MYC Modulation around the CDK2/p27/SKP2 Axis
Abstract
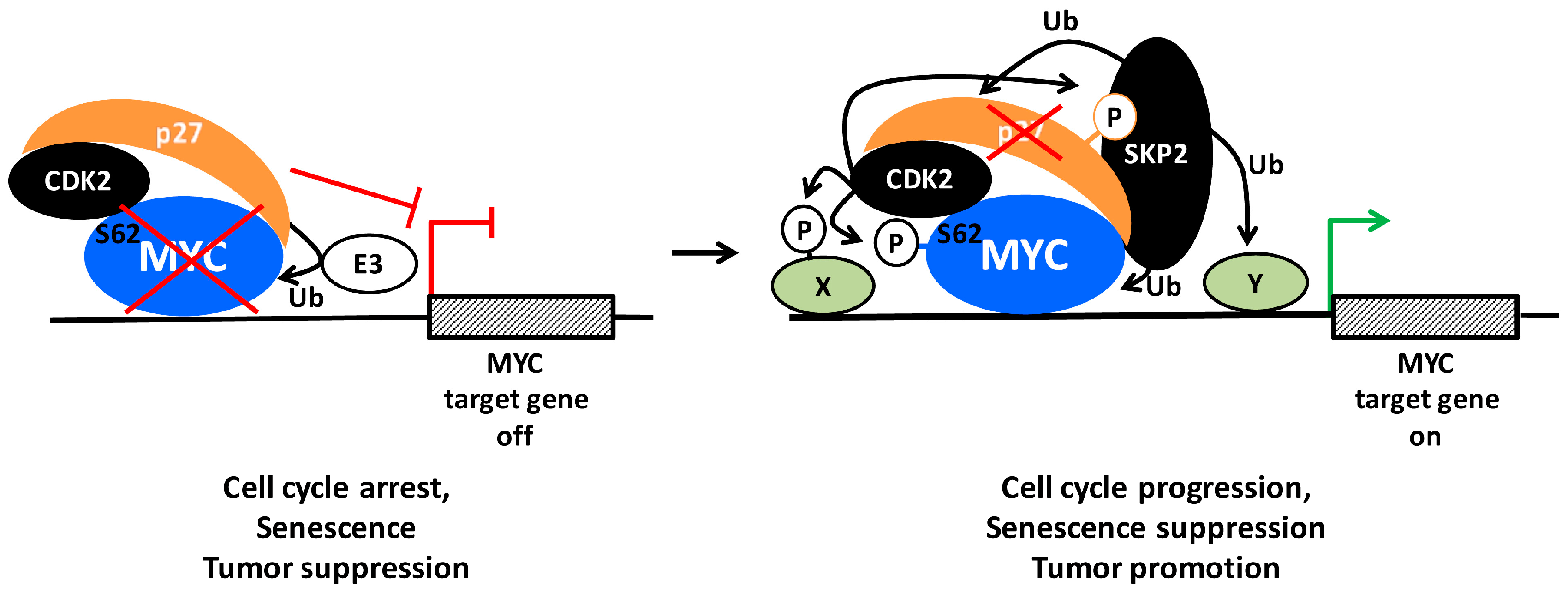
:1. Introduction
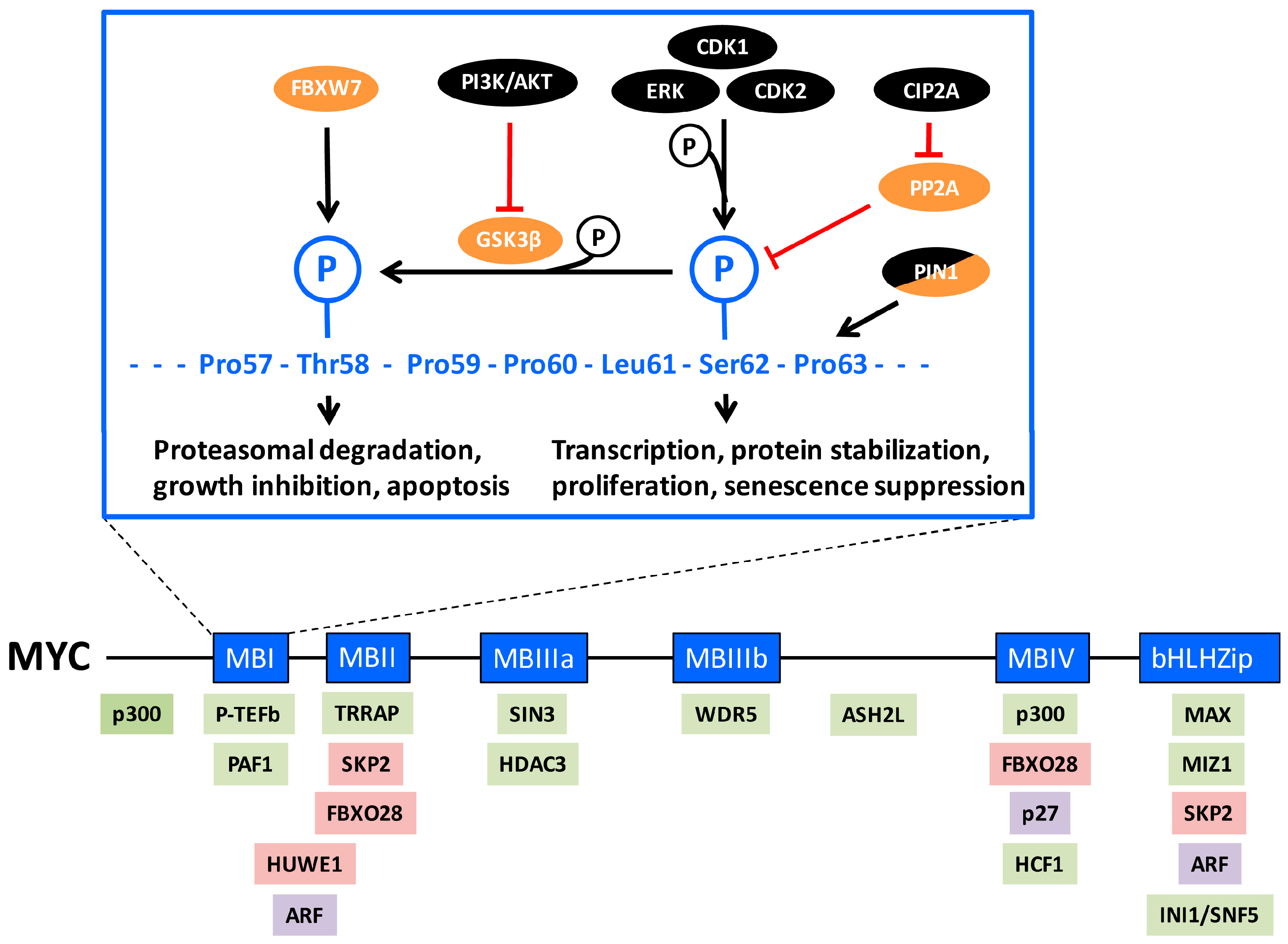
2. MYC Regulation by Phosphorylations in MYC-Box 1
3. Role of Ser-62 Phosphorylation in the Regulation of MYC’s Biological Activity
4. Role of Ser-62 Phosphorylation in Stabilization of MYC
5. Role of Ser-62 Phosphorylation in MYC-Regulated Transcription
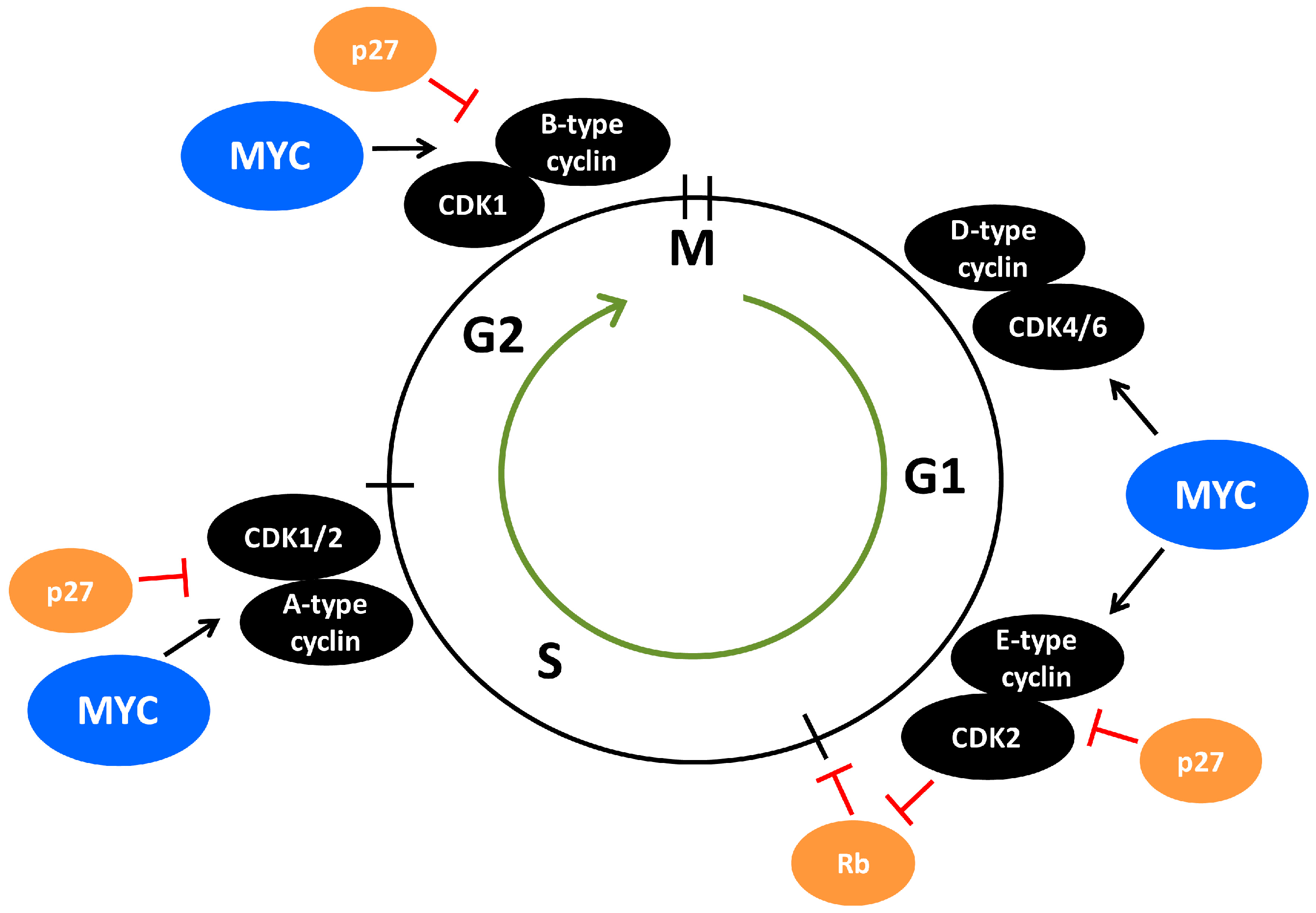
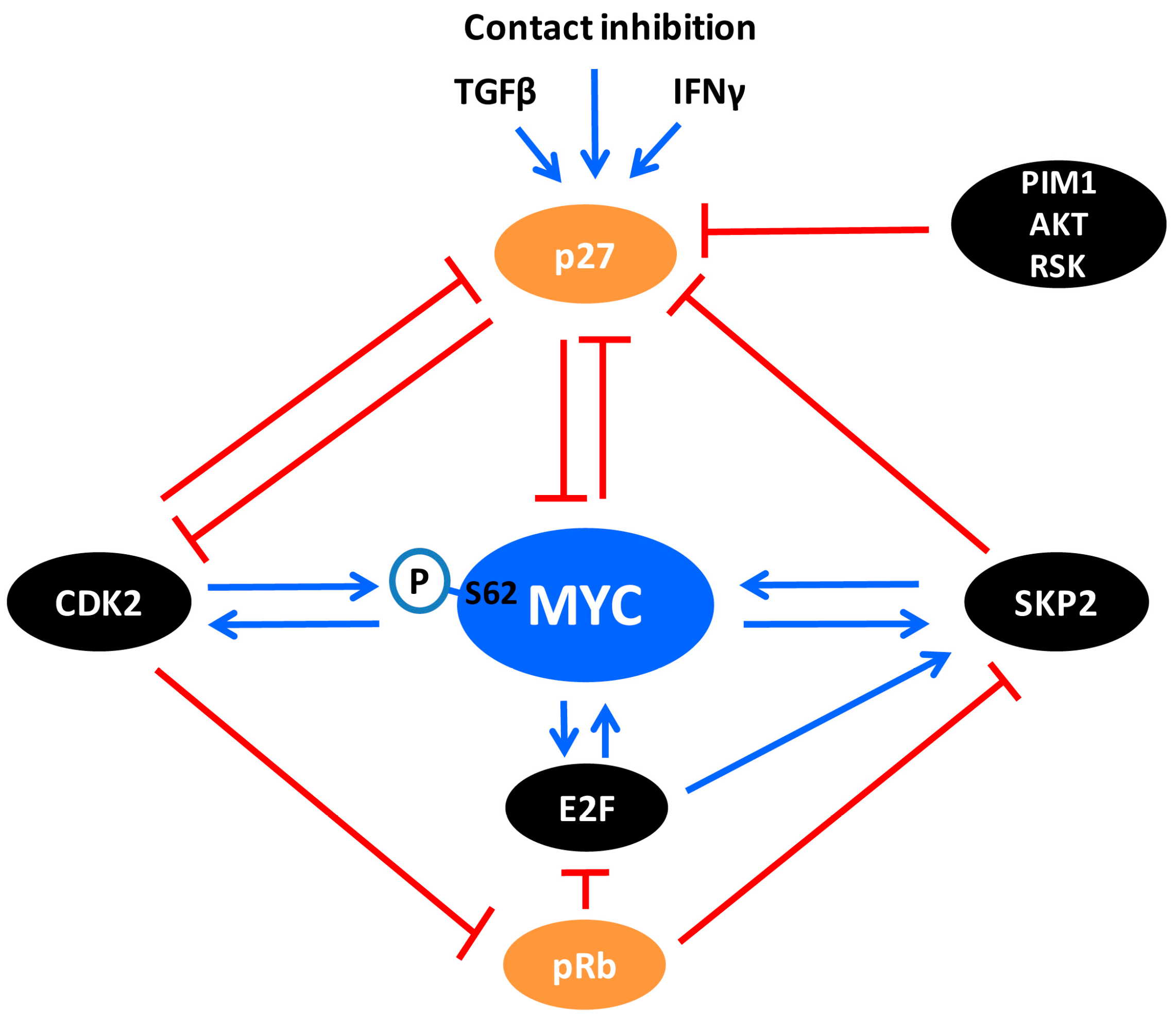
6. The Crosstalk between MYC and CDK2
7. The CDK2/p27/SKP2 Triangle
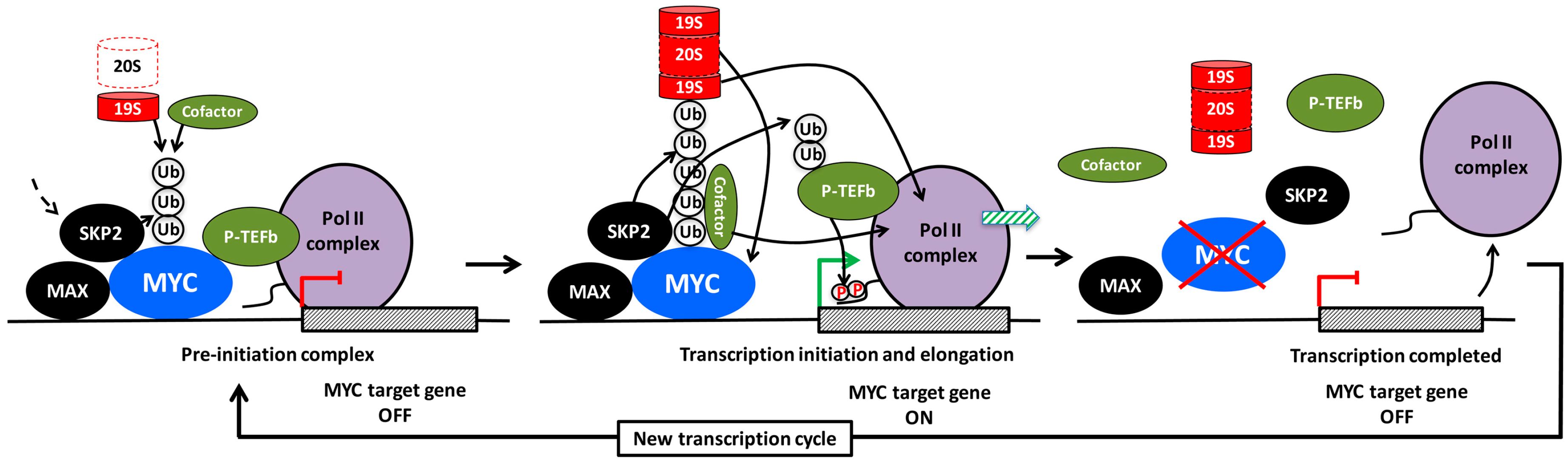
8. The Crosstalk between MYC and SKP2
9. The Role of Ubiquitylation for MYC-Driven Transcription
10. The Crosstalk between MYC and p27
11. Targeting the MYC/CDK2/SKP2/p27 Axis in Cancer
12. Concluding Remarks and Future Perspectives
Acknowledgments
Conflicts of Interest
References
- Blackwood, E.M.; Eisenman, R.N. Max: A helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with myc. Science 1991, 251, 1211–1217. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V. MYC on the path to cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Eilers, M.; Eisenman, R.N. MYC’s broad reach. Genes Dev. 2008, 22, 2755–2766. [Google Scholar] [CrossRef] [PubMed]
- Larsson, L.G.; Henriksson, M.A. The yin and yang functions of the MYC oncoprotein in cancer development and as targets for therapy. Exp. Cell Res. 2010, 316, 1429–1437. [Google Scholar] [CrossRef] [PubMed]
- Meyer, N.; Penn, L.Z. Reflecting on 25 years with MYC. Nat. Rev. Cancer 2008, 8, 976–990. [Google Scholar] [CrossRef] [PubMed]
- Tu, W.B.; Helander, S.; Pilstal, R.; Hickman, K.A.; Lourenco, C.; Jurisica, I.; Raught, B.; Wallner, B.; Sunnerhagen, M.; Penn, L.Z. MYC and its interactors take shape. Biochim. Biophys. Acta 2015, 1849, 469–483. [Google Scholar] [CrossRef] [PubMed]
- McMahon, S.B.; Van Buskirk, H.A.; Dugan, K.A.; Copeland, T.D.; Cole, M.D. The novel ATM-related protein TRRAP is an essential cofactor for the c-myc and e2f oncoproteins. Cell 1998, 94, 363–374. [Google Scholar] [CrossRef]
- Adhikary, S.; Marinoni, F.; Hock, A.; Hulleman, E.; Popov, N.; Beier, R.; Bernard, S.; Quarto, M.; Capra, M.; Goettig, S.; et al. The ubiquitin ligase hectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell 2005, 123, 409–421. [Google Scholar] [CrossRef] [PubMed]
- Cepeda, D.; Ng, H.F.; Sharifi, H.R.; Mahmoudi, S.; Cerrato, V.S.; Fredlund, E.; Magnusson, K.; Nilsson, H.; Malyukova, A.; Rantala, J.; et al. Cdk-mediated activation of the scf(fbxo) (28) ubiquitin ligase promotes myc-driven transcription and tumourigenesis and predicts poor survival in breast cancer. EMBO Mol. Med. 2013, 5, 1067–1086. [Google Scholar] [CrossRef] [PubMed]
- Vervoorts, J.; Luscher, B. Post-translational regulation of the tumor suppressor p27(kip1). Cell. Mol. Life Sci. 2008, 65, 3255–3264. [Google Scholar] [CrossRef] [PubMed]
- Menssen, A.; Hydbring, P.; Kapelle, K.; Vervoorts, J.; Diebold, J.; Luscher, B.; Larsson, L.G.; Hermeking, H. The c-myc oncoprotein, the nampt enzyme, the sirt1-inhibitor dbc1, and the sirt1 deacetylase form a positive feedback loop. Proc. Natl. Acad. Sci. USA 2012, 109, E187–E196. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Faiola, F.; Martinez, E. Six lysine residues on c-myc are direct substrates for acetylation by p300. Biochem. Biophys. Res. Commun. 2005, 336, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Carey, M.; Workman, J.L. The role of chromatin during transcription. Cell 2007, 128, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Thomas, L.R.; Wang, Q.; Grieb, B.C.; Phan, J.; Foshage, A.M.; Sun, Q.; Olejniczak, E.T.; Clark, T.; Dey, S.; Lorey, S.; et al. Interaction with wdr5 promotes target gene recognition and tumorigenesis by myc. Mol. Cell 2015, 58, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Ullius, A.; Luscher-Firzlaff, J.; Costa, I.G.; Walsemann, G.; Forst, A.H.; Gusmao, E.G.; Kapelle, K.; Kleine, H.; Kremmer, E.; Vervoorts, J.; et al. The interaction of myc with the trithorax protein ash2l promotes gene transcription by regulating h3k27 modification. Nucleic Acids Res. 2014, 42, 6901–6920. [Google Scholar] [CrossRef] [PubMed]
- Shilatifard, A. The compass family of histone H3K4 methylases: Mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 2012, 81, 65–95. [Google Scholar] [CrossRef] [PubMed]
- Eberhardy, S.R.; Farnham, P.J. Myc recruits p-tefb to mediate the final step in the transcriptional activation of the cad promoter. J. Biol. Chem. 2002, 277, 40156–40162. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, K.; Enokida, H.; Tachiwada, T.; Nishiyama, K.; Seki, N.; Nakagawa, M. Increased SKP2 and CKS1 gene expression contributes to the progression of human urothelial carcinoma. J. Urol. 2007, 178, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Rahl, P.B.; Lin, C.Y.; Seila, A.C.; Flynn, R.A.; McCuine, S.; Burge, C.B.; Sharp, P.A.; Young, R.A. C-myc regulates transcriptional pause release. Cell 2010, 141, 432–445. [Google Scholar] [CrossRef] [PubMed]
- Jaenicke, L.A.; von Eyss, B.; Carstensen, A.; Wolf, E.; Xu, W.; Greifenberg, A.K.; Geyer, M.; Eilers, M.; Popov, N. Ubiquitin-dependent turnover of myc antagonizes myc/paf1c complex accumulation to drive transcriptional elongation. Mol. Cell 2016, 61, 54–67. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.H.; Liang, Y.Y.; Liang, M.; Zhai, W.; Lin, X. Direct interaction of c-Myc with Smad2 and Smad3 to inhibit tgf-beta-mediated induction of the cdk inhibitor p15(ink4b). Mol. Cell 2002, 9, 133–143. [Google Scholar] [CrossRef]
- Gartel, A.L.; Ye, X.; Goufman, E.; Shianov, P.; Hay, N.; Najmabadi, F.; Tyner, A.L. Myc represses the p21(waf1/cip1) promoter and interacts with sp1/sp3. Proc. Natl. Acad. Sci. USA 2001, 98, 4510–4515. [Google Scholar] [CrossRef] [PubMed]
- Peukert, K.; Staller, P.; Schneider, A.; Carmichael, G.; Hanel, F.; Eilers, M. An alternative pathway for gene regulation by myc. EMBO J. 1997, 16, 5672–5686. [Google Scholar] [CrossRef] [PubMed]
- Seoane, J.; Pouponnot, C.; Staller, P.; Schader, M.; Eilers, M.; Massague, J. Tgfbeta influences myc, miz-1 and smad to control the cdk inhibitor p15ink4b. Nat. Cell Biol. 2001, 3, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Staller, P.; Peukert, K.; Kiermaier, A.; Seoane, J.; Lukas, J.; Karsunky, H.; Moroy, T.; Bartek, J.; Massague, J.; Hanel, F.; et al. Repression of p15ink4b expression by myc through association with miz-1. Nat. Cell Biol. 2001, 3, 392–399. [Google Scholar] [CrossRef] [PubMed]
- Walz, S.; Lorenzin, F.; Morton, J.; Wiese, K.E.; von Eyss, B.; Herold, S.; Rycak, L.; Dumay-Odelot, H.; Karim, S.; Bartkuhn, M.; et al. Activation and repression by oncogenic myc shape tumour-specific gene expression profiles. Nature 2014, 511, 483–487. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Cetinkaya, C.; Munoz-Alonso, M.J.; von der Lehr, N.; Bahram, F.; Beuger, V.; Eilers, M.; Leon, J.; Larsson, L.G. Myc represses differentiation-induced p21cip1 expression via miz-1-dependent interaction with the p21 core promoter. Oncogene 2003, 22, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Sanz, P.; Quintanilla, A.; Lafita, M.C.; Moreno-Bueno, G.; Garcia-Gutierrez, L.; Tabor, V.; Varela, I.; Shiio, Y.; Larsson, L.G.; Portillo, F.; et al. Sin3b interacts with myc and decreases myc levels. J. Biol. Chem. 2014, 289, 22221–22236. [Google Scholar] [CrossRef] [PubMed]
- Kurland, J.F.; Tansey, W.P. Myc-mediated transcriptional repression by recruitment of histone deacetylase. Cancer Res. 2008, 68, 3624–3629. [Google Scholar] [CrossRef] [PubMed]
- Kress, T.R.; Sabo, A.; Amati, B. Myc: Connecting selective transcriptional control to global RNA production. Nat. Rev. Cancer 2015, 15, 593–607. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.Y.; Loven, J.; Rahl, P.B.; Paranal, R.M.; Burge, C.B.; Bradner, J.E.; Lee, T.I.; Young, R.A. Transcriptional amplification in tumor cells with elevated c-myc. Cell 2012, 151, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Nie, Z.; Hu, G.; Wei, G.; Cui, K.; Yamane, A.; Resch, W.; Wang, R.; Green, D.R.; Tessarollo, L.; Casellas, R.; et al. C-myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 2012, 151, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Sabo, A.; Kress, T.R.; Pelizzola, M.; de Pretis, S.; Gorski, M.M.; Tesi, A.; Morelli, M.J.; Bora, P.; Doni, M.; Verrecchia, A.; et al. Selective transcriptional regulation by myc in cellular growth control and lymphomagenesis. Nature 2014, 511, 488–492. [Google Scholar] [CrossRef] [PubMed]
