Proteomic Analysis of Lysine Acetylation and Succinylation to Investigate the Pathogenicity of Virulent Pseudomonas syringae pv. tomato DC3000 and Avirulent Line Pseudomonas syringae pv. tomato DC3000 avrRpm1 on Arabidopsis thaliana
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Inoculation of A. thaliana with Pst DC3000
2.3. LC-MS/MS Analysis of Lysine Acetylation and Succinylation
2.4. Motif Identification and Residues Heat Map
2.5. Functional Enrichment Analysis
3. Results
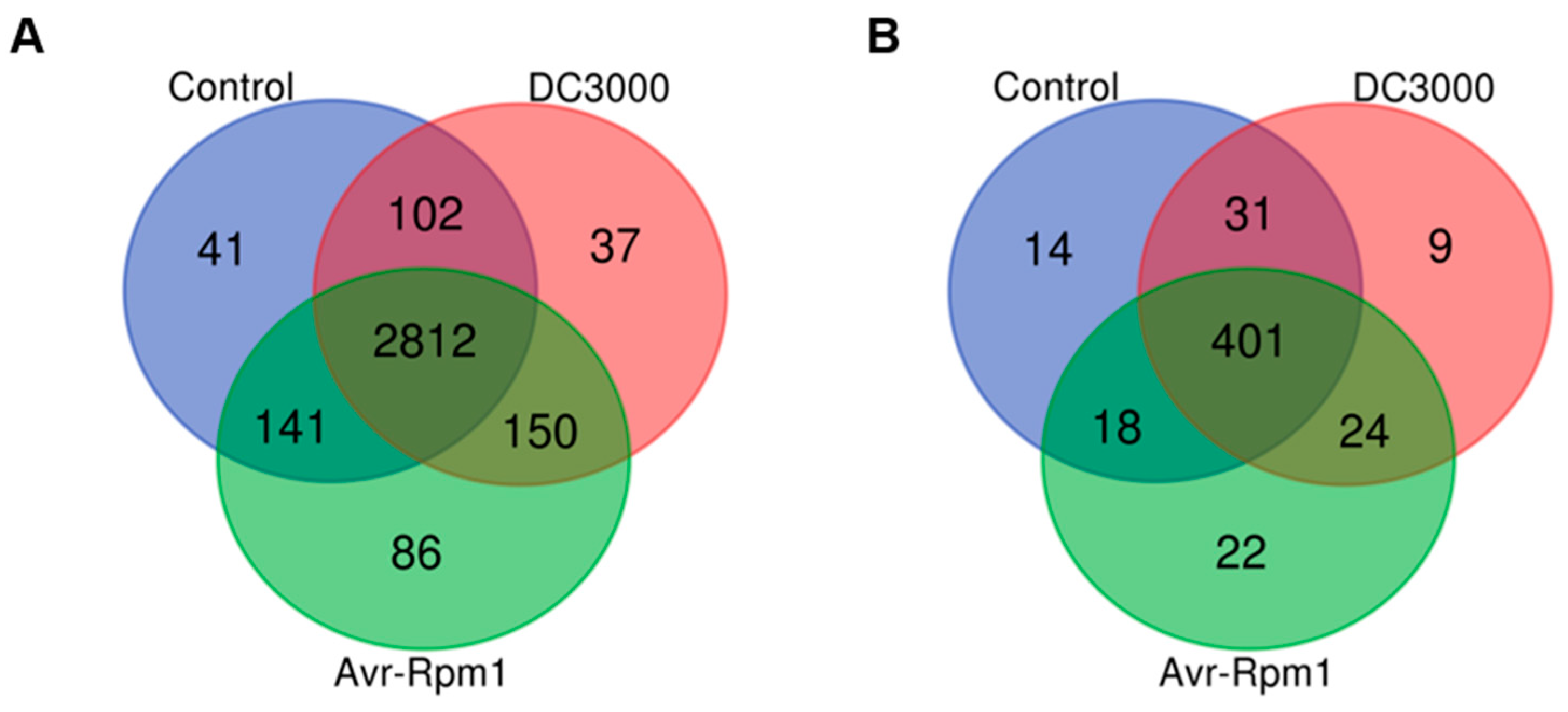
3.1. Systematic Identification of Lysine Acetylome and Succinylation in A. thaliana
3.2. Motif Analysis of Lysine Acetylation and Succinylation Sites
- A−1\F−1\G−1\L−1\P−1\V−1\Y−1KacN+1
- KacF+1\N+1\S+1\T+1\Y+1\H+1\R+1\R+2
- KacK+1\+5\+7
- A-2E-2KacP+2
- KacK+1A+2
- Y−1Kac.
