Identification of Bradyrhizobium elkanii Genes Involved in Incompatibility with Vigna radiata
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Transposon Mutagenesis of B. elkanii USDA61
2.3. Plant Cultivation, Mutant Screening, and Inoculation Tests
2.4. Nucleotide Sequence Analysis of Tn5-Flanking Regions
2.5. Microscopy
2.6. Nucleotide Sequence Accession Numbers
3. Results
3.1. Isolation of the Transposon Mutants of B. elkanii
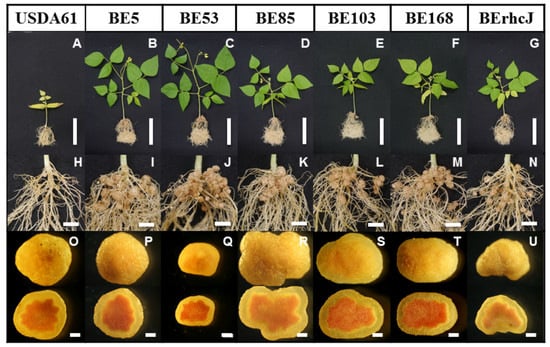
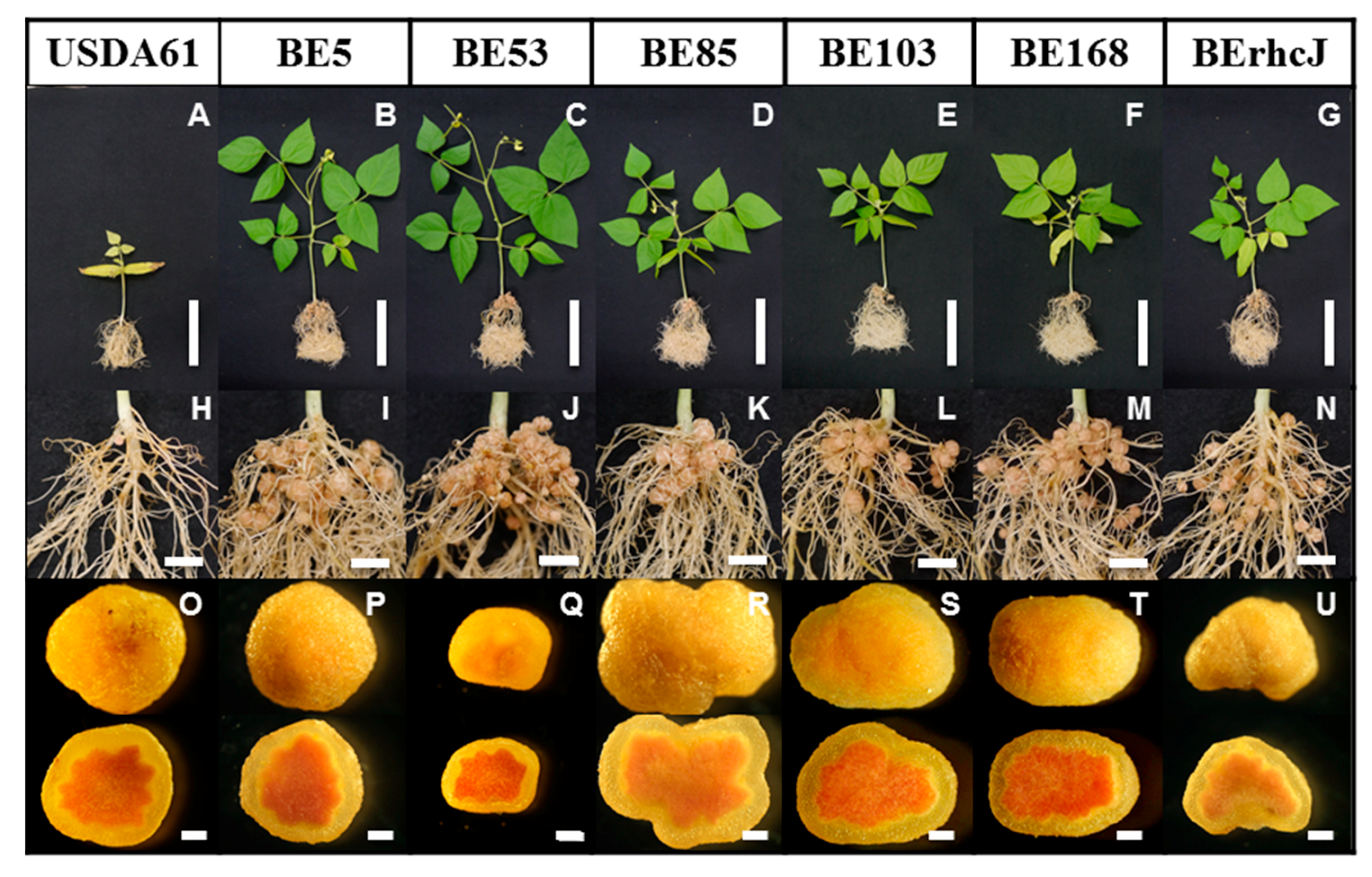
3.2. Symbiotic Phenotypes of the Transposon Mutants on V. radiata cv. KPS1 Plants
3.3. Symbiotic Phenotypes of the KPS1-Nodulating Mutants on Rj4 Soybean
3.4. B. elkanii Genes Responsible for the Symbiotic Incompatibility
4. Discussion
5. Concluding Remarks
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cullimore, J.V.; Ranjeva, R.; Bono, J.-J. Perception of lipo-chitooligosaccharidic Nod factors in legumes. Trends Plant Sci. 2001, 6, 24–30. [Google Scholar] [CrossRef]
- Radutoiu, S.; Madsen, L.H.; Madsen, E.B.; Jurkiewicz, A.; Fukai, E.; Quistgaard, E.M.H.; Albrektsen, A.S.; James, E.K.; Thirup, S.; Stougaard, J. LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J. 2007, 26, 3923–3935. [Google Scholar] [CrossRef] [PubMed]
- Lerouge, P.; Roche, P.; Faucher, C.; Maillet, F.; Truchet, G.; Promé, J.C.; Dénarié, J. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 1990, 344, 781–784. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, S.; Kaneko, T.; Sato, S.; Saeki, K. Hijacking of leguminous nodulation signaling by the rhizobial type III secretion system. Proc. Natl. Acad. Sci. USA 2013, 110, 17131–17136. [Google Scholar] [CrossRef] [PubMed]
- Rumjanek, N.G.; Dobert, R.C.; van Berkum, P.; Triplett, E.W. Common soybean inoculant strains in Brazil are members of Bradyrhizobium elkanii. Appl. Environ. Microbiol. 1993, 59, 4371–4373. [Google Scholar] [PubMed]
- Hayashi, M.; Saeki, Y.; Haga, M.; Harada, K.; Kouchi, H.; Umehara, Y. Rj (rj) genes involved in nitrogen-fixing root nodule formation in soybean. Breed. Sci. 2012, 61, 544–553. [Google Scholar] [CrossRef] [PubMed]
- Sadowsky, M.J.; Cregan, P.B. The Soybean Rj4 Allele Restricts Nodulation by Bradyrhizobium japonicum Serogroup 123 Strains. Appl. Environ. Microbiol. 1992, 58, 720–723. [Google Scholar] [PubMed]
- Faruque, O.M.; Miwa, H.; Yasuda, M.; Fujii, Y.; Kaneko, T.; Sato, S.; Okazaki, S. Identification of Bradyrhizobium elkanii Genes Involved in Incompatibility with Soybean Plants Carrying the Rj4 Allele. Appl. Environ. Microbiol. 2015, 81, 6710–6717. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Tang, F.; Gao, M.; Krishnan, H.B.; Zhu, H. R gene-controlled host specificity in the legume-rhizobia symbiosis. Proc. Natl. Acad. Sci. USA 2010, 107, 18735–18740. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Yang, S.; Liu, J.; Zhu, H. Rj4, a Gene Controlling Nodulation Specificity in Soybeans, Encodes a Thaumatin-Like Protein But Not the One Previously Reported. Plant Physiol. 2016, 170, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Loh, J.; Stacey, G. Nodulation gene regulation in Bradyrhizobium japonicum: A unique integration of global regulatory circuits. Appl. Environ. Microbiol. 2003, 69, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, S.; Zehner, S.; Hempel, J.; Lang, K.; Göttfert, M. Genetic organization and functional analysis of the type III secretion system of Bradyrhizobium elkanii. FEMS Microbiol. Lett. 2009, 295, 88–95. [Google Scholar] [CrossRef] [PubMed]
- De Lyra Mdo, C.; Lopez-Baena, F.J.; Madinabeitia, N.; Vinardell, J.M.; Espuny Mdel, R.; Cubo, M.T.; Belloguin, R.A.; Ruiz-Sainz, J.E.; Ollero, F.J. Inactivation of the Sinorhizobium fredii HH103 rhcJ gene abolishes nodulation outer proteins (Nops) secretion and decreases the symbiotic capacity with soybean. Int. Microbiol. 2006, 9, 125–133. [Google Scholar] [PubMed]
- Göttfert, M.; Röthlisberger, S.; Kündig, C.; Beck, C.; Marty, R.; Hennecke, H. Potential symbiosis-specific genes uncovered by sequencing a 410-kilobase DNA region of the Bradyrhizobium japonicum chromosome. J. Bacteriol. 2001, 183, 1405–1412. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, H.B.; Lorio, J.; Kim, W.S.; Jiang, G.; Kim, K.Y.; DeBoer, M.; Pueppke, S.G. Extracellular proteins involved in soybean cultivar-specific nodulation are associated with pilus-like surface appendages and exported by a type III protein secretion system in Sinorhizobium fredii USDA257. Mol. Plant Microbe Interact. 2003, 16, 617–625. [Google Scholar] [CrossRef] [PubMed]
- Viprey, V.; Del Greco, A.; Golinowski, W.; Broughton, W.J.; Perret, X. Symbiotic implications of type III protein secretion machinery in Rhizobium. Mol. Microbiol. 1998, 28, 1381–1389. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, T.; Nakamura, Y.; Sato, S.; Asamizu, E.; Kato, T.; Sasamoto, S.; Watanabe, A.; Idesawa, K.; Ishikawa, A.; Kawashima, K.; et al. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 2000, 7, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Sadowsky, M.J.; Tully, R.E.; Cregan, P.B.; Keyser, H.H. Genetic Diversity in Bradyrhizobium japonicum Serogroup 123 and Its Relation to Genotype-Specific Nodulation of Soybean. Appl. Environ. Microbiol. 1987, 53, 2624–2630. [Google Scholar] [PubMed]
- Green, M.R.; Sambrook, J. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2012. [Google Scholar]
- Simon, R.; Priefer, U.; Pühler, A. A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Nat. Biotechnol. 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Herrero, M.; de Lorenzo, V.; Timmis, K.N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 1990, 172, 6557–6567. [Google Scholar] [CrossRef] [PubMed]