- Beroukhim, R.; Mermel, C.H.; Porter, D.; Wei, G.; Raychaudhuri, S.; Donovan, J.; Barretina, J.; Boehm, J.S.; Dobson, J.; Urashima, M.; et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010, 463, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Levens, D. You don’t muck with Myc. Genes Cancer 2010, 1, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Loven, J.; Hoke, H.A.; Lin, C.Y.; Lau, A.; Orlando, D.A.; Vakoc, C.R.; Bradner, J.E.; Lee, T.I.; Young, R.A. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013, 153, 320–334. [Google Scholar] [CrossRef] [PubMed]
- Sur, I.K.; Hallikas, O.; Vaharautio, A.; Yan, J.; Turunen, M.; Enge, M.; Taipale, M.; Karhu, A.; Aaltonen, L.A.; Taipale, J. Mice lacking a myc enhancer that includes human snp rs6983267 are resistant to intestinal tumors. Science 2012, 338, 1360–1363. [Google Scholar] [CrossRef] [PubMed]
- Wierstra, I.; Alves, J. The c-myc promoter: Still mystery and challenge. Adv. Cancer Res. 2008, 99, 113–333. [Google Scholar] [PubMed]
- Zhang, X.; Choi, P.S.; Francis, J.M.; Imielinski, M.; Watanabe, H.; Cherniack, A.D.; Meyerson, M. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat. Genet. 2016, 48, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Farrell, A.S.; Sears, R.C. Myc Degradation. Cold Spring Harb. Perspect. Med. 2014, 4, a014365. [Google Scholar] [CrossRef] [PubMed]
- Kalkat, M.; Chan, P.K.; Wasylishen, A.R.; Srikumar, T.; Kim, S.S.; Ponzielli, R.; Bazett-Jones, D.P.; Raught, B.; Penn, L.Z. Identification of c-myc sumoylation by mass spectrometry. PLoS ONE 2014, 9, e115337. [Google Scholar] [CrossRef] [PubMed]
- Luscher, B.; Vervoorts, J. Regulation of gene transcription by the oncoprotein myc. Gene 2012, 494, 145–160. [Google Scholar] [CrossRef] [PubMed]
- Sabo, A.; Doni, M.; Amati, B. Sumoylation of myc-family proteins. PLoS ONE 2014, 9, e91072. [Google Scholar] [CrossRef] [PubMed]
- Thomas, L.R.; Tansey, W.P. Proteolytic control of the oncoprotein transcription factor myc. Adv. Cancer Res. 2011, 110, 77–106. [Google Scholar] [PubMed]
- Bhat, M.; Robichaud, N.; Hulea, L.; Sonenberg, N.; Pelletier, J.; Topisirovic, I. Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 2015, 14, 261–278. [Google Scholar] [CrossRef] [PubMed]
- Castell, A.; Larsson, L.G. Targeting myc translation in colorectal cancer. Cancer Discov. 2015, 5, 701–703. [Google Scholar] [CrossRef] [PubMed]
- Wiegering, A.; Uthe, F.W.; Jamieson, T.; Ruoss, Y.; Huttenrauch, M.; Kuspert, M.; Pfann, C.; Nixon, C.; Herold, S.; Walz, S.; et al. Targeting translation initiation bypasses signaling crosstalk mechanisms that maintain high myc levels in colorectal cancer. Cancer Discov. 2015, 5, 768–781. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, A.L.; Singh, K.; Zhong, Y.; Drewe, P.; Rajasekhar, V.K.; Sanghvi, V.R.; Mavrakis, K.J.; Jiang, M.; Roderick, J.E.; Van der Meulen, J.; et al. Rna g-quadruplexes cause eif4a-dependent oncogene translation in cancer. Nature 2014, 513, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Hann, S.R. Role of post-translational modifications in regulating c-myc proteolysis, transcriptional activity and biological function. Semin. Cancer Biol. 2006, 16, 288–302. [Google Scholar] [CrossRef] [PubMed]
- Benassi, B.; Fanciulli, M.; Fiorentino, F.; Porrello, A.; Chiorino, G.; Loda, M.; Zupi, G.; Biroccio, A. C-myc phosphorylation is required for cellular response to oxidative stress. Mol. Cell 2006, 21, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Lutterbach, B.; Hann, S.R. Hierarchical phosphorylation at N-terminal transformation-sensitive sites in c-myc protein is regulated by mitogens and in mitosis. Mol. Cell. Biol. 1994, 14, 5510–5522. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, K.; Kitanaka, C.; Yamana, H.; Kokubu, A.; Mochizuki, T.; Kuchino, Y. Regulation of c-myc through phosphorylation at Ser-62 and Ser-71 by c-Jun N-terminal kinase. J. Biol. Chem. 1999, 274, 32580–32587. [Google Scholar] [CrossRef] [PubMed]
- Sears, R.; Nuckolls, F.; Haura, E.; Taya, Y.; Tamai, K.; Nevins, J.R. Multiple ras-dependent phosphorylation pathways regulate myc protein stability. Genes Dev. 2000, 14, 2501–2514. [Google Scholar] [CrossRef] [PubMed]
- Seth, A.; Alvarez, E.; Gupta, S.; Davis, R.J. A phosphorylation site located in the NH2-terminal domain of c-myc increases transactivation of gene expression. J. Biol. Chem. 1991, 266, 23521–23524. [Google Scholar] [PubMed]
- Watnick, R.S.; Rodriguez, R.K.; Wang, S.; Blois, A.L.; Rangarajan, A.; Ince, T.; Weinberg, R.A. Thrombospondin-1 repression is mediated via distinct mechanisms in fibroblasts and epithelial cells. Oncogene 2015, 34, 2823–2835. [Google Scholar] [CrossRef] [PubMed]
- Yeh, E.; Cunningham, M.; Arnold, H.; Chasse, D.; Monteith, T.; Ivaldi, G.; Hahn, W.C.; Stukenberg, P.T.; Shenolikar, S.; Uchida, T.; et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat. Cell Biol. 2004, 6, 308–318. [Google Scholar] [CrossRef] [PubMed]
- Hoang, A.T.; Lutterbach, B.; Lewis, B.C.; Yano, T.; Chou, T.Y.; Barrett, J.F.; Raffeld, M.; Hann, S.R.; Dang, C.V. A link between increased transforming activity of lymphoma-derived myc mutant alleles, their defective regulation by p107, and altered phosphorylation of the c-Myc transactivation domain. Mol. Cell. Biol. 1995, 15, 4031–4042. [Google Scholar] [CrossRef] [PubMed]
- Hydbring, P.; Bahram, F.; Su, Y.; Tronnersjo, S.; Hogstrand, K.; von der Lehr, N.; Sharifi, H.R.; Lilischkis, R.; Hein, N.; Wu, S.; et al. Phosphorylation by Cdk2 is required for Myc to repress Ras-induced senescence in cotransformation. Proc. Natl. Acad. Sci. USA 2010, 107, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.R.; Kim, J.; Bae, S.; Soh, J.W.; Lee, Y.S. Cdk5-mediated phosphorylation of c-Myc on Ser-62 is essential in transcriptional activation of cyclin b1 by cyclin g1. J. Biol. Chem. 2008, 283, 15601–15610. [Google Scholar] [CrossRef] [PubMed]
- Sjostrom, S.K.; Finn, G.; Hahn, W.C.; Rowitch, D.H.; Kenney, A.M. The cdk1 complex plays a prime role in regulating n-myc phosphorylation and turnover in neural precursors. Dev. Cell 2005, 9, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, D.; Camarda, R.; Zhou, A.Y.; Yau, C.; Momcilovic, O.; Balakrishnan, S.; Corella, A.N.; Eyob, H.; Kessenbrock, K.; Lawson, D.A.; et al. Pim1 kinase inhibition as a targeted therapy against triple-negative breast tumors with elevated myc expression. Nat. Med. 2016, 22, 1321–1329. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Z.; Li, X.; Magnuson, N.S. Pim kinase-dependent inhibition of c-Myc degradation. Oncogene 2008, 27, 4809–4819. [Google Scholar] [CrossRef] [PubMed]
- Gregory, M.A.; Qi, Y.; Hann, S.R. Phosphorylation by glycogen synthase kinase-3 controls c-Myc proteolysis and subnuclear localization. J. Biol. Chem. 2003, 278, 51606–51612. [Google Scholar] [CrossRef] [PubMed]
- Welcker, M.; Orian, A.; Jin, J.; Grim, J.E.; Harper, J.W.; Eisenman, R.N.; Clurman, B.E. The fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. USA 2004, 101, 9085–9090. [Google Scholar] [CrossRef] [PubMed]
- Yada, M.; Hatakeyama, S.; Kamura, T.; Nishiyama, M.; Tsunematsu, R.; Imaki, H.; Ishida, N.; Okumura, F.; Nakayama, K.; Nakayama, K.I. Phosphorylation-dependent degradation of c-Myc is mediated by the f-box protein fbw7. EMBO J. 2004, 23, 2116–2125. [Google Scholar] [CrossRef] [PubMed]
- Cardozo, T.; Pagano, M. The scf ubiquitin ligase: Insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 2004, 5, 739–751. [Google Scholar] [CrossRef] [PubMed]
- Bahram, F.; von der Lehr, N.; Cetinkaya, C.; Larsson, L.G. C-myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood 2000, 95, 2104–2110. [Google Scholar] [PubMed]
