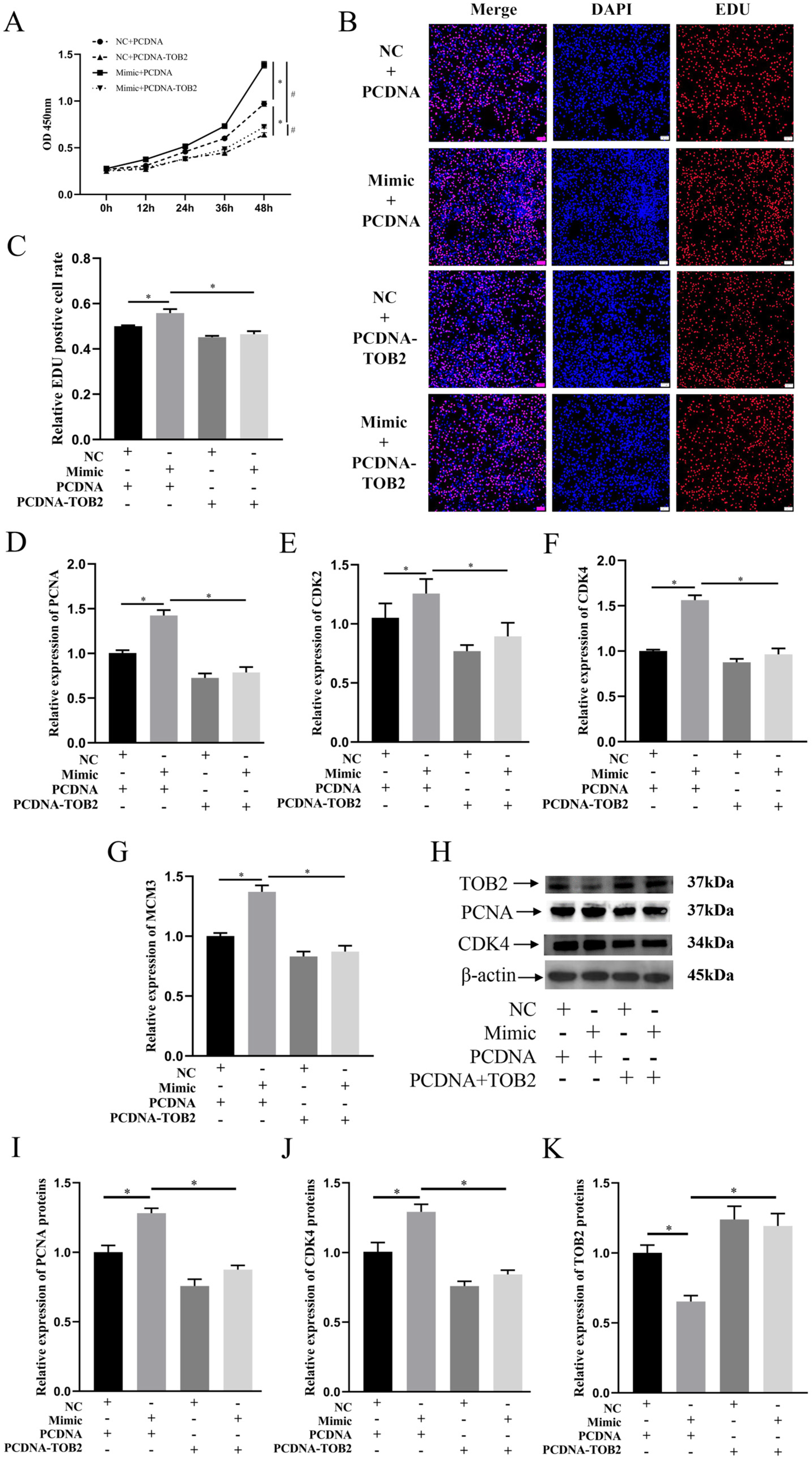
3.5. TOB2 Attenuated the Proliferation Effect of miR-425-5p on BMECs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rauner, G.; Barash, I. Cell hierarchy and lineage commitment in the bovine mammary gland. PLoS ONE 2012, 7, e3011. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, H.; Zhang, Y.; Zhang, J.; Qi, S.; Zhang, Y.; Gao, M.-Q. Overexpression of lncRNA H19 changes basic characteristics and affects immune response of bovine mammary epithelial cells. PeerJ 2019, 7, 6715. [Google Scholar] [CrossRef] [PubMed]
- Tao, S.; Orellana, R.M.; Weng, X.; Marins, T.N.; Dahl, G.E.; Bernard, J.K. Symposium review: The influences of heat stress on bovine mammary gland function. J. Dairy Sci. 2018, 101, 5642–5654. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Yong, Y.; Ju, X. Effect of heat stress on growth and production performance of livestock and poultry: Mechanism to prevention. J. Therm. Biol. 2021, 99, 103019. [Google Scholar] [CrossRef] [PubMed]
- West, J.W.; Mullinix, B.G.; Bernard, J.K. Effects of hot, humid weather on milk temperature, dry matter intake, and milk yield of lactating dairy cows. J. Dairy Sci. 2003, 86, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Ammer, S.; Lambertz, C.; von Soosten, D.; Zimmer, K.; Meyer, U.; Dänicke, S.; Gauly, M. Impact of diet composition and temperature-humidity index on water and dry matter intake of high-yielding dairy cows. J. Anim. Physiol. Anim. Nutr. 2017, 102, 103–113. [Google Scholar] [CrossRef]
- Ravagnolo, O.; Misztal, I.; Hoogenboom, G. Genetic Component of Heat Stress in Dairy Cattle, Development of Heat Index Function. J. Dairy Sci. 2000, 83, 2120–2125. [Google Scholar] [CrossRef]
- Dovolou, E.; Giannoulis, T.; Nanas, I.; Amiridis, G.S. Heat Stress: A Serious Disruptor of the Reproductive Physiology of Dairy Cows. Animals 2023, 13, 1846. [Google Scholar] [CrossRef]
- Bagath, M.; Krishnan, G.; Devaraj, C.; Rashamol, V.P.; Pragna, P.; Lees, A.M.; Sejian, V. The impact of heat stress on the immune system in dairy cattle: A review. Res. Vet. Sci. 2019, 126, 94–102. [Google Scholar] [CrossRef]
- Ibáñez-Ventoso, C.; Vora, M.; Driscoll, M. Sequence relationships among C. elegans, D. melanogaster and human microRNAs highlight the extensive conservation of microRNAs in biology. PLoS ONE 2008, 3, e2818. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Yalcin, A.; Meyer, J.; Lendeckel, W.; Tuschl, T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002, 12, 735–739. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, C.C.; Cheng, H.H.; Tewari, M. MicroRNA profiling: Approaches and considerations. Nat. Rev. Genet. 2012, 13, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.; Reinhart, B.J.; Bartel, D.P.; Zamore, P.D. A biochemical framework for RNA silencing in plants. Genes Dev. 2003, 17, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Toscano-Garibay, J.D.; Aquino-Jarquin, G. Transcriptional regulation mechanism mediated by miRNA-DNA•DNA triplex structure stabilized by Argonaute. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2014, 1839, 1079–1083. [Google Scholar] [CrossRef] [PubMed]