3.3. Functional Annotation of Lysine-Acetylated and Lysine-Succinylated Proteins
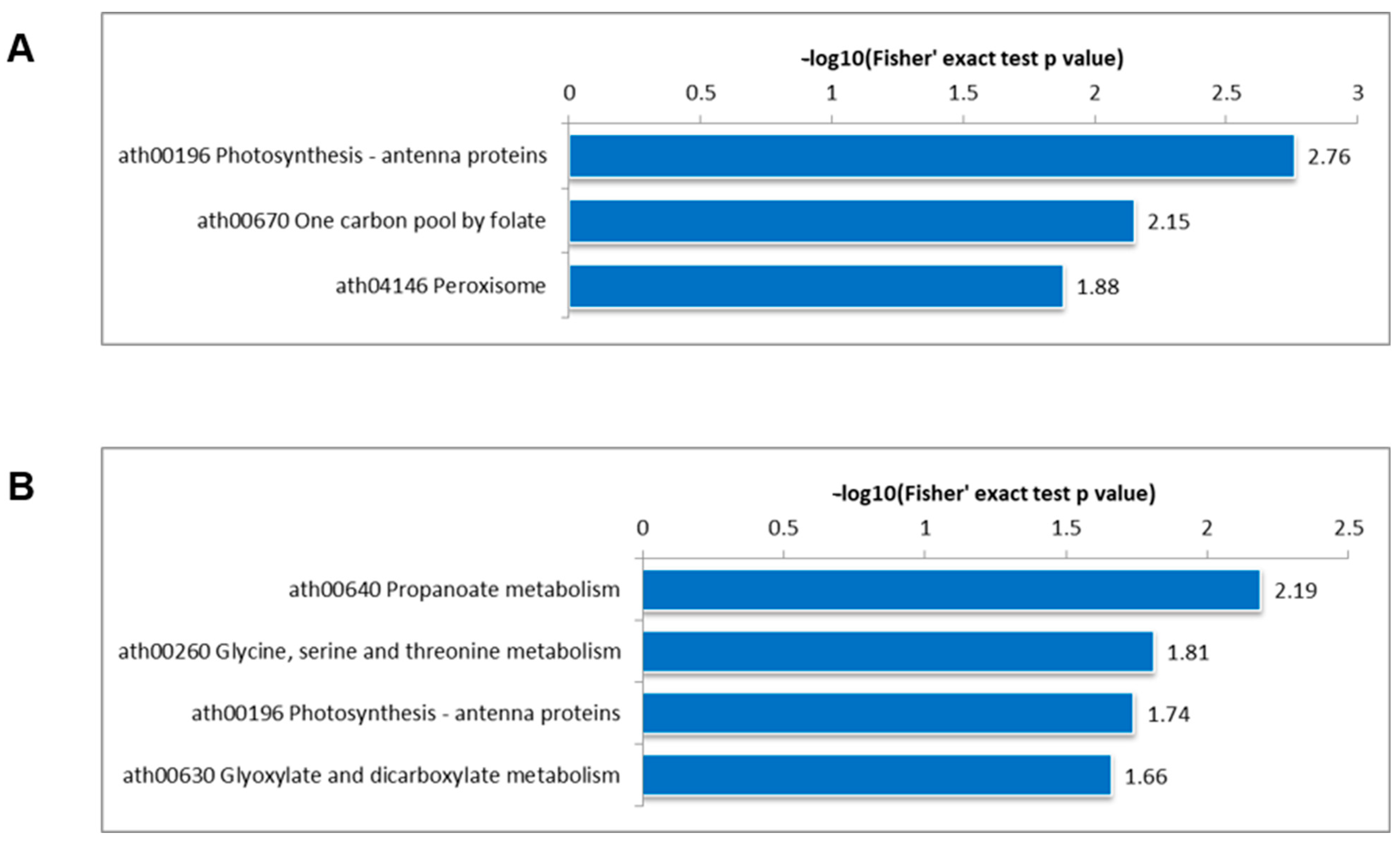
3.4. KEGG Pathway Enrichment Analysis
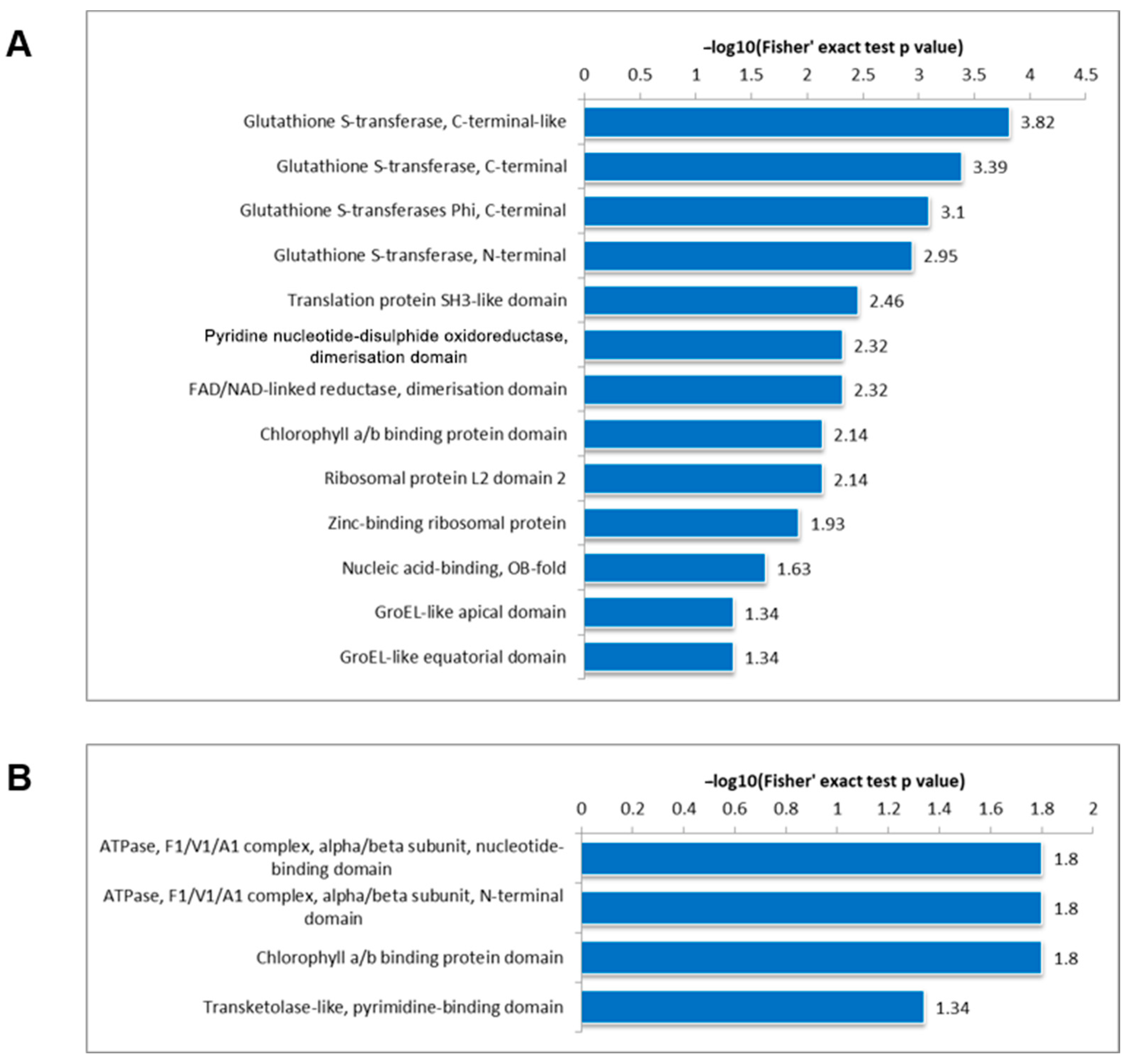
3.5. Protein Domain Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fisher, M.C.; Henk, D.A.; Briggs, C.J.; Brownstein, J.S.; Madoff, L.C.; McCraw, S.L.; Gurr, S.J. Emerging Fungal Threats to Animal, Plant and Ecosystem Health. Nature 2012, 484, 186–194. [Google Scholar] [CrossRef]
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 Plant Pathogenic Bacteria in Molecular Plant Pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef] [PubMed]
- Bent, A.F.; Mackey, D. Elicitors, Effectors, and R Genes: The New Paradigm and A Lifetime Supply of Questions. Annu. Rev. Phytopathol. 2007, 45, 399–436. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.W.; Luan, S.; Lee, S.C. A Prominent Role for RCAR3-mediated ABA Signaling in Response to Pseudomonas syringae pv. tomato DC3000 Infection in Arabidopsis. Plant Cell Physiol. 2014, 55, 1691–1703. [Google Scholar]
- de la Torre, F.; Gutiérrez-Beltrán, E.; Pareja-Jaime, Y.; Chakravarthy, S.; Martin, G.B.; del Pozo, O. The Tomato Calcium Sensor Cbl10 and Its Interacting Protein Kinase Cipk6 Define a Signaling Pathway in Plant Immunity. Plant Cell 2013, 25, 2748–2764. [Google Scholar] [CrossRef] [PubMed]
- Alfano, J.R.; Collmer, A. Type III Secretion System Effector Proteins: Double Agents in Bacterial Disease and Plant Defense. Annu. Rev. Phytopathol. 2004, 42, 385–414. [Google Scholar] [CrossRef] [PubMed]
- Thomma, B.P.; Nurnberger, T.; Joosten, M.H. Of PAMPs and Effectors: The Blurred PTI-ETI Dichotomy. Plant Cell 2011, 23, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Eitas, T.K.; Dangl, J.L. NB-LRR Proteins: Pairs, Pieces, Perception, Partners, and Pathways. Curr. Opin. Plant Biol. 2010, 13, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Yoon, M.; Middleditch, M.J.; Rikkerink, E.H.A. A Conserved Glutamate Residue in RPM1-INTERACTING PROTEIN4 is ADP-ribosylated by the Pseudomonas Effector AvrRpm2 to activate RPM1-mediated plant resistance. Plant Cell 2022, 34, 4950–4972. [Google Scholar] [CrossRef]