- Gregory, M.A.; Hann, S.R. C-myc proteolysis by the ubiquitin-proteasome pathway: Stabilization of c-myc in burkitt’s lymphoma cells. Mol. Cell. Biol. 2000, 20, 2423–2435. [Google Scholar] [CrossRef] [PubMed]
- Hemann, M.T.; Bric, A.; Teruya-Feldstein, J.; Herbst, A.; Nilsson, J.A.; Cordon-Cardo, C.; Cleveland, J.L.; Tansey, W.P.; Lowe, S.W. Evasion of the p53 tumour surveillance network by tumour-derived Myc mutants. Nature 2005, 436, 807–811. [Google Scholar] [CrossRef] [PubMed]
- Salghetti, S.E.; Kim, S.Y.; Tansey, W.P. Destruction of myc by ubiquitin-mediated proteolysis: Cancer-associated and transforming mutations stabilize Myc. EMBO J. 1999, 18, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Akhoondi, S.; Sun, D.; von der Lehr, N.; Apostolidou, S.; Klotz, K.; Maljukova, A.; Cepeda, D.; Fiegl, H.; Dafou, D.; Marth, C.; et al. Fbxw7/hcdc4 is a general tumor suppressor in human cancer. Cancer Res. 2007, 67, 9006–9012. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.J.; Welcker, M.; Clurman, B.E. Tumor suppression by the Fbw7 ubiquitin ligase: Mechanisms and opportunities. Cancer Cell 2014, 26, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Manning, B.D.; Toker, A. Akt/pkb signaling: Navigating the network. Cell 2017, 169, 381–405. [Google Scholar] [CrossRef] [PubMed]
- Kenney, A.M.; Cole, M.D.; Rowitch, D.H. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development 2003, 130, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Chesler, L.; Schlieve, C.; Goldenberg, D.D.; Kenney, A.; Kim, G.; McMillan, A.; Matthay, K.K.; Rowitch, D.; Weiss, W.A. Inhibition of phosphatidylinositol 3-kinase destabilizes mycn protein and blocks malignant progression in neuroblastoma. Cancer Res. 2006, 66, 8139–8146. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Marques, M.; Carrera, A.C. Phosphoinositide 3-kinase activation in late G1 is required for c-Myc stabilization and s phase entry. Mol. Cell. Biol. 2006, 26, 9116–9125. [Google Scholar] [CrossRef] [PubMed]
- Arnold, H.K.; Zhang, X.; Daniel, C.J.; Tibbitts, D.; Escamilla-Powers, J.; Farrell, A.; Tokarz, S.; Morgan, C.; Sears, R.C. The axin1 scaffold protein promotes formation of a degradation complex for c-Myc. EMBO J. 2009, 28, 500–512. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Farrell, A.S.; Daniel, C.J.; Arnold, H.; Scanlan, C.; Laraway, B.J.; Janghorban, M.; Lum, L.; Chen, D.; Troxell, M.; et al. Mechanistic insight into myc stabilization in breast cancer involving aberrant axin1 expression. Proc. Natl. Acad. Sci. USA 2012, 109, 2790–2795. [Google Scholar] [CrossRef] [PubMed]
- Junttila, M.R.; Puustinen, P.; Niemela, M.; Ahola, R.; Arnold, H.; Bottzauw, T.; Ala-aho, R.; Nielsen, C.; Ivaska, J.; Taya, Y.; et al. Cip2a inhibits pp2a in human malignancies. Cell 2007, 130, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Khanna, A.; Pimanda, J.E.; Westermarck, J. Cancerous inhibitor of protein phosphatase 2A, an emerging human oncoprotein and a potential cancer therapy target. Cancer Res. 2013, 73, 6548–6553. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.W.; Claassen, G.F.; Hann, S.R.; Cole, M.D. The c-Myc transactivation domain is a direct modulator of apoptotic versus proliferative signals. Mol. Cell. Biol. 2000, 20, 4309–4319. [Google Scholar] [CrossRef] [PubMed]
- Pulverer, B.J.; Fisher, C.; Vousden, K.; Littlewood, T.; Evan, G.; Woodgett, J.R. Site-specific modulation of c-Myc cotransformation by residues phosphorylated in vivo. Oncogene 1994, 9, 59–70. [Google Scholar] [PubMed]
- Henriksson, M.; Bakardjiev, A.; Klein, G.; Luscher, B. Phosphorylation sites mapping in the N-terminal domain of c-Myc modulate its transforming potential. Oncogene 1993, 8, 3199–3209. [Google Scholar] [PubMed]
- Wang, X.; Cunningham, M.; Zhang, X.; Tokarz, S.; Laraway, B.; Troxell, M.; Sears, R.C. Phosphorylation regulates c-myc’s oncogenic activity in the mammary gland. Cancer Res. 2011, 71, 925–936. [Google Scholar] [CrossRef] [PubMed]
- Myant, K.; Qiao, X.; Halonen, T.; Come, C.; Laine, A.; Janghorban, M.; Partanen, J.I.; Cassidy, J.; Ogg, E.L.; Cammareri, P.; et al. Serine 62-phosphorylated myc associates with nuclear lamins and its regulation by cip2a is essential for regenerative proliferation. Cell Rep. 2015, 12, 1019–1031. [Google Scholar] [CrossRef] [PubMed]
- Popov, N.; Herold, S.; Llamazares, M.; Schulein, C.; Eilers, M. Fbw7 and usp28 regulate myc protein stability in response to DNA damage. Cell Cycle 2007, 6, 2327–2331. [Google Scholar] [CrossRef] [PubMed]
- Chou, T.Y.; Hart, G.W.; Dang, C.V. C-myc is glycosylated at threonine 58, a known phosphorylation site and a mutational hot spot in lymphomas. J. Biol. Chem. 1995, 270, 18961–18965. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Eisenman, R.N. Regulation of c-myc protein abundance by a protein phosphatase 2a-glycogen synthase kinase 3beta-negative feedback pathway. Genes Cancer 2012, 3, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Seth, A.; Gupta, S.; Davis, R.J. Cell cycle regulation of the c-myc transcriptional activation domain. Mol. Cell. Biol. 1993, 13, 4125–4136. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Seth, A.; Davis, R.J. Transactivation of gene expression by myc is inhibited by mutation at the phosphorylation sites thr-58 and ser-62. Proc. Natl. Acad. Sci. USA 1993, 90, 3216–3220. [Google Scholar] [CrossRef] [PubMed]
- Farrell, A.S.; Pelz, C.; Wang, X.; Daniel, C.J.; Wang, Z.; Su, Y.; Janghorban, M.; Zhang, X.; Morgan, C.; Impey, S.; et al. Pin1 regulates the dynamics of c-myc DNA binding to facilitate target gene regulation and oncogenesis. Mol. Cell. Biol. 2013, 33, 2930–2949. [Google Scholar] [CrossRef] [PubMed]
- Pineda-Lucena, A.; Ho, C.S.; Mao, D.Y.; Sheng, Y.; Laister, R.C.; Muhandiram, R.; Lu, Y.; Seet, B.T.; Katz, S.; Szyperski, T.; et al. A structure-based model of the c-myc/bin1 protein interaction shows alternative splicing of bin1 and c-myc phosphorylation are key binding determinants. J. Mol. Biol. 2005, 351, 182–194. [Google Scholar] [CrossRef] [PubMed]
- Elliott, K.; Sakamuro, D.; Basu, A.; Du, W.; Wunner, W.; Staller, P.; Gaubatz, S.; Zhang, H.; Prochownik, E.; Eilers, M.; et al. Bin1 functionally interacts with myc and inhibits cell proliferation via multiple mechanisms. Oncogene 1999, 18, 3564–3573. [Google Scholar] [CrossRef] [PubMed]
- DuHadaway, J.B.; Sakamuro, D.; Ewert, D.L.; Prendergast, G.C. Bin1 mediates apoptosis by c-myc in transformed primary cells. Cancer Res. 2001, 61, 3151–3156. [Google Scholar] [PubMed]
- Pavri, R.; Zhu, B.; Li, G.; Trojer, P.; Mandal, S.; Shilatifard, A.; Reinberg, D. Histone h2b monoubiquitination functions cooperatively with fact to regulate elongation by RNA polymerase II. Cell 2006, 125, 703–717. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, C.; Marquardt, J.; Bras, A.; Medema, R.H.; Eilers, M. Myc-induced proliferation and transformation require akt-mediated phosphorylation of foxo proteins. EMBO J. 2004, 23, 2830–2840. [Google Scholar] [CrossRef] [PubMed]
- Rahl, P.B.; Young, R.A. Myc and transcription elongation. Cold Spring Harb. Perspect. Med. 2014, 4, a020990. [Google Scholar] [CrossRef] [PubMed]
- Kind, J.; Pagie, L.; Ortabozkoyun, H.; Boyle, S.; de Vries, S.S.; Janssen, H.; Amendola, M.; Nolen, L.D.; Bickmore, W.A.; van Steensel, B. Single-cell dynamics of genome-nuclear lamina interactions. Cell 2013, 153, 178–192. [Google Scholar] [CrossRef] [PubMed]
- Ashton, G.H.; Morton, J.P.; Myant, K.; Phesse, T.J.; Ridgway, R.A.; Marsh, V.; Wilkins, J.A.; Athineos, D.; Muncan, V.; Kemp, R.; et al. Focal adhesion kinase is required for intestinal regeneration and tumorigenesis downstream of wnt/c-myc signaling. Dev. Cell 2010, 19, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Athineos, D.; Sansom, O.J. Myc heterozygosity attenuates the phenotypes of apc deficiency in the small intestine. Oncogene 2010, 29, 2585–2590. [Google Scholar] [CrossRef] [PubMed]