- Huntzinger, E.; Izaurralde, E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011, 12, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Ipsaro, J.J.; Joshua-Tor, L. From guide to target: Molecular insights into eukaryotic RNA-interference machinery. Nat. Struct. Mol. Biol. 2015, 22, 20–28. [Google Scholar] [CrossRef]
- Xu, W.; San Lucas, A.; Wang, Z.; Liu, Y. Identifying microRNA targets in different gene regions. BMC Bioinform. 2014, 15, S4. [Google Scholar] [CrossRef]
- Winkler, G.S. The mammalian anti-proliferative BTG/Tob protein family. J. Cell. Physiol. 2010, 222, 66–72. [Google Scholar] [CrossRef]
- Ge, Y.; Li, J.; Hao, Y.; Hu, Y.; Chen, D.; Wu, B.; Fang, F. MicroRNA-543 functions as an osteogenesis promoter in human periodontal ligament-derived stem cells by inhibiting transducer of ERBB2, 2. J. Periodontal Res. 2018, 53, 832–841. [Google Scholar] [CrossRef]
- Bao, N.; Zhang, P.; Zhu, Y.; Du, P.; Jin, G.; Wu, B.; Ding, T. miR-378a-3p promotes renal cell carcinoma proliferation, migration, and invasion by targeting TOB2. Clin. Transl. Oncol. 2023, 25, 748–757. [Google Scholar] [CrossRef] [PubMed]
- Quan, J.; Li, Y.; Pan, X.; Lai, Y.; He, T.; Lin, C.; Zhou, L.; Zhao, L.; Sun, S.; Ding, Y.; et al. Oncogenic miR-425-5p is associated with cellular migration, proliferation and apoptosis in renal cell carcinoma. Oncol. Lett. 2018, 16, 2175–2184. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Guo, J.; Zhang, Y.; Liu, J.; Ma, H.; Tang, Y. MiR-425-5p accelerated the proliferation, migration, and invasion of ovarian cancer cells via targeting AFF4. J. Ovarian Res. 2021, 14, 138. [Google Scholar] [CrossRef] [PubMed]
- Rode, M.; Silva, A.; Cisilotto, J.; Rosolen, D.; Creczynski-Pasa, T.B. miR-425-5p as an exosomal biomarker for metastatic prostate cancer. Cell. Signal. 2021, 87, 110113. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Xiong, Y.; Peng, Y.; Gao, Y.; Qin, J.; Chu, G.; Pang, W.; Yang, G.S. miR-425-5p Inhibits Differentiation and Proliferation in Porcine Intramuscular Preadipocytes. Int. J. Mol. Sci. 2017, 18, 2101. [Google Scholar] [CrossRef]
- Chen, G.; Sun, W.; Li, Y.; Li, M.; Jia, X.; Wang, J.; Lai, S. CDKN1BmiR-196a Promotes Proliferation of Mammary Epithelial Cells by Targeting. Animals 2023, 13, 3682. [Google Scholar] [CrossRef]
- Curtis, A.; Scharf, B.; Eichen, P.; Spiers, D.E. Relationships between ambient conditions, thermal status, and feed intake of cattle during summer heat stress with access to shade. J. Therm. Biol. 2017, 63, 104–111. [Google Scholar] [CrossRef]
- Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef]
- Cui, L.; Xue, R.; Zhang, X.; Chen, S.; Wan, Y.; Wu, W. Sleep deprivation inhibits proliferation of adult hippocampal neural progenitor cells by a mechanism involving IL-17 and p38 MAPK. Brain Res. 2019, 1714, 81–87. [Google Scholar] [CrossRef]
- Hu, S.; Cai, J.; Fang, H.; Chen, Z.; Zhang, J.; Cai, R. RPS14 promotes the development and progression of glioma via p53 signaling pathway. Exp. Cell Res. 2022, 423, 113451. [Google Scholar] [CrossRef]
- Li, N.; Xiao, Y.; Wang, H.; Zhong, Y.; Yang, H.; Huang, K. Insulin desensitization and cell senescence induced by heat stress in pig testicular cell model. Anim. Biotechnol. 2023, 19, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Collier, R.; Stiening, C.; Pollard, B.; VanBaale, M.; Baumgard, L.; Gentry, P.; Coussens, P.M. Use of gene expression microarrays for evaluating environmental stress tolerance at the cellular level in cattle. J. Anim. Sci. 2006, 84, E1–E13. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Yang, J.; Qi, Z.; Wu, H.; Wang, B.; Zou, F.; Mei, H.; Liu, J.; Wang, W.; Liu, Q. Heat shock proteins: Biological functions, pathological roles, and therapeutic opportunities. MedComm 2022, 3, e161. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.O.; Song, N.-H.; Park, K.-J.; Park, D.-H.; Kim, S.-H.; Chae, Y.-S.; Chung, Y.-G.; Chi, S.-G.; Kang, S.-H. Romo1 is associated with ROS production and cellular growth in human gliomas. J. Neuro-Oncol. 2014, 121, 73–81. [Google Scholar] [CrossRef]
- Ran, T.; Ke, S.; Song, X.; Ma, T.; Xu, Y.; Wang, M. WIPI1 promotes osteosarcoma cell proliferation by inhibiting CDKN1A. Gene 2021, 782, 145537. [Google Scholar] [CrossRef]
- Wu, F.; Ji, A.; Zhang, Z.; Li, J.; Li, P. miR-491-5p inhibits the proliferation and migration of A549 cells by FOXP4. Exp. Ther. Med. 2021, 21, 622. [Google Scholar] [CrossRef]
- Chen, Y.; Meng, F.; Liu, Y.; Zhu, C.; Ling, Y.; Liu, C.; Li, L.; Liu, Y.; He, X.; Cao, J.; et al. Effects of resveratrol on DLD and NDUFB9 decrease in frozen semen of Mongolian sheep. Cryobiology 2023, 114, 104791. [Google Scholar] [CrossRef]
- Wang, J.; Shi, Y.; Elzo, M.A.; Dang, S.; Jia, X.; Lai, S. Genetic diversity of ATP8 and ATP6 genes is associated with high-altitude adaptation in yak. Mitochondrial DNA Part A 2017, 114, 104791. [Google Scholar] [CrossRef]
- Čunátová, K.; Reguera, D.P.; Vrbacký, M.; Fernández-Vizarra, E.; Ding, S.; Fearnley, I.M.; Zeviani, M.; Houštěk, J.; Mráček, T.; Pecina, P. Loss of COX4I1 Leads to Combined Respiratory Chain Deficiency and Impaired Mitochondrial Protein Synthesis. Cells 2021, 10, 369. [Google Scholar] [CrossRef]