- Figurski, D.H.; Helinski, D.R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 1979, 76, 1648–1652. [Google Scholar] [CrossRef] [PubMed]
- Krause, A.; Doerfel, A.; Göttfert, M. Mutational and transcriptional analysis of the type III secretion system of Bradyrhizobium japonicum. Mol. Plant Microbe Interact. 2002, 15, 1228–1235. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.M.; Ricke, S.C. Efficient amplification of multiple transposon-flanking sequences. J. Microbiol. Methods 2000, 41, 195–199. [Google Scholar] [CrossRef]
- Kaneko, T.; Faculty of Life Sciences, Kyoto Sangyo University, Motoyama, Kamigamo, Kita-Ku, Kyoto, Japan. Complete genome sequence of Bradyrhizobium elkanii USDA61. Unpublished work. 2015. [Google Scholar]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000. [Google Scholar] [CrossRef]
- Marie, C.; Deakin, W.J.; Ojanen-Reuhs, T.; Diallo, E.; Reuhs, B.; Broughton, W.J.; Perret, X. TtsI, a Key Regulator of Rhizobium Species NGR234 Is Required for Type III-Dependent Protein Secretion and Synthesis of Rhamnose-Rich Polysaccharides. Mol. Plant Microbe Interact. 2004, 958, 958–966. [Google Scholar] [CrossRef] [PubMed]
- Hempel, J.; Zehner, S.; Göttfert, M.; Patschkowski, T. Analysis of the secretome of the soybean symbiont Bradyrhizobium japonicum. J. Biotechnol. 2009, 140, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, S.; Okabe, S.; Higashi, M.; Shimoda, Y.; Sato, S.; Tabata, S.; Hashiguchi, M.; Akashi, R.; Gottfert, M.; Saeki, K. Identification and functional analysis of type III effector proteins in Mesorhizobium loti. Mol. Plant Microbe Interact. 2010, 23, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Marie, C.; Deakin, W.J.; Viprey, V.; Kopciñska, J.; Golinowski, W.; Krishnan, H.B.; Perret, X.; Broughton, W.J. Characterization of Nops, nodulation outer proteins, secreted via the type III secretion system of NGR234. Mol. Plant Microbe Interact. 2003, 16, 743–751. [Google Scholar] [CrossRef] [PubMed]
- Meinhardt, L.W.; Krishnan, H.B.; Balatti, P.A.; Pueppke, S.G. Molecular cloning and characterization of a sym plasmid locus that regulates cultivar-specific nodulation of soybean by Rhizobium fredii USDA257. Mol. Microbiol. 1993, 9, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Wassem, R.; Kobayashi, H.; Kambara, K.; Le Quéré, A.; Walker, G.C.; Broughton, W.J.; Deakin, W.J. TtsI regulates symbiotic genes in Rhizobium species NGR234 by binding to tts boxes. Mol. Microbiol. 2008, 68, 736–748. [Google Scholar] [CrossRef] [PubMed]
- Zehner, S.; Schober, G.; Wenzel, M.; Lang, K.; Göttfert, M. Expression of the Bradyrhizobium japonicum type III secretion system in legume nodules and analysis of the associated tts box promoter. Mol. Plant Microbe Interact. 2008, 21, 1087–1093. [Google Scholar] [CrossRef] [PubMed]