- Hydbring, P.; Malumbres, M.; Sicinski, P. Non-canonical functions of cell cycle cyclins and cyclin-dependent kinases. Nat. Rev. Mol. Cell Biol. 2016, 17, 280–292. [Google Scholar] [CrossRef] [PubMed]
- Pelengaris, S.; Khan, M.; Evan, G. C-myc: More than just a matter of life and death. Nat. Rev. Cancer 2002, 2, 764–776. [Google Scholar] [CrossRef] [PubMed]
- Perez-Roger, I.; Solomon, D.L.; Sewing, A.; Land, H. Myc activation of cyclin e/cdk2 kinase involves induction of cyclin e gene transcription and inhibition of p27(kip1) binding to newly formed complexes. Oncogene 1997, 14, 2373–2381. [Google Scholar] [CrossRef] [PubMed]
- Prall, O.W.; Rogan, E.M.; Musgrove, E.A.; Watts, C.K.; Sutherland, R.L. C-myc or cyclin d1 mimics estrogen effects on cyclin e-cdk2 activation and cell cycle reentry. Mol. Cell. Biol. 1998, 18, 4499–4508. [Google Scholar] [CrossRef] [PubMed]
- Beier, R.; Burgin, A.; Kiermaier, A.; Fero, M.; Karsunky, H.; Saffrich, R.; Moroy, T.; Ansorge, W.; Roberts, J.; Eilers, M. Induction of cyclin e-cdk2 kinase activity, e2f-dependent transcription and cell growth by myc are genetically separable events. EMBO J. 2000, 19, 5813–5823. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, C.; Thieke, K.; Maier, A.; Saffrich, R.; Hanley-Hyde, J.; Ansorge, W.; Reed, S.; Sicinski, P.; Bartek, J.; Eilers, M. Direct induction of cyclin d2 by myc contributes to cell cycle progression and sequestration of p27. EMBO J. 1999, 18, 5321–5333. [Google Scholar] [CrossRef] [PubMed]
- Hermeking, H.; Rago, C.; Schuhmacher, M.; Li, Q.; Barrett, J.F.; Obaya, A.J.; O’Connell, B.C.; Mateyak, M.K.; Tam, W.; Kohlhuber, F.; et al. Identification of cdk4 as a target of c-myc. Proc. Natl. Acad. Sci. USA 2000, 97, 2229–2234. [Google Scholar] [CrossRef] [PubMed]
- Perez-Roger, I.; Kim, S.H.; Griffiths, B.; Sewing, A.; Land, H. Cyclins d1 and d2 mediate myc-induced proliferation via sequestration of p27(kip1) and p21(cip1). EMBO J. 1999, 18, 5310–5320. [Google Scholar] [CrossRef] [PubMed]
- Bretones, G.; Acosta, J.C.; Caraballo, J.M.; Ferrandiz, N.; Gomez-Casares, M.T.; Albajar, M.; Blanco, R.; Ruiz, P.; Hung, W.C.; Albero, M.P.; et al. Skp2 oncogene is a direct myc target gene and myc down-regulates p27(kip1) through skp2 in human leukemia cells. J. Biol. Chem. 2011, 286, 9815–9825. [Google Scholar] [CrossRef] [PubMed]
- Keller, U.B.; Old, J.B.; Dorsey, F.C.; Nilsson, J.A.; Nilsson, L.; MacLean, K.H.; Chung, L.; Yang, C.; Spruck, C.; Boyd, K.; et al. Myc targets cks1 to provoke the suppression of p27kip1, proliferation and lymphomagenesis. EMBO J. 2007, 26, 2562–2574. [Google Scholar] [CrossRef] [PubMed]
- O’Hagan, R.C.; Ohh, M.; David, G.; de Alboran, I.M.; Alt, F.W.; Kaelin, W.G., Jr.; DePinho, R.A. Myc-enhanced expression of cul1 promotes ubiquitin-dependent proteolysis and cell cycle progression. Genes Dev. 2000, 14, 2185–2191. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.; Welcker, M.; Hizli, A.A.; Posakony, J.J.; Aebersold, R.; Clurman, B.E. Identification of cdk2 substrates in human cell lysates. Genome Biol. 2008, 9, R149. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.C.; Clurman, B.E. Cyclin e in normal and neoplastic cell cycles. Oncogene 2005, 24, 2776–2786. [Google Scholar] [CrossRef] [PubMed]
- Odajima, J.; Saini, S.; Jung, P.; Ndassa-Colday, Y.; Ficaro, S.; Geng, Y.; Marco, E.; Michowski, W.; Wang, Y.E.; DeCaprio, J.A.; et al. Proteomic landscape of tissue-specific cyclin e functions in vivo. PLoS Genet. 2016, 12, e1006429. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Faha, B.; Dembski, M.; Tsai, L.H.; Harlow, E.; Dyson, N. Independent binding of the retinoblastoma protein and p107 to the transcription factor e2f. Nature 1992, 355, 176–179. [Google Scholar] [CrossRef] [PubMed]
- Classon, M.; Harlow, E. The retinoblastoma tumour suppressor in development and cancer. Nat. Rev. Cancer 2002, 2, 910–917. [Google Scholar] [CrossRef] [PubMed]
- Dyson, N.J. Rb1: A prototype tumor suppressor and an enigma. Genes Dev. 2016, 30, 1492–1502. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Eaton, E.N.; Picon, M.; Roberts, J.M.; Lundberg, A.S.; Gifford, A.; Sardet, C.; Weinberg, R.A. Regulation of cyclin e transcription by e2fs and retinoblastoma protein. Oncogene 1996, 12, 1173–1180. [Google Scholar] [PubMed]
- Knudsen, E.S.; Wang, J.Y. Differential regulation of retinoblastoma protein function by specific cdk phosphorylation sites. J. Biol. Chem. 1996, 271, 8313–8320. [Google Scholar] [CrossRef] [PubMed]
- Munro, S.; Carr, S.M.; La Thangue, N.B. Diversity within the prb pathway: Is there a code of conduct? Oncogene 2012, 31, 4343–4352. [Google Scholar] [CrossRef] [PubMed]
- Zarkowska, T.; Mittnacht, S. Differential phosphorylation of the retinoblastoma protein by g1/s cyclin-dependent kinases. J. Biol. Chem. 1997, 272, 12738–12746. [Google Scholar] [CrossRef] [PubMed]
- Montagnoli, A.; Fiore, F.; Eytan, E.; Carrano, A.C.; Draetta, G.F.; Hershko, A.; Pagano, M. Ubiquitination of p27 is regulated by cdk-dependent phosphorylation and trimeric complex formation. Genes Dev. 1999, 13, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Sheaff, R.J.; Groudine, M.; Gordon, M.; Roberts, J.M.; Clurman, B.E. Cyclin e-cdk2 is a regulator of p27kip1. Genes Dev. 1997, 11, 1464–1478. [Google Scholar] [CrossRef] [PubMed]
- Vlach, J.; Hennecke, S.; Amati, B. Phosphorylation-dependent degradation of the cyclin-dependent kinase inhibitor p27. EMBO J. 1997, 16, 5334–5344. [Google Scholar] [CrossRef] [PubMed]
- Bahram, F.; Hydbring, P.; Tronnersjo, S.; Zakaria, S.M.; Frings, O.; Fahlen, S.; Nilsson, H.; Goodwin, J.; von der Lehr, N.; Su, Y.; et al. Interferon-gamma-induced p27kip1 binds to and targets myc for proteasome-mediated degradation. Oncotarget 2016, 7, 2837–2854. [Google Scholar] [PubMed]
- Malumbres, M.; Barbacid, M. Cell cycle, cdks and cancer: A changing paradigm. Nat. Rev. Cancer 2009, 9, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Ishimi, Y.; Komamura-Kohno, Y.; You, Z.; Omori, A.; Kitagawa, M. Inhibition of mcm4,6,7 helicase activity by phosphorylation with cyclin a/cdk2. J. Biol. Chem. 2000, 275, 16235–16241. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Wells, N.J.; Hunter, T. Multistep regulation of DNA replication by cdk phosphorylation of hscdc6. Proc. Natl. Acad. Sci. USA 1999, 96, 6193–6198. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Deng, M.; Wei, Q.; Liu, T.; Tong, X.; Ye, X. Phosphorylation of mcm3 protein by cyclin e/cyclin-dependent kinase 2 (cdk2) regulates its function in cell cycle. J. Biol. Chem. 2011, 286, 39776–39785. [Google Scholar] [CrossRef] [PubMed]
- Petersen, B.O.; Lukas, J.; Sorensen, C.S.; Bartek, J.; Helin, K. Phosphorylation of mammalian cdc6 by cyclin a/cdk2 regulates its subcellular localization. EMBO J. 1999, 18, 396–410. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Yu, Q.; Sicinska, E.; Das, M.; Schneider, J.E.; Bhattacharya, S.; Rideout, W.M.; Bronson, R.T.; Gardner, H.; Sicinski, P. Cyclin e ablation in the mouse. Cell 2003, 114, 431–443. [Google Scholar] [CrossRef]
- Chuang, L.C.; Teixeira, L.K.; Wohlschlegel, J.A.; Henze, M.; Yates, J.R.; Mendez, J.; Reed, S.I. Phosphorylation of mcm2 by cdc7 promotes pre-replication complex assembly during cell-cycle re-entry. Mol. Cell 2009, 35, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Sola, D.; Ying, C.Y.; Grandori, C.; Ruggiero, L.; Chen, B.; Li, M.; Galloway, D.A.; Gu, W.; Gautier, J.; Dalla-Favera, R. Non-transcriptional control of DNA replication by c-myc. Nature 2007, 448, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Campaner, S.; Doni, M.; Hydbring, P.; Verrecchia, A.; Bianchi, L.; Sardella, D.; Schleker, T.; Perna, D.; Tronnersjo, S.; Murga, M.; et al. Cdk2 suppresses cellular senescence induced by the c-myc oncogene. Nat. Cell Biol. 2010, 12, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Felsher, D.W.; Bishop, J.M. Transient excess of myc activity can elicit genomic instability and tumorigenesis. Proc. Natl. Acad. Sci. USA 1999, 96, 3940–3944. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Dang, C.V. C-myc overexpression uncouples DNA replication from mitosis. Mol. Cell. Biol. 1999, 19, 5339–5351. [Google Scholar] [CrossRef] [PubMed]