- Carabetta, V.J.; Greco, T.M.; Cristea, I.M.; Dubnau, D. YfmK is An Nε-lysine Acetyltransferase That Directly Acetylates the Histone-like Protein Hbsu in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2019, 116, 3752–3757. [Google Scholar] [CrossRef]
- Ren, J.; Sang, Y.; Lu, J.; Yao, Y.F. Protein Acetylation and Its Role in Bacterial Virulence. Trends Microbiol. 2017, 25, 768–779. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ma, X.; Wang, C.; Liu, S.; Yu, G.; Gao, M.; Qian, H.; Liu, M.; Luisi, B.F.; Gabriel, D.W.; et al. Acetylation of A Fungal Effector that Translocates Host PR1 Facilitates Virulence. eLife 2022, 11, e82628. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Li, W.; Xie, J. Prokaryotic Nε-lysine Acetylomes and Implications for New Antibiotics. J. Cell. Biochem. 2012, 113, 3601–3609. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.I.; Lima, B.P.; Wolfe, A.J. Bacterial Protein Acetylation: The Dawning of a New Age. Mol. Microbiol. 2010, 77, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Narita, T.; Weinert, B.T.; Choudhary, C. Functions and Mechanisms of Non-histone Protein Acetylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 156–174. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.J.; Seto, E. Lysine Acetylation: Codified Crosstalk with Other Posttranslational Modifications. Mol. Cell 2008, 31, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Weinert, B.T.; Schölz, C.; Wagner, S.A.; Iesmantavicius, V.; Su, D.; Daniel, J.A.; Choudhary, C. Lysine Succinylation is A Frequently Occurring Modification in Prokaryotes and Eukaryotes and Extensively Overlaps with Acetylation. Cell Rep. 2013, 4, 842–851. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.A.; Beli, P.; Weinert, B.T.; Nielsen, M.L.; Cox, J.; Mann, M.; Choudhary, C. A Proteome-wide, Quantitative Survey of in Vivo Ubiquitylation Sites Reveals Widespread Regulatory Roles. Mol. Cell. Proteom. 2011, 10, M111.013284. [Google Scholar] [CrossRef]