- Danielsen, S.; Kilstrup, M.; Barilla, K.; Jochimsen, B.; Neuhard, J. Characterization of the Escherichia coli codBA operon encoding cytosine permease and cytosine deaminase. Mol. Microbiol. 1992, 6, 1335–1344. [Google Scholar] [CrossRef] [PubMed]
- Bagnarol, E.; Popovici, J.; Alloisio, N.; Maréchal, J.; Pujic, P.; Normand, P.; Fernandez, M.P. Differential Frankia protein patterns induced by phenolic extracts from Myricaceae seeds. Physiol. Plant. 2007, 130, 380–390. [Google Scholar] [CrossRef]
- Higgins, C.F. ABC transporters: Physiology, structure and mechanism—An overview. Res. Microbiol. 2001, 152, 205–210. [Google Scholar] [CrossRef]
- Abouhamad, W.N.; Manson, M.D. The dipeptide permease of Escherichia coli closely resembles other bacterial transport systems and shows growth-phase-dependent expression. Mol. Microbiol. 1994, 14, 1077–1092. [Google Scholar] [CrossRef] [PubMed]
- Doeven, M.K.; van den Bogaart, G.; Krasnikov, V.; Poolman, B. Probing receptor-translocator interactions in the oligopeptide ABC transporter by fluorescence correlation spectroscopy. Biophys. J. 2008, 94, 3956–3965. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.-S.; Franck, W.L.; Cytryn, E.; Jeong, S.; Joshi, T.; Emerich, D.W.; Sadowsky, M.J.; Xu, D.; Stacey, G. An Oligonucleotide Microarray Resource for Transcriptional Profiling of Bradyrhizobium japonicum. Mol. Plant Microbe Interact. 2007. [Google Scholar] [CrossRef] [PubMed]
- Pessi, G.; Ahrens, C.H.; Rehrauer, H.; Lindemann, A.; Hauser, F.; Fischer, H.-M.; Hennecke, H. Genome-Wide Transcript Analysis of Bradyrhizobium japonicum Bacteroids in Soybean Root Nodules. Mol. Plant Microbe Interact. 2007, 20, 1353–1363. [Google Scholar] [CrossRef] [PubMed]
- Vogt, S.L.; Green, C.; Stevens, K.M.; Day, B.; Erickson, D.L.; Woods, D.E.; Storey, D.G. The stringent response is essential for Pseudomonas aeruginosa virulence in the rat lung agar bead and Drosophila melanogaster feeding models of infection. Infect. Immun. 2011, 79, 4094–4104. [Google Scholar] [CrossRef] [PubMed]
- Calderón-Flores, A.; Du Pont, G.; Huerta-Saquero, A.; Merchant-Larios, H.; Servín-González, L.; Durán, S. The stringent response is required for amino acid and nitrate utilization, nod factor regulation, nodulation, and nitrogen fixation in Rhizobium etli. J. Bacteriol. 2005, 187, 5075–5083. [Google Scholar] [CrossRef] [PubMed]
- Chatterji, D.; Kumar Ojha, A. Revisiting the stringent response, ppGpp and starvation signaling. Curr. Opin. Microbiol. 2001, 4, 160–165. [Google Scholar] [CrossRef]
- Moris, M.; Braeken, K.; Schoeters, E.; Verreth, C.; Beullens, S.; Vanderleyden, J.; Michiels, J. Effective symbiosis between Rhizobium etli and Phaseolus vulgaris requires the alarmone ppGpp. J. Bacteriol. 2005, 187, 5460–5469. [Google Scholar] [CrossRef] [PubMed]
- Wells, D.H.; Long, S.R. Mutations in rpoBC suppress the defects of a Sinorhizobium meliloti relA mutant. J. Bacteriol. 2003, 185, 5602–5610. [Google Scholar] [CrossRef] [PubMed]