- Maya-Mendoza, A.; Ostrakova, J.; Kosar, M.; Hall, A.; Duskova, P.; Mistrik, M.; Merchut-Maya, J.M.; Hodny, Z.; Bartkova, J.; Christensen, C.; et al. Myc and ras oncogenes engage different energy metabolism programs and evoke distinct patterns of oxidative and DNA replication stress. Mol. Oncol. 2015, 9, 601–616. [Google Scholar] [CrossRef] [PubMed]
- Reimann, M.; Loddenkemper, C.; Rudolph, C.; Schildhauer, I.; Teichmann, B.; Stein, H.; Schlegelberger, B.; Dorken, B.; Schmitt, C.A. The myc-evoked DNA damage response accounts for treatment resistance in primary lymphomas in vivo. Blood 2007, 110, 2996–3004. [Google Scholar] [CrossRef] [PubMed]
- Robinson, K.; Asawachaicharn, N.; Galloway, D.A.; Grandori, C. C-myc accelerates s-phase and requires wrn to avoid replication stress. PLoS ONE 2009, 4, e5951. [Google Scholar] [CrossRef] [PubMed]
- Vafa, O.; Wade, M.; Kern, S.; Beeche, M.; Pandita, T.K.; Hampton, G.M.; Wahl, G.M. C-myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: A mechanism for oncogene-induced genetic instability. Mol. Cell 2002, 9, 1031–1044. [Google Scholar] [CrossRef]
- Branzei, D.; Foiani, M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008, 9, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Buis, J.; Stoneham, T.; Spehalski, E.; Ferguson, D.O. Mre11 regulates ctip-dependent double-strand break repair by interaction with cdk2. Nat. Struct. Mol. Biol. 2012, 19, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Deans, A.J.; Khanna, K.K.; McNees, C.J.; Mercurio, C.; Heierhorst, J.; McArthur, G.A. Cyclin-dependent kinase 2 functions in normal DNA repair and is a therapeutic target in brca1-deficient cancers. Cancer Res. 2006, 66, 8219–8226. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Regan, K.M.; Lou, Z.; Chen, J.; Tindall, D.J. Cdk2-dependent phosphorylation of foxo1 as an apoptotic response to DNA damage. Science 2006, 314, 294–297. [Google Scholar] [CrossRef] [PubMed]
- Huertas, P.; Jackson, S.P. Human ctip mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 2009, 284, 9558–9565. [Google Scholar] [CrossRef] [PubMed]
- Polato, F.; Callen, E.; Wong, N.; Faryabi, R.; Bunting, S.; Chen, H.T.; Kozak, M.; Kruhlak, M.J.; Reczek, C.R.; Lee, W.H.; et al. Ctip-mediated resection is essential for viability and can operate independently of brca1. J. Exp. Med. 2014, 211, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Chen, J. DNA damage-induced cell cycle checkpoint control requires ctip, a phosphorylation-dependent binding partner of brca1 c-terminal domains. Mol. Cell. Biol. 2004, 24, 9478–9486. [Google Scholar] [CrossRef] [PubMed]
- Kuilman, T.; Michaloglou, C.; Mooi, W.J.; Peeper, D.S. The essence of senescence. Genes Dev. 2010, 24, 2463–2479. [Google Scholar] [CrossRef] [PubMed]
- Larsson, L.G. Oncogene- and tumor suppressor gene-mediated suppression of cellular senescence. Semin. Cancer Biol. 2011, 21, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Espin, D.; Serrano, M. Cellular senescence: From physiology to pathology. Nat. Rev. Mol. Cell Biol. 2014, 15, 482–496. [Google Scholar] [CrossRef] [PubMed]
- Luoto, K.R.; Meng, A.X.; Wasylishen, A.R.; Zhao, H.; Coackley, C.L.; Penn, L.Z.; Bristow, R.G. Tumor cell kill by c-myc depletion: Role of myc-regulated genes that control DNA double-strand break repair. Cancer Res. 2010, 70, 8748–8759. [Google Scholar] [CrossRef] [PubMed]
- Guerra, L.; Albihn, A.; Tronnersjo, S.; Yan, Q.; Guidi, R.; Stenerlow, B.; Sterzenbach, T.; Josenhans, C.; Fox, J.G.; Schauer, D.B.; et al. Myc is required for activation of the atm-dependent checkpoints in response to DNA damage. PLoS ONE 2010, 5, e8924. [Google Scholar] [CrossRef] [PubMed]
- Sherr, C.J.; Roberts, J.M. Cdk inhibitors: Positive and negative regulators of g1-phase progression. Genes Dev. 1999, 13, 1501–1512. [Google Scholar] [CrossRef] [PubMed]
- Blain, S.W.; Montalvo, E.; Massague, J. Differential interaction of the cyclin-dependent kinase (cdk) inhibitor p27kip1 with cyclin a-cdk2 and cyclin d2-cdk4. J. Biol. Chem. 1997, 272, 25863–25872. [Google Scholar] [CrossRef] [PubMed]
- Polyak, K.; Lee, M.H.; Erdjument-Bromage, H.; Koff, A.; Roberts, J.M.; Tempst, P.; Massague, J. Cloning of p27kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell 1994, 78, 59–66. [Google Scholar] [CrossRef]
- Polyak, K.; Kato, J.Y.; Solomon, M.J.; Sherr, C.J.; Massague, J.; Roberts, J.M.; Koff, A. P27kip1, a cyclin-cdk inhibitor, links transforming growth factor-beta and contact inhibition to cell cycle arrest. Genes Dev. 1994, 8, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Toyoshima, H.; Hunter, T. P27, a novel inhibitor of g1 cyclin-cdk protein kinase activity, is related to p21. Cell 1994, 78, 67–74. [Google Scholar] [CrossRef]
- Chu, I.M.; Hengst, L.; Slingerland, J.M. The cdk inhibitor p27 in human cancer: Prognostic potential and relevance to anticancer therapy. Nat. Rev. Cancer 2008, 8, 253–267. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Hunter, T. Ubiquitylation and proteasomal degradation of the p21(cip1), p27(kip1) and p57(kip2) cdk inhibitors. Cell Cycle 2010, 9, 2342–2352. [Google Scholar] [CrossRef] [PubMed]
- Slingerland, J.; Pagano, M. Regulation of the cdk inhibitor p27 and its deregulation in cancer. J. Cell. Physiol. 2000, 183, 10–17. [Google Scholar] [CrossRef]
- Carrano, A.C.; Eytan, E.; Hershko, A.; Pagano, M. Skp2 is required for ubiquitin-mediated degradation of the cdk inhibitor p27. Nat. Cell Biol. 1999, 1, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Sutterluty, H.; Chatelain, E.; Marti, A.; Wirbelauer, C.; Senften, M.; Muller, U.; Krek, W. P45skp2 promotes p27kip1 degradation and induces s phase in quiescent cells. Nat. Cell Biol. 1999, 1, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Kobayashi, R.; Galaktionov, K.; Beach, D. P19skp1 and p45skp2 are essential elements of the cyclin a-cdk2 s phase kinase. Cell 1995, 82, 915–925. [Google Scholar] [CrossRef]
- Lisztwan, J.; Marti, A.; Sutterluty, H.; Gstaiger, M.; Wirbelauer, C.; Krek, W. Association of human cul-1 and ubiquitin-conjugating enzyme cdc34 with the f-box protein p45(skp2): Evidence for evolutionary conservation in the subunit composition of the cdc34-scf pathway. EMBO J. 1998, 17, 368–383. [Google Scholar] [CrossRef] [PubMed]
- Lyapina, S.A.; Correll, C.C.; Kipreos, E.T.; Deshaies, R.J. Human cul1 forms an evolutionarily conserved ubiquitin ligase complex (scf) with skp1 and an f-box protein. Proc. Natl. Acad. Sci. USA 1998, 95, 7451–7456. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.K.; Gervais, J.L.; Zhang, H. Human cul-1 associates with the skp1/skp2 complex and regulates p21(cip1/waf1) and cyclin d proteins. Proc. Natl. Acad. Sci. USA 1998, 95, 11324–11329. [Google Scholar] [CrossRef] [PubMed]
- Frescas, D.; Pagano, M. Deregulated proteolysis by the f-box proteins skp2 and beta-trcp: Tipping the scales of cancer. Nat. Rev. Cancer 2008, 8, 438–449. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Regan, K.M.; Wang, F.; Wang, D.; Smith, D.I.; van Deursen, J.M.; Tindall, D.J. Skp2 inhibits foxo1 in tumor suppression through ubiquitin-mediated degradation. Proc. Natl. Acad. Sci. USA 2005, 102, 1649–1654. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.W.; Li, C.F.; Jin, G.; Cai, Z.; Han, F.; Chan, C.H.; Yang, W.L.; Li, B.K.; Rezaeian, A.H.; Li, H.Y.; et al. Skp2-dependent ubiquitination and activation of lkb1 is essential for cancer cell survival under energy stress. Mol. Cell 2015, 57, 1022–1033. [Google Scholar] [CrossRef] [PubMed]