- Sun, L.; Yao, Z.; Guo, Z.; Zhang, L.; Wang, Y.; Mao, R.; Lin, Y.; Fu, Y.; Lin, X. Comprehensive Analysis of the Lysine Acetylome in Aeromonas hydrophila Reveals Cross-talk between Lysine Acetylation and Succinylation in LuxS. Emerg. Microbes Infect. 2019, 8, 1229–1239. [Google Scholar] [CrossRef]
- Zhang, Z.; Tan, M.; Xie, Z.; Dai, L.; Chen, Y.; Zhao, Y. Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 2011, 7, 58–63. [Google Scholar] [CrossRef]
- Zhen, S.; Deng, X.; Wang, J.; Zhu, G.; Cao, H.; Yuan, L.; Yan, Y. First Comprehensive Proteome Analyses of Lysine Acetylation and Succinylation in Seedling Leaves of Brachypodium distachyon L. Sci. Rep. 2016, 6, 31576. [Google Scholar] [CrossRef]
- Zhou, X.; Qian, G.; Yi, X.; Li, X.; Liu, W. Systematic Analysis of the Lysine Acetylome in Candida albicans. J. Proteom. Res. 2016, 15, 2525–2536. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; He, Y.; Zhou, X.; Qian, G.; Lv, G.; Shen, Y.; Liu, J.; Li, D.; Li, X.; Liu, W. Systematic Analysis of the Lysine Succinylome in Candida albicans. J. Proteom. Res. 2016, 15, 3793–3801. [Google Scholar] [CrossRef]
- Zhang, W.; Jiang, L.H.; Huang, J.; Ding, Y.Q.; Liu, Z.B. Loss of Proton/calcium Exchange 1 Results in the Activation of Plant Defense and Accelerated Senescence in Arabidopsis. Plant Sci. 2020, 296, 110472. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Kir, J.; Liu, D.; Bryant, D.; Guo, Y.J.; Stephens, R.; Baseler, M.W.; Lane, H.C.; et al. DAVID Bioinformatics Resources: Expanded Annotation Database and Novel Algorithms to Better Extract Biology from Large Gene Lists. Nucleic Acids Res. 2007, 35, W169–W175. [Google Scholar] [CrossRef] [PubMed]