- Hubber, A.; Vergunst, A.C.; Sullivan, J.T.; Hooykaas, P.J.J.; Ronson, C.W. Symbiotic phenotypes and translocated effector proteins of the Mesorhizobium loti strain R7A VirB/D4 type IV secretion system. Mol. Microbiol. 2004, 54, 561–574. [Google Scholar] [CrossRef] [PubMed]
- Kelly, S.J.; Muszyński, A.; Kawaharada, Y.; Hubber, A.M.; Sullivan, J.T.; Sandal, N.; Carlson, R.W.; Stougaard, J.; Ronson, C.W. Conditional Requirement for Exopolysaccharide in the Mesorhizobium-Lotus Symbiosis. Mol. Plant Microbe Interact. 2013, 26, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Price, P.A.; Tanner, H.R.; Dillon, B.A.; Shabab, M.; Walker, G.C.; Griffitts, J.S. Rhizobial peptidase HrrP cleaves host-encoded signaling peptides and mediates symbiotic compatibility. Proc. Natl. Acad. Sci. USA 2015. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.-J.; Zeng, Y.; Xie, Z.-P.; Staehelin, C. Symbiosis-Promoting and Deleterious Effects of NopT, a Novel Type 3 Effector of Rhizobium sp. Strain NGR234. J. Bacteriol. 2008, 190, 5101–5110. [Google Scholar] [CrossRef] [PubMed]
- Tsukui, T.; Eda, S.; Kaneko, T.; Sato, S.; Okazaki, S.; Kakizaki-Chiba, K.; Itakura, M.; Mitsui, H.; Yamashita, A.; Terasawa, K.; et al. The type III secretion system of Bradyrhizobium japonicum USDA122 mediates symbiotic incompatibility with Rj2 soybean plants. Appl. Environ. Microbiol. 2013, 79, 1048–1051. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, M.; Friedrich, L.; Göttfert, M.; Zehner, S. The type III-secreted protein NopE1 affects symbiosis and exhibits a calcium-dependent autocleavage activity. Mol. Plant Microbe Interact. 2010, 23, 124–129. [Google Scholar] [CrossRef] [PubMed]



| Strain, Plasmid or Oligonucleotide | Characteristics or Sequence a | Reference or Source |
|---|---|---|
| Bacterial strains | ||
| Bradyrhizobium elkanii | ||
| USDA61 | Wild-type strain, Polr | Keyser b |
| BErhcJ | USDA61 derivative harboring insertion in rhcJ region, Polr, Kmr, Tcr | [12] |
| BE5 | Tn5 mutant of USDA61, Polr, Kmr | This study |
| BE53 | Tn5 mutant of USDA61, Polr, Kmr | This study |
| BE85 | Tn5 mutant of USDA61, Polr, Kmr | This study |
| BE103 | Tn5 mutant of USDA61, Polr, Kmr | This study |
| BE168 | Tn5 mutant of USDA61, Polr, Kmr | This study |
| BE53S | USDA61 derivative containing an insertion of plasmid pSUPSCAKm:int innB, Polr, Kmr | This study |
| Escherichia coli | ||
| HB101 | recA, hsdR, hsdM, pro, Smr | Invitrogen, Carlsbad, CA, USA |
| S17-1 | pro recA RP4-2(Tcs:Mu) (Kms:Tn7); Mob+ | [20] |
| DH10B | Cloning strain | Invitrogen |
| Plasmids | ||
| pUTKm | Transposon delivery vector; Apr, Kmr | [21] |
| pRK2013 | Helper plasmid, ColE1 replicon carrying RK2 transfer genes; Kmr, tra | [22] |
| pSUPSCAKm | Derivative of pSUPPOL2SCA [23] with a kanamycin resistance gene in the DraI site, oriT of RP4, Tcr, Kmr | This study |