- Jin, G.; Lee, S.W.; Zhang, X.; Cai, Z.; Gao, Y.; Chou, P.C.; Rezaeian, A.H.; Han, F.; Wang, C.Y.; Yao, J.C.; et al. Skp2-mediated raga ubiquitination elicits a negative feedback to prevent amino-acid-dependent mtorc1 hyperactivation by recruiting gator1. Mol. Cell 2015, 58, 989–1000. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhang, X.; Zhang, L.; Wu, C.Y.; Rezaeian, A.H.; Chan, C.H.; Li, J.M.; Wang, J.; Gao, Y.; Han, F.; et al. Skp2 e3 ligase integrates atm activation and homologous recombination repair by ubiquitinating nbs1. Mol. Cell 2012, 46, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Bauzon, F.; Fu, H.; Lu, Z.; Cui, J.; Nakayama, K.; Nakayama, K.I.; Locker, J.; Zhu, L. Skp2 deletion unmasks a p27 safeguard that blocks tumorigenesis in the absence of prb and p53 tumor suppressors. Cancer Cell 2013, 24, 645–659. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, K.; Nagahama, H.; Minamishima, Y.A.; Miyake, S.; Ishida, N.; Hatakeyama, S.; Kitagawa, M.; Iemura, S.; Natsume, T.; Nakayama, K.I. Skp2-mediated degradation of p27 regulates progression into mitosis. Dev. Cell 2004, 6, 661–672. [Google Scholar] [CrossRef]
- Kossatz, U.; Dietrich, N.; Zender, L.; Buer, J.; Manns, M.P.; Malek, N.P. Skp2-dependent degradation of p27kip1 is essential for cell cycle progression. Genes Dev. 2004, 18, 2602–2607. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, K.; Nagahama, H.; Minamishima, Y.A.; Matsumoto, M.; Nakamichi, I.; Kitagawa, K.; Shirane, M.; Tsunematsu, R.; Tsukiyama, T.; Ishida, N.; et al. Targeted disruption of skp2 results in accumulation of cyclin e and p27(kip1), polyploidy and centrosome overduplication. EMBO J. 2000, 19, 2069–2081. [Google Scholar] [CrossRef] [PubMed]
- Aleem, E.; Kiyokawa, H.; Kaldis, P. Cdc2-cyclin e complexes regulate the g1/s phase transition. Nat. Cell Biol. 2005, 7, 831–836. [Google Scholar] [CrossRef] [PubMed]
- Signoretti, S.; Di Marcotullio, L.; Richardson, A.; Ramaswamy, S.; Isaac, B.; Rue, M.; Monti, F.; Loda, M.; Pagano, M. Oncogenic role of the ubiquitin ligase subunit skp2 in human breast cancer. J. Clin. Investig. 2002, 110, 633–641. [Google Scholar] [CrossRef] [PubMed]
- Gstaiger, M.; Jordan, R.; Lim, M.; Catzavelos, C.; Mestan, J.; Slingerland, J.; Krek, W. Skp2 is oncogenic and overexpressed in human cancers. Proc. Natl. Acad. Sci. USA 2001, 98, 5043–5048. [Google Scholar] [CrossRef] [PubMed]
- Chiappetta, G.; De Marco, C.; Quintiero, A.; Califano, D.; Gherardi, S.; Malanga, D.; Scrima, M.; Montero-Conde, C.; Cito, L.; Monaco, M.; et al. Overexpression of the s-phase kinase-associated protein 2 in thyroid cancer. Endocr. Relat. Cancer 2007, 14, 405–420. [Google Scholar] [CrossRef] [PubMed]
- Slotky, M.; Shapira, M.; Ben-Izhak, O.; Linn, S.; Futerman, B.; Tsalic, M.; Hershko, D.D. The expression of the ubiquitin ligase subunit cks1 in human breast cancer. Breast Cancer Res. 2005, 7, R737–R744. [Google Scholar] [CrossRef] [PubMed]
- Ganoth, D.; Bornstein, G.; Ko, T.K.; Larsen, B.; Tyers, M.; Pagano, M.; Hershko, A. The cell-cycle regulatory protein cks1 is required for scf(skp2)-mediated ubiquitinylation of p27. Nat. Cell Biol. 2001, 3, 321–324. [Google Scholar] [CrossRef] [PubMed]
- Spruck, C.; Strohmaier, H.; Watson, M.; Smith, A.P.; Ryan, A.; Krek, T.W.; Reed, S.I. A cdk-independent function of mammalian cks1: Targeting of scf(skp2) to the cdk inhibitor p27kip1. Mol. Cell 2001, 7, 639–650. [Google Scholar] [CrossRef]
- Bondar, T.; Kalinina, A.; Khair, L.; Kopanja, D.; Nag, A.; Bagchi, S.; Raychaudhuri, P. Cul4a and ddb1 associate with skp2 to target p27kip1 for proteolysis involving the cop9 signalosome. Mol. Cell. Biol. 2006, 26, 2531–2539. [Google Scholar] [CrossRef] [PubMed]
- Vernell, R.; Helin, K.; Muller, H. Identification of target genes of the p16ink4a-prb-e2f pathway. J. Biol. Chem. 2003, 278, 46124–46137. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, C. F-box protein skp2: A novel transcriptional target of e2f. Oncogene 2006, 25, 2615–2627. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Ayad, N.G.; Wan, Y.; Zhang, G.J.; Kirschner, M.W.; Kaelin, W.G., Jr. Degradation of the scf component skp2 in cell-cycle phase g1 by the anaphase-promoting complex. Nature 2004, 428, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Binne, U.K.; Classon, M.K.; Dick, F.A.; Wei, W.; Rape, M.; Kaelin, W.G., Jr.; Naar, A.M.; Dyson, N.J. Retinoblastoma protein and anaphase-promoting complex physically interact and functionally cooperate during cell-cycle exit. Nat. Cell Biol. 2007, 9, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Ji, P.; Jiang, H.; Rekhtman, K.; Bloom, J.; Ichetovkin, M.; Pagano, M.; Zhu, L. An rb-skp2-p27 pathway mediates acute cell cycle inhibition by rb and is retained in a partial-penetrance rb mutant. Mol. Cell 2004, 16, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Jamal, A.; Swarnalatha, M.; Sultana, S.; Joshi, P.; Panda, S.K.; Kumar, V. The g1 phase e3 ubiquitin ligase truss that gets deregulated in human cancers is a novel substrate of the s-phase e3 ubiquitin ligase skp2. Cell Cycle 2015, 14, 2688–2700. [Google Scholar] [CrossRef] [PubMed]
- Von der Lehr, N.; Johansson, S.; Wu, S.; Bahram, F.; Castell, A.; Cetinkaya, C.; Hydbring, P.; Weidung, I.; Nakayama, K.; Nakayama, K.I.; et al. The f-box protein skp2 participates in c-myc proteosomal degradation and acts as a cofactor for c-myc-regulated transcription. Mol. Cell 2003, 11, 1189–1200. [Google Scholar] [CrossRef]
- Kim, S.Y.; Herbst, A.; Tworkowski, K.A.; Salghetti, S.E.; Tansey, W.P. Skp2 regulates myc protein stability and activity. Mol. Cell 2003, 11, 1177–1188. [Google Scholar] [CrossRef]
- Adler, A.S.; Lin, M.; Horlings, H.; Nuyten, D.S.; van de Vijver, M.J.; Chang, H.Y. Genetic regulators of large-scale transcriptional signatures in cancer. Nat. Genet. 2006, 38, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Fujii, M.; Lyakh, L.A.; Bracken, C.P.; Fukuoka, J.; Hayakawa, M.; Tsukiyama, T.; Soll, S.J.; Harris, M.; Rocha, S.; Roche, K.C.; et al. Snip1 is a candidate modifier of the transcriptional activity of c-myc on e box-dependent target genes. Mol. Cell 2006, 24, 771–783. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.H.; Lee, S.W.; Wang, J.; Lin, H.K. Regulation of skp2 expression and activity and its role in cancer progression. ScientificWorldJournal 2010, 10, 1001–1015. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Ji, L.H.; Liu, W.; Zhao, G.; Wu, Z.Y. Skp2-rnai suppresses proliferation and migration of gallbladder carcinoma cells by enhancing p27 expression. World J. Gastroenterol. 2013, 19, 4917–4924. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, D.; Mannava, S.; Grachtchouk, V.; Tang, W.H.; Patil, S.; Wawrzyniak, J.A.; Berman, A.E.; Giordano, T.J.; Prochownik, E.V.; Soengas, M.S.; et al. C-myc overexpression is required for continuous suppression of oncogene-induced senescence in melanoma cells. Oncogene 2008, 27, 6623–6634. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.K.; Chen, Z.; Wang, G.; Nardella, C.; Lee, S.W.; Chan, C.H.; Yang, W.L.; Wang, J.; Egia, A.; Nakayama, K.I.; et al. Skp2 targeting suppresses tumorigenesis by arf-p53-independent cellular senescence. Nature 2010, 464, 374–379. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Spears, E.; Boone, D.N.; Li, Z.; Gregory, M.A.; Hann, S.R. Domain-specific c-myc ubiquitylation controls c-myc transcriptional and apoptotic activity. Proc. Natl. Acad. Sci. USA 2013, 110, 978–983. [Google Scholar] [CrossRef] [PubMed]
- Larsson, L.G. Snip1: Myc’s new helper in transcriptional activation. Mol. Cell 2006, 24, 811–812. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.; Tyers, M. Transcriptional regulation: Kamikaze activators. Curr. Biol. 2000, 10, R341–R343. [Google Scholar] [CrossRef]
- Lipford, J.R.; Deshaies, R.J. Diverse roles for ubiquitin-dependent proteolysis in transcriptional activation. Nat. Cell Biol. 2003, 5, 845–850. [Google Scholar] [CrossRef] [PubMed]
- Muratani, M.; Tansey, W.P. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 2003, 4, 192–201. [Google Scholar] [CrossRef] [PubMed]
- von der Lehr, N.; Johansson, S.; Larsson, L.G. Implication of the ubiquitin/proteasome system in myc-regulated transcription. Cell Cycle 2003, 2, 403–407. [Google Scholar] [PubMed]
- Geng, F.; Wenzel, S.; Tansey, W.P. Ubiquitin and proteasomes in transcription. Annu. Rev. Biochem. 2012, 81, 177–201. [Google Scholar] [CrossRef] [PubMed]
