Boron Tolerance in Aspergillus nidulans Is Sustained by the SltA Pathway Through the SLC-Family Transporters SbtA and SbtB
Abstract
:1. Introduction
2. Materials and Methods
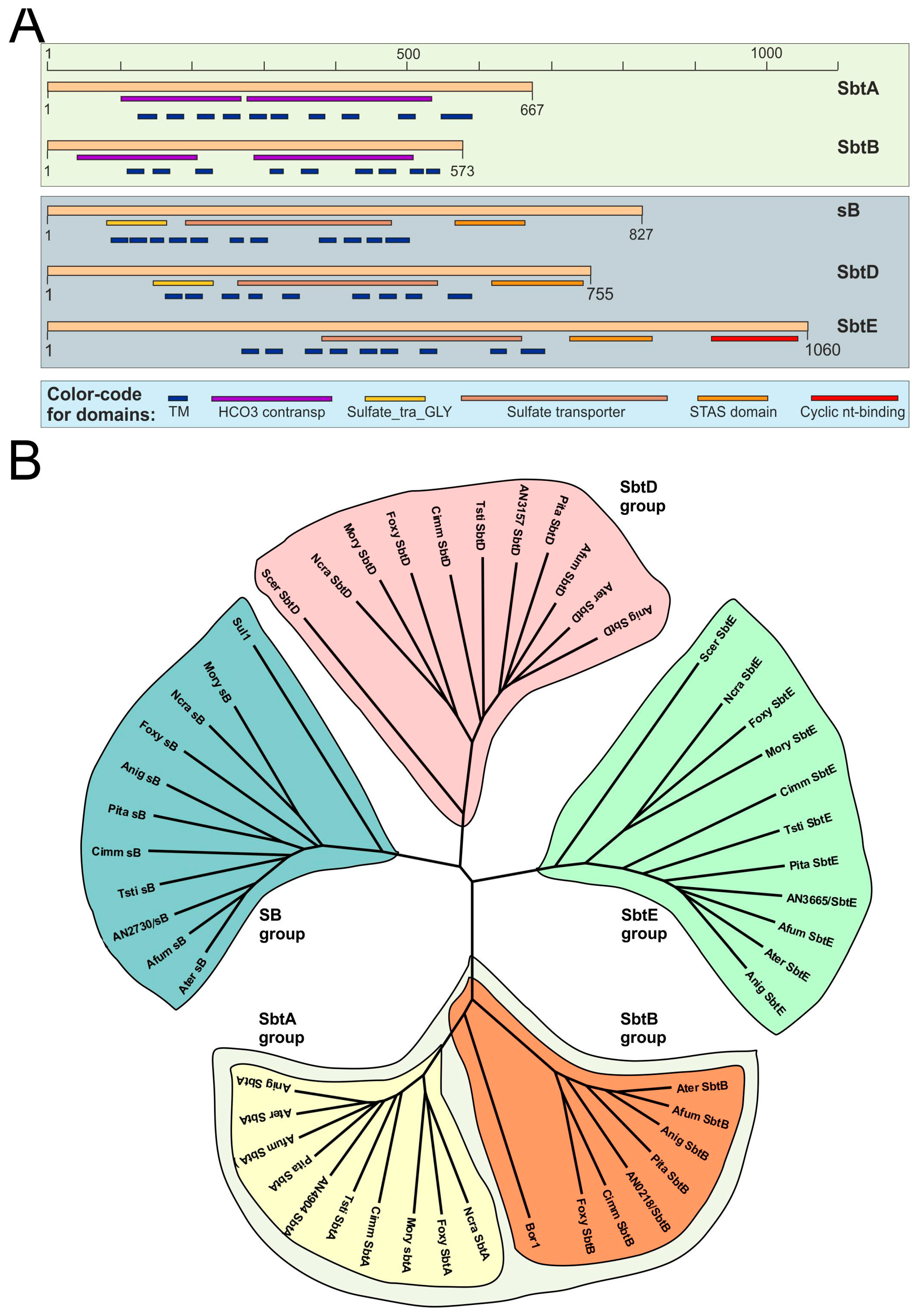