| pSUPSCAKm:int innB | pSUPSCAKm carrying a 0.5-kb DNA fragment containing internal sequence of innB, Kmr | This study |
| Oligonucleotides | ||
| Linker 1 | 5′-TTTCTGCTCGAATTCAAGCTTCTAACGATGTACGGGGACACATG-3′ | [24] |
| Linker 2 | 5′-TGTCCCCGTACATCGTTAGAACTACTCGTACCATCCACAT-3′ | [24] |
| Y-linker primer | 5′-CTGCTCGAATTCAAGCTTCT-3′ | [24] |
| Tn5 primer | 5′-GGCCAGATCTGATCAAGAGA-3′ | [24] |
| Psuppol-F | 5′-ATAAACCAGCCAGCCGGAA-3′ | This study |
| Psuppol-R | 5′-TTCTGACAACGATCGGAGGA-3′ | This study |
| BE5-F | 5′-TCATGCAGGTGAATGTCGAT-3′ | This study |
| BE5-R | 5′-CTATCCGCAGGAGTTGAACG-3′ | This study |
| BE53-F | 5′-AGATTGATGTTGCCGAGGAC-3′ | This study |
| BE53-R | 5′-TGAAAAAGCTCCGTGAGGTC -3′ | This study |
| BE85-F | 5′-GCGCGGATATTGACATTGAT-3′ | This study |
| BE85-R | 5′-AGGCCGTCGATCTCTATCAC-3′ | This study |
| BE103-F | 5′-ACAAGAAGATGTCGGCCAAG-3′ | This study |
| BE103-R | 5′-TGCTCGCAGAATACAACTGC-3′ | This study |
| BE168-F | 5′-TCGAAAGCGCACTAGATTGA-3′ | This study |
| BE168-R | 5′-AGCCGTAAATATCGGACAGC-3′ | This study |
| InnBXbaI-intF | 5′-CGGTGGCGGCGGCCGCTCTAGAAAAATGCGCAACTGGAAGAT-3′ | This study |
| InnBEcoRI-intR | 5′-CGATAAGCTTGATATCGAATTCATCTGCTCACCAAGCCAATC-3′ | This study |
| Gene with Tn5 Insertion | ||||||
|---|---|---|---|---|---|---|
| Strains | Nodulation on Rj4 Soybean a | Designated Symbol/GenBank Accession No. b | Length (bp) | Deduced Gene Products | tts Box c | Strain and Locus Tag/Gene (GenBank Accession No., % Identity by BLASTP Analysis) |
| BE5 | + | innA/KX499540 | 1212 | Cytosine deaminase | - |
|
| BE53 | - | innB/KX499541 | 2280 | Hypothetical protein | + |
|
| BE85 | + | innC/KX499542 | 504 | TerB family tellurite resistance protein | - |
|
| BE103 | + | innD/KX499543 | 1089 | ABC transporter substrate-binding protein | - |
|
| BE168 | + | innE/KX499544 | 2292 | GTP pyrophosphokinase | - |
|
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, H.P.; Miwa, H.; Kaneko, T.; Sato, S.; Okazaki, S. Identification of Bradyrhizobium elkanii Genes Involved in Incompatibility with Vigna radiata. Genes 2017, 8, 374. https://doi.org/10.3390/genes8120374
Nguyen HP, Miwa H, Kaneko T, Sato S, Okazaki S. Identification of Bradyrhizobium elkanii Genes Involved in Incompatibility with Vigna radiata. Genes. 2017; 8(12):374. https://doi.org/10.3390/genes8120374
Chicago/Turabian StyleNguyen, Hien P., Hiroki Miwa, Takakazu Kaneko, Shusei Sato, and Shin Okazaki. 2017. "Identification of Bradyrhizobium elkanii Genes Involved in Incompatibility with Vigna radiata" Genes 8, no. 12: 374. https://doi.org/10.3390/genes8120374
APA StyleNguyen, H. P., Miwa, H., Kaneko, T., Sato, S., & Okazaki, S. (2017). Identification of Bradyrhizobium elkanii Genes Involved in Incompatibility with Vigna radiata. Genes, 8(12), 374. https://doi.org/10.3390/genes8120374