- Salghetti, S.E.; Muratani, M.; Wijnen, H.; Futcher, B.; Tansey, W.P. Functional overlap of sequences that activate transcription and signal ubiquitin-mediated proteolysis. Proc. Natl. Acad. Sci. USA 2000, 97, 3118–3123. [Google Scholar] [CrossRef] [PubMed]
- Collins, G.A.; Goldberg, A.L. The logic of the 26s proteasome. Cell 2017, 169, 792–806. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Heng, J.I.; Guardavaccaro, D.; Jiang, R.; Pagano, M.; Guillemot, F.; Iavarone, A.; Lasorella, A. The hect-domain ubiquitin ligase huwe1 controls neural differentiation and proliferation by destabilizing the n-myc oncoprotein. Nat. Cell Biol. 2008, 10, 643–653. [Google Scholar] [CrossRef] [PubMed]
- Kurosu, T.; Peterlin, B.M. Vp16 and ubiquitin; binding of p-tefb via its activation domain and ubiquitin facilitates elongation of transcription of target genes. Curr. Biol. 2004, 14, 1112–1116. [Google Scholar] [CrossRef] [PubMed]
- Barboric, M.; Zhang, F.; Besenicar, M.; Plemenitas, A.; Peterlin, B.M. Ubiquitylation of cdk9 by skp2 facilitates optimal tat transactivation. J. Virol. 2005, 79, 11135–11141. [Google Scholar] [CrossRef] [PubMed]
- Ferdous, A.; Gonzalez, F.; Sun, L.; Kodadek, T.; Johnston, S.A. The 19s regulatory particle of the proteasome is required for efficient transcription elongation by rna polymerase ii. Mol. Cell 2001, 7, 981–991. [Google Scholar] [CrossRef]
- Gonzalez, F.; Delahodde, A.; Kodadek, T.; Johnston, S.A. Recruitment of a 19s proteasome subcomplex to an activated promoter. Science 2002, 296, 548–550. [Google Scholar] [CrossRef] [PubMed]
- Chaves, S.; Baskerville, C.; Yu, V.; Reed, S.I. Cks1, cdk1, and the 19s proteasome collaborate to regulate gene induction-dependent nucleosome eviction in yeast. Mol. Cell. Biol. 2010, 30, 5284–5294. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.C.; Kaiser, P.; Rudyak, S.; Baskerville, C.; Watson, M.H.; Reed, S.I. Cks1-dependent proteasome recruitment and activation of cdc20 transcription in budding yeast. Nature 2003, 423, 1009–1013. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Ezhkova, E.; Li, B.; Pattenden, S.G.; Tansey, W.P.; Workman, J.L. The proteasome regulatory particle alters the saga coactivator to enhance its interactions with transcriptional activators. Cell 2005, 123, 423–436. [Google Scholar] [CrossRef] [PubMed]
- Malik, S.; Shukla, A.; Sen, P.; Bhaumik, S.R. The 19s proteasome subcomplex establishes a specific protein interaction network at the promoter for stimulated transcriptional initiation in vivo. J. Biol. Chem. 2009, 284, 35714–35724. [Google Scholar] [CrossRef] [PubMed]
- Arabi, A.; Wu, S.; Ridderstrale, K.; Bierhoff, H.; Shiue, C.; Fatyol, K.; Fahlen, S.; Hydbring, P.; Soderberg, O.; Grummt, I.; et al. C-myc associates with ribosomal DNA and activates rna polymerase i transcription. Nat. Cell Biol. 2005, 7, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Reid, G.; Hubner, M.R.; Metivier, R.; Brand, H.; Denger, S.; Manu, D.; Beaudouin, J.; Ellenberg, J.; Gannon, F. Cyclic, proteasome-mediated turnover of unliganded and liganded eralpha on responsive promoters is an integral feature of estrogen signaling. Mol. Cell 2003, 11, 695–707. [Google Scholar] [CrossRef]
- Zhang, H.; Sun, L.; Liang, J.; Yu, W.; Zhang, Y.; Wang, Y.; Chen, Y.; Li, R.; Sun, X.; Shang, Y. The catalytic subunit of the proteasome is engaged in the entire process of estrogen receptor-regulated transcription. EMBO J. 2006, 25, 4223–4233. [Google Scholar] [CrossRef] [PubMed]
- Ostendorff, H.P.; Peirano, R.I.; Peters, M.A.; Schluter, A.; Bossenz, M.; Scheffner, M.; Bach, I. Ubiquitination-dependent cofactor exchange on lim homeodomain transcription factors. Nature 2002, 416, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Perissi, V.; Aggarwal, A.; Glass, C.K.; Rose, D.W.; Rosenfeld, M.G. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell 2004, 116, 511–526. [Google Scholar] [CrossRef]
- Wu, R.C.; Feng, Q.; Lonard, D.M.; O’Malley, B.W. Src-3 coactivator functional lifetime is regulated by a phospho-dependent ubiquitin time clock. Cell 2007, 129, 1125–1140. [Google Scholar] [CrossRef] [PubMed]
- Howard, G.C.; Tansey, W.P. Interaction of gcn4 with target gene chromatin is modulated by proteasome function. Mol. Biol. Cell 2016, 27, 2735–2741. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, C.; Dittrich, O.; Kiermaier, A.; Dohmann, K.; Menkel, A.; Eilers, M.; Luscher, B. Regulation of cyclin d2 gene expression by the myc/max/mad network: Myc-dependent trrap recruitment and histone acetylation at the cyclin d2 promoter. Genes Dev. 2001, 15, 2042–2047. [Google Scholar] [CrossRef] [PubMed]
- Amente, S.; Bertoni, A.; Morano, A.; Lania, L.; Avvedimento, E.V.; Majello, B. Lsd1-mediated demethylation of histone h3 lysine 4 triggers myc-induced transcription. Oncogene 2010, 29, 3691–3702. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Liu, S.; Li, B.; Xie, Y.; Adhiambo, C.; Yang, Q.; Ballard, B.R.; Nakayama, K.I.; Matusik, R.J.; Chen, Z. Skp2 inactivation suppresses prostate tumorigenesis by mediating jarid1b ubiquitination. Oncotarget 2015, 6, 771–788. [Google Scholar] [CrossRef] [PubMed]
- Secombe, J.; Li, L.; Carlos, L.; Eisenman, R.N. The trithorax group protein lid is a trimethyl histone h3k4 demethylase required for dmyc-induced cell growth. Genes Dev. 2007, 21, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Li, C.F.; Zhang, X.; Gong, Z.; Chan, C.H.; Lee, S.W.; Jin, G.; Rezaeian, A.H.; Han, F.; Wang, J.; et al. Skp2-macroh2a1-cdk8 axis orchestrates g2/m transition and tumorigenesis. Nat. Commun. 2015, 6, 6641. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D. Remodeling of chromatin structure in senescent cells and its potential impact on tumor suppression and aging. Gene 2007, 397, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Chandramohan, V.; Mineva, N.D.; Burke, B.; Jeay, S.; Wu, M.; Shen, J.; Yang, W.; Hann, S.R.; Sonenshein, G.E. C-myc represses foxo3a-mediated transcription of the gene encoding the p27(kip1) cyclin dependent kinase inhibitor. J. Cell. Biochem. 2008, 104, 2091–2106. [Google Scholar] [CrossRef] [PubMed]
- Fujita, N.; Sato, S.; Tsuruo, T. Phosphorylation of p27kip1 at threonine 198 by p90 ribosomal protein s6 kinases promotes its binding to 14-3-3 and cytoplasmic localization. J. Biol. Chem. 2003, 278, 49254–49260. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Zubovitz, J.; Petrocelli, T.; Kotchetkov, R.; Connor, M.K.; Han, K.; Lee, J.H.; Ciarallo, S.; Catzavelos, C.; Beniston, R.; et al. Pkb/akt phosphorylates p27, impairs nuclear import of p27 and opposes p27-mediated g1 arrest. Nat. Med. 2002, 8, 1153–1160. [Google Scholar] [CrossRef] [PubMed]
- Morishita, D.; Katayama, R.; Sekimizu, K.; Tsuruo, T.; Fujita, N. Pim kinases promote cell cycle progression by phosphorylating and down-regulating p27kip1 at the transcriptional and posttranscriptional levels. Cancer Res. 2008, 68, 5076–5085. [Google Scholar] [CrossRef] [PubMed]
- Cowling, V.H.; Chandriani, S.; Whitfield, M.L.; Cole, M.D. A conserved myc protein domain, mbiv, regulates DNA binding, apoptosis, transformation, and g2 arrest. Mol. Cell. Biol. 2006, 26, 4226–4239. [Google Scholar] [CrossRef] [PubMed]
- Vervoorts, J.; Luscher-Firzlaff, J.; Luscher, B. The ins and outs of myc regulation by posttranslational mechanisms. J. Biol. Chem. 2006, 281, 34725–34729. [Google Scholar] [CrossRef] [PubMed]
- Wasylishen, A.R.; Chan-Seng-Yue, M.; Bros, C.; Dingar, D.; Tu, W.B.; Kalkat, M.; Chan, P.K.; Mullen, P.J.; Huang, L.; Meyer, N.; et al. Myc phosphorylation at novel regulatory regions suppresses transforming activity. Cancer Res. 2013, 73, 6504–6515. [Google Scholar] [CrossRef] [PubMed]
- Wasylishen, A.R.; Kalkat, M.; Kim, S.S.; Pandyra, A.; Chan, P.K.; Oliveri, S.; Sedivy, E.; Konforte, D.; Bros, C.; Raught, B.; et al. Myc activity is negatively regulated by a c-terminal lysine cluster. Oncogene 2014, 33, 1066–1072. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Chiao, C.Y.; Enzer, K.G.; Stankiewicz, A.J.; Faller, D.V.; Dai, Y. Sirt1 inactivation evokes antitumor activities in nsclc through the tumor suppressor p27. Mol. Cancer Res. 2015, 13, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Luscher, B.; Kuenzel, E.A.; Krebs, E.G.; Eisenman, R.N. Myc oncoproteins are phosphorylated by casein kinase ii. EMBO J. 1989, 8, 1111–1119. [Google Scholar] [PubMed]