- Zhou, J.; Yang, Z.; Cheng, S.; Yu, J.; Huang, C.; Feng, Q. miRNA-425-5p enhances lung cancer growth via the PTEN/PI3K/AKT signaling axis. BMC Pulm. Med. 2020, 20, 223. [Google Scholar] [CrossRef] [PubMed]
- González-Magaña, A.; Blanco, F.J. Human PCNA Structure, Function and Interactions. Biomolecules 2020, 10, 570. [Google Scholar] [CrossRef]
- Lei, M. The MCM complex: Its role in DNA replication and implications for cancer therapy. Curr. Cancer Drug Targets 2005, 5, 365–380. [Google Scholar] [CrossRef] [PubMed]
- Bačević, K.; Lossaint, G.; Achour, T.; Georget, V.; Fisher, D.; Dulić, V. Cdk2 strengthens the intra-S checkpoint and counteracts cell cycle exit induced by DNA damage. Sci. Rep. 2017, 7, 13429. [Google Scholar] [CrossRef]
- Goel, S.; Bergholz, J.; Zhao, J.J. Targeting CDK4 and CDK6 in cancer. Nat. Rev. Cancer 2022, 22, 356–372. [Google Scholar] [CrossRef]
- Wang, Y.; Li, S.; Yuan, S.; Pan, Y.; Hua, Y.; Liu, J.Y. MiR-375 promotes human periodontal ligament stem cells proliferation and osteogenic differentiation by targeting transducer of ERBB2, 2. Arch. Oral Biol. 2020, 117, 104818. [Google Scholar] [CrossRef]
- Ikematsu, N.; Yoshida, Y.; Kawamura-Tsuzuku, J.; Ohsugi, M.; Onda, M.; Hirai, M.; Fujimoto, J.; Yamamoto, T. Tob2, a novel anti-proliferative Tob/BTG1 family member, associates with a component of the CCR4 transcriptional regulatory complex capable of binding cyclin-dependent kinases. Oncogene 1999, 18, 7432–7441. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Strouz, K.; Huang, K.; Shyu, A.B. Tob2 phosphorylation regulates global mRNA turnover to reshape transcriptome and impact cell proliferation. RNA 2020, 26, 1143–1159. [Google Scholar] [CrossRef]
- Ogami, K.; Hosoda, N.; Funakoshi, Y.; Hoshino, S. Antiproliferative protein Tob directly regulates c-myc proto-oncogene expression through cytoplasmic polyadenylation element-binding protein CPEB. Oncogene 2014, 33, 55–64. [Google Scholar] [CrossRef]
- Chen, G.; Huang, G.; Lin, H.; Wu, X.; Tan, X.; Chen, Z. MicroRNA-425-5p modulates osteoporosis by targeting annexin A2. Immun. Ageing 2021, 18, 45. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Chen, G.; Xu, S.; Xia, S.; Sun, W.; Wang, J.; Chen, S.; Lai, S.; Jia, X. miR-425-5p Regulates Proliferation of Bovine Mammary Epithelial Cells by Targeting TOB2. Genes 2024, 15, 174. https://doi.org/10.3390/genes15020174
Li Y, Chen G, Xu S, Xia S, Sun W, Wang J, Chen S, Lai S, Jia X. miR-425-5p Regulates Proliferation of Bovine Mammary Epithelial Cells by Targeting TOB2. Genes. 2024; 15(2):174. https://doi.org/10.3390/genes15020174
Chicago/Turabian StyleLi, Yuchao, Guanhe Chen, Shuxiang Xu, Siqi Xia, Wenqiang Sun, Jie Wang, Shiyi Chen, Songjia Lai, and Xianbo Jia. 2024. "miR-425-5p Regulates Proliferation of Bovine Mammary Epithelial Cells by Targeting TOB2" Genes 15, no. 2: 174. https://doi.org/10.3390/genes15020174