- Xing, E.; Fan, X.; Jiang, F.; Zhang, Y. Advancements in Research on Prevention and Control Strategies for Maize White Spot Disease. Genes 2023, 14, 2061. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, L.; Ji, H.; Song, W.; Zhong, Z.; Jiang, M.; Zhang, Y.; Li, Q.; Cheng, L.; Kou, M. RNA-Sequencing Analysis Revealed Genes Associated with Sweet Potato (Ipomoea batatas (L.) Lam.) Responses to Stem Rot during Different Infection Stages. Genes 2023, 14, 2215. [Google Scholar] [CrossRef]
- Ronald, P.C.; Beutler, B. Plant and Animal Sensors of Conserved Microbial Signatures. Science 2010, 330, 1061–1064. [Google Scholar] [CrossRef]
- DeFalco, T.A.; Zipfel, C. Molecular Mechanisms of Early Plant Pattern-triggered Immune Signaling. Mol. Cell 2021, 81, 3449–3467. [Google Scholar] [CrossRef]
- Xia, L.; Kong, X.; Song, H.; Han, Q.; Zhang, S. Advances in Proteome-wide Analysis of Plant Lysine Acetylation. Plant Commun. 2021, 3, 100266. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, G.; Song, L.; Mu, P.; Wang, S.; Liang, W.; Lin, Q. Global Analysis of Protein Lysine Succinylation Profiles in Common Wheat. BMC Genom. 2017, 18, 309. [Google Scholar] [CrossRef] [PubMed]
- Melo-Braga, M.N.; Verano-Braga, T.; Leon, I.R.; Antonacci, D.; Nogueira, F.C.S.; Thelen, J.J.; Larsen, M.R.; Palmisano, G. Modulation of Protein Phosphorylation, N-glycosylation and Lys-acetylation in Grape (Vitis vinifera) Mesocarp and Exocarp Owing to Lobesia botrana Infection. Mol. Cell. Proteom. 2012, 11, 945–956. [Google Scholar] [CrossRef] [PubMed]
- Walley, J.W.; Shen, Z.; McReynolds, M.R.; Schmelz, E.A.; Briggs, S.P. Fungal-Induced Protein Hyperacetylation in Maize Identified by Acetylome Profiling. Proc. Natl. Acad. Sci. USA 2018, 115, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Fan, G.; Wang, Z.; Gu, Z. Phytoplasma-induced Changes in the Acetylome and Succinylome of Paulownia tomentosa Provide Evidence for Involvement of Acetylated Proteins in Witches’ Broom disease. Mol. Cell. Proteom. 2019, 18, 1703. [Google Scholar] [CrossRef] [PubMed]