- Hauck, L.; Harms, C.; An, J.; Rohne, J.; Gertz, K.; Dietz, R.; Endres, M.; von Harsdorf, R. Protein kinase ck2 links extracellular growth factor signaling with the control of p27(kip1) stability in the heart. Nat. Med. 2008, 14, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Pippa, R.; Espinosa, L.; Gundem, G.; Garcia-Escudero, R.; Dominguez, A.; Orlando, S.; Gallastegui, E.; Saiz, C.; Besson, A.; Pujol, M.J.; et al. P27kip1 represses transcription by direct interaction with p130/e2f4 at the promoters of target genes. Oncogene 2012, 31, 4207–4220. [Google Scholar] [CrossRef] [PubMed]
- McArthur, G.A.; Foley, K.P.; Fero, M.L.; Walkley, C.R.; Deans, A.J.; Roberts, J.M.; Eisenman, R.N. Mad1 and p27(kip1) cooperate to promote terminal differentiation of granulocytes and to inhibit myc expression and cyclin e-cdk2 activity. Mol. Cell. Biol. 2002, 22, 3014–3023. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.C.; Martins, C.P.; Bronkhorst, Y.; Randel, E.; Berns, A.; Fero, M.; Clurman, B.E. Identification of oncogenes collaborating with p27kip1 loss by insertional mutagenesis and high-throughput insertion site analysis. Proc. Natl. Acad. Sci. USA 2002, 99, 11293–11298. [Google Scholar] [CrossRef] [PubMed]
- Seviour, E.G.; Sehgal, V.; Lu, Y.; Luo, Z.; Moss, T.; Zhang, F.; Hill, S.M.; Liu, W.; Maiti, S.N.; Cooper, L.; et al. Functional proteomics identifies mirnas to target a p27/myc/phospho-rb signature in breast and ovarian cancer. Oncogene 2016, 35, 691–701. [Google Scholar] [CrossRef] [PubMed]
- Caraballo, J.M.; Acosta, J.C.; Cortes, M.A.; Albajar, M.; Gomez-Casares, M.T.; Batlle-Lopez, A.; Cuadrado, M.A.; Onaindia, A.; Bretones, G.; Llorca, J.; et al. High p27 protein levels in chronic lymphocytic leukemia are associated to low myc and skp2 expression, confer resistance to apoptosis and antagonize myc effects on cell cycle. Oncotarget 2014, 5, 4694–4708. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.S.; Meitner, P.; Konkin, T.A.; Cho, Y.S.; Resnick, M.B.; Moss, S.F. Altered expression of skp2, c-myc and p27 proteins but not mrna after h. Pylori eradication in chronic gastritis. Mod. Pathol. 2006, 19, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Besson, A.; Assoian, R.K.; Roberts, J.M. Regulation of the cytoskeleton: An oncogenic function for cdk inhibitors? Nat. Rev. Cancer 2004, 4, 948–955. [Google Scholar] [CrossRef] [PubMed]
- Denicourt, C.; Saenz, C.C.; Datnow, B.; Cui, X.S.; Dowdy, S.F. Relocalized p27kip1 tumor suppressor functions as a cytoplasmic metastatic oncogene in melanoma. Cancer Res. 2007, 67, 9238–9243. [Google Scholar] [CrossRef] [PubMed]
- Asghar, U.; Witkiewicz, A.K.; Turner, N.C.; Knudsen, E.S. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat. Rev. Drug Discov. 2015, 14, 130–146. [Google Scholar] [CrossRef] [PubMed]
- Goga, A.; Yang, D.; Tward, A.D.; Morgan, D.O.; Bishop, J.M. Inhibition of cdk1 as a potential therapy for tumors over-expressing myc. Nat. Med. 2007, 13, 820–827. [Google Scholar] [CrossRef] [PubMed]
- Molenaar, J.J.; Ebus, M.E.; Geerts, D.; Koster, J.; Lamers, F.; Valentijn, L.J.; Westerhout, E.M.; Versteeg, R.; Caron, H.N. Inactivation of cdk2 is synthetically lethal to mycn over-expressing cancer cells. Proc. Natl. Acad. Sci. USA 2009, 106, 12968–12973. [Google Scholar] [CrossRef] [PubMed]
- Hydbring, P.; Wang, Y.; Fassl, A.; Li, X.; Matia, V.; Otto, T.; Choi, Y.J.; Sweeney, K.E.; Suski, J.M.; Yin, H.; et al. Cell-cycle-targeting micrornas as therapeutic tools against refractory cancers. Cancer Cell 2017, 31, 576–590. [Google Scholar] [CrossRef] [PubMed]
- Schiewer, M.J.; Knudsen, K.E. Not so fast: Cultivating mirs as kinks in the chain of the cell cycle. Cancer Cell 2017, 31, 471–473. [Google Scholar] [CrossRef] [PubMed]
- Janghorban, M.; Farrell, A.S.; Allen-Petersen, B.L.; Pelz, C.; Daniel, C.J.; Oddo, J.; Langer, E.M.; Christensen, D.J.; Sears, R.C. Targeting c-myc by antagonizing pp2a inhibitors in breast cancer. Proc. Natl. Acad. Sci. USA 2014, 111, 9157–9162. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Grigoryan, A.V.; Li, Y.; Hao, B.; Pagano, M.; Cardozo, T.J. Specific small molecule inhibitors of skp2-mediated p27 degradation. Chem. Biol. 2012, 19, 1515–1524. [Google Scholar] [CrossRef] [PubMed]
- Ooi, L.C.; Watanabe, N.; Futamura, Y.; Sulaiman, S.F.; Darah, I.; Osada, H. Identification of small molecule inhibitors of p27(kip1) ubiquitination by high-throughput screening. Cancer Sci. 2013, 104, 1461–1467. [Google Scholar] [CrossRef] [PubMed]
- Pavlides, S.C.; Huang, K.T.; Reid, D.A.; Wu, L.; Blank, S.V.; Mittal, K.; Guo, L.; Rothenberg, E.; Rueda, B.; Cardozo, T.; et al. Inhibitors of scf-skp2/cks1 e3 ligase block estrogen-induced growth stimulation and degradation of nuclear p27kip1: Therapeutic potential for endometrial cancer. Endocrinology 2013, 154, 4030–4045. [Google Scholar] [CrossRef] [PubMed]
- Ludwik, K.A.; Lannigan, D.A. Ribosomal s6 kinase (rsk) modulators: A patent review. Expert Opin. Ther. Pat. 2016, 26, 1061–1078. [Google Scholar] [CrossRef] [PubMed]
- Pretre, V.; Wicki, A. Inhibition of akt and other agc kinases: A target for clinical cancer therapy? Semin. Cancer Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- McKeown, M.R.; Bradner, J.E. Therapeutic strategies to inhibit myc. Cold Spring Harb. Perspect. Med. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, S.; Prochownik, E.V. Small-molecule inhibitors of the myc oncoprotein. Biochim. Biophys. Acta 2015, 1849, 525–543. [Google Scholar] [CrossRef] [PubMed]
- Prochownik, E.V.; Vogt, P.K. Therapeutic targeting of myc. Genes Cancer 2010, 1, 650–659. [Google Scholar] [CrossRef] [PubMed]
- Cetinkaya, C.; Hultquist, A.; Su, Y.; Wu, S.; Bahram, F.; Pahlman, S.; Guzhova, I.; Larsson, L.G. Combined ifn-gamma and retinoic acid treatment targets the n-myc/max/mad1 network resulting in repression of n-myc target genes in mycn-amplified neuroblastoma cells. Mol. Cancer Ther. 2007, 6, 2634–2641. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.H.; Maher, S.G.; Young, H.A. Clinical use of interferon-gamma. Ann. N. Y. Acad. Sci. 2009, 1182, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, M.R.; Merlino, G. The two faces of interferon-gamma in cancer. Clin. Cancer Res. 2011, 17, 6118–6124. [Google Scholar] [CrossRef] [PubMed]
- Finn, O.J. Cancer immunology. N. Engl. J. Med. 2008, 358, 2704–2715. [Google Scholar] [CrossRef] [PubMed]
- Braumuller, H.; Wieder, T.; Brenner, E.; Assmann, S.; Hahn, M.; Alkhaled, M.; Schilbach, K.; Essmann, F.; Kneilling, M.; Griessinger, C.; et al. T-helper-1-cell cytokines drive cancer into senescence. Nature 2013, 494, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Rakhra, K.; Bachireddy, P.; Zabuawala, T.; Zeiser, R.; Xu, L.; Kopelman, A.; Fan, A.C.; Yang, Q.; Braunstein, L.; Crosby, E.; et al. Cd4(+) T cells contribute to the remodeling of the microenvironment required for sustained tumor regression upon oncogene inactivation. Cancer Cell 2010, 18, 485–498. [Google Scholar] [CrossRef] [PubMed]
- Mellman, I.; Coukos, G.; Dranoff, G. Cancer immunotherapy comes of age. Nature 2011, 480, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Abraham, W.D.; Zheng, Y.; Bustamante Lopez, S.C.; Luo, S.S.; Irvine, D.J. Active targeting of chemotherapy to disseminated tumors using nanoparticle-carrying t cells. Sci. Transl. Med. 2015, 7, 291ra94. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hydbring, P.; Castell, A.; Larsson, L.-G. MYC Modulation around the CDK2/p27/SKP2 Axis. Genes 2017, 8, 174. https://doi.org/10.3390/genes8070174
Hydbring P, Castell A, Larsson L-G. MYC Modulation around the CDK2/p27/SKP2 Axis. Genes. 2017; 8(7):174. https://doi.org/10.3390/genes8070174
Chicago/Turabian StyleHydbring, Per, Alina Castell, and Lars-Gunnar Larsson. 2017. "MYC Modulation around the CDK2/p27/SKP2 Axis" Genes 8, no. 7: 174. https://doi.org/10.3390/genes8070174