- Yuan, B.; Liu, T.; Cheng, Y.; Gao, S.; Li, L.; Cai, L.; Yang, J.; Chen, J.; Zhong, K. Comprehensive Proteomic Analysis of Lysine Acetylation in Nicotiana benthamiana after Sensing CWMV Infection. Front. Microbiol. 2021, 12, 672559. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, C.; Kumar, C.; Gnad, F.; Nielsen, M.L.; Rehman, M.; Walther, T.C.; Olsen, J.V.; Mann, M. Lysine Acetylation Targets Protein Complexes and Co-regulates Major Cellular Functions. Science 2009, 325, 834–840. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Yang, C.; Xiong, H.; Lin, Y.; Yao, J.; Li, H.; Xie, L.; Zhao, W.; Yao, Y.; et al. Acetylation of Metabolic Enzymes Coordinates Carbon Source Utilization and Metabolic Flux. Science 2010, 327, 1004–1007. [Google Scholar] [CrossRef]
- Zhao, S.; Xu, W.; Jiang, W.; Yu, W.; Lin, Y.; Zhang, T.; Yao, J.; Zhou, L.; Zeng, Y.; Li, H.; et al. Regulation of Cellular Metabolism by Protein Lysine Acetylation. Science 2010, 327, 1000–1004. [Google Scholar] [CrossRef]
- Lehtimaki, N.; Koskela, M.M.; Mulo, P. Posttranslational modifications of chloroplast proteins: An emerging field. Plant Physiol. 2015, 168, 768–775. [Google Scholar] [CrossRef]
- He, D.; Wang, Q.; Li, M.; Damaris, R.N.; Yi, X.; Cheng, Z.; Yang, P. Global Proteome Analyses of Lysine Acetylation and Succinylation Reveal the Widespread Involvement of both Modification in Metabolism in the Embryo of Germinating Rice Seed. J. Proteom. Res. 2016, 15, 879–890. [Google Scholar] [CrossRef]
- Xu, Y.X.; Chen, W.; Ma, C.L.; Shen, S.Y.; Zhou, Y.Y.; Zhou, L.Q.; Chen, L. Proteome and Acetyl-proteome Profiling of Camellia sinensis cv. ‘Anjin Baicha’ during Periodic Albinism Reveals Alterations in Photosynthetic and Secondary Metabolite Biosynthetic Pathways. Front. Plant Sci. 2017, 8, 2104. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Ding, Y.; Wang, W.; Zhao, X.; Liu, Y.; Timko, M.P.; Zhang, Z.; Zhang, H. Insights into Gene Regulation of Jasmonate-Induced Whole-Plant Senescence of Tobacco under Non-Starvation Conditions. Plant Cell Physiol. 2022, 63, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Kanayama, Y.; Zheng, M.S.; Kusano, T.; Hase, S.; Ikegami, M.; Shah, J. Antagonistic Interactions between the SA and JA Signaling Pathways in Arabidopsis Modulate Expression of Defense Genes and Gene-for-gene Resistance to Cucumber Mosaic Virus. Plant Cell Physiol. 2004, 45, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Banday, Z.Z.; Nandi, A.K. Arabidopsis thaliana GLUTATHIONE-S-TRANSFERASE THETA 2 Interacts with RSI1/FLD to Activate Systemic Acquired Resistance. Mol. Plant Pathol. 2018, 19, 464–475. [Google Scholar] [CrossRef]
- Zhou, H.; Finkemeier, I.; Guan, W.; Tossounian, M.A.; Wei, B.; Young, D.; Huang, J.; Messens, J.; Yang, X.; Zhu, J.; et al. Oxidative Stress-triggered Interactions between the Succinyl- and Acetyl-proteomes of Rice Leaves. Plant Cell Environ. 2018, 41, 1139–1153. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, Y.; Liu, Y.; Yang, K.; Zhao, Y.; Wen, C.; Yang, Y.; Zhang, W. Proteomic Analysis of Lysine Acetylation and Succinylation to Investigate the Pathogenicity of Virulent Pseudomonas syringae pv. tomato DC3000 and Avirulent Line Pseudomonas syringae pv. tomato DC3000 avrRpm1 on Arabidopsis thaliana. Genes 2024, 15, 499. https://doi.org/10.3390/genes15040499
Ding Y, Liu Y, Yang K, Zhao Y, Wen C, Yang Y, Zhang W. Proteomic Analysis of Lysine Acetylation and Succinylation to Investigate the Pathogenicity of Virulent Pseudomonas syringae pv. tomato DC3000 and Avirulent Line Pseudomonas syringae pv. tomato DC3000 avrRpm1 on Arabidopsis thaliana. Genes. 2024; 15(4):499. https://doi.org/10.3390/genes15040499
Chicago/Turabian StyleDing, Yongqiang, Yangxuan Liu, Kexin Yang, Yiran Zhao, Chun Wen, Yi Yang, and Wei Zhang. 2024. "Proteomic Analysis of Lysine Acetylation and Succinylation to Investigate the Pathogenicity of Virulent Pseudomonas syringae pv. tomato DC3000 and Avirulent Line Pseudomonas syringae pv. tomato DC3000 avrRpm1 on Arabidopsis thaliana" Genes 15, no. 4: 499. https://doi.org/10.3390/genes15040499