2.1. Sequence Analyses
2.2. Strains, Oligonucleotides, Culture Conditions and Protoplast Transformation
2.3. RNA Isolation and Northern-Blot Analyses
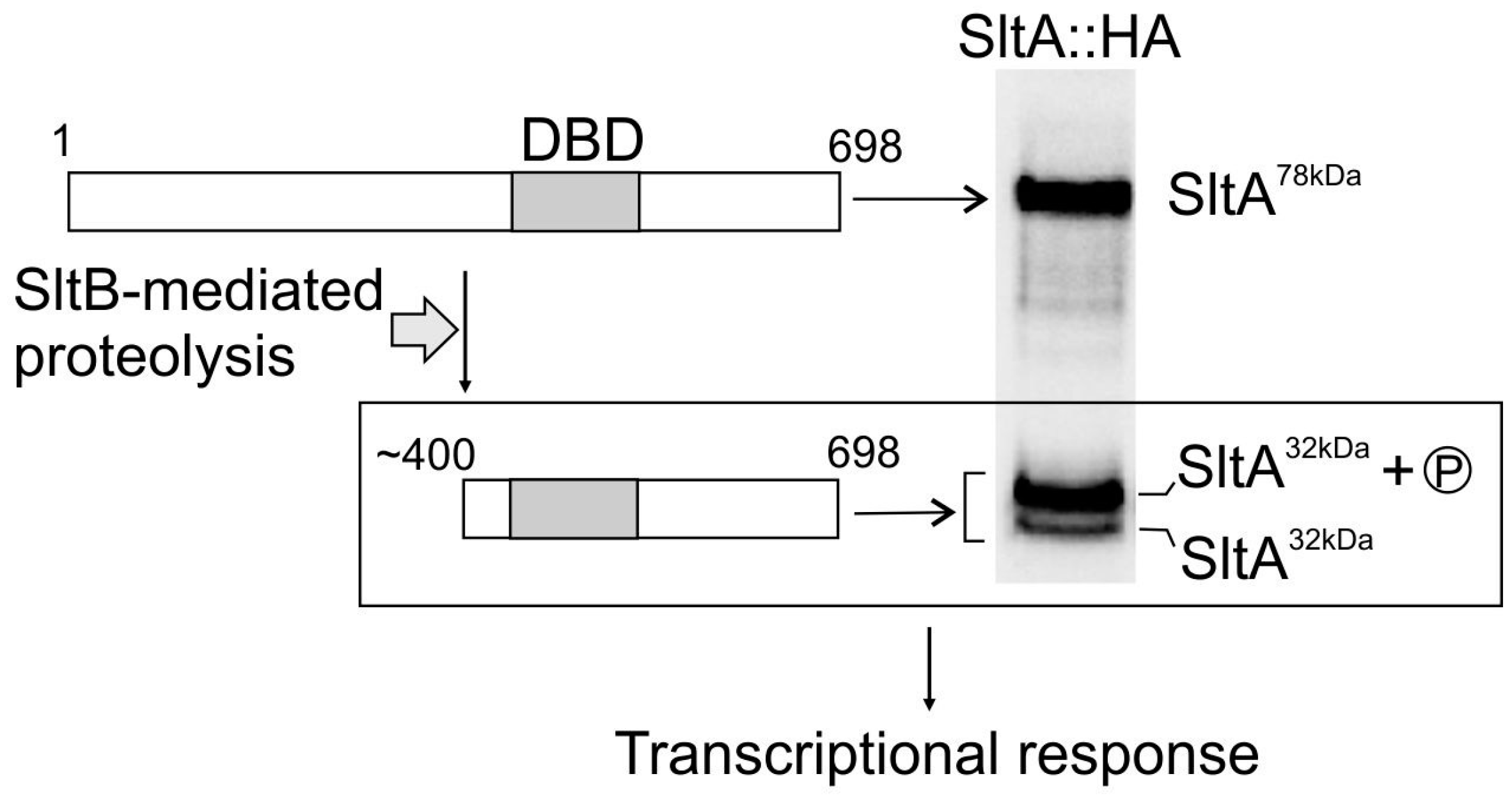
3. Results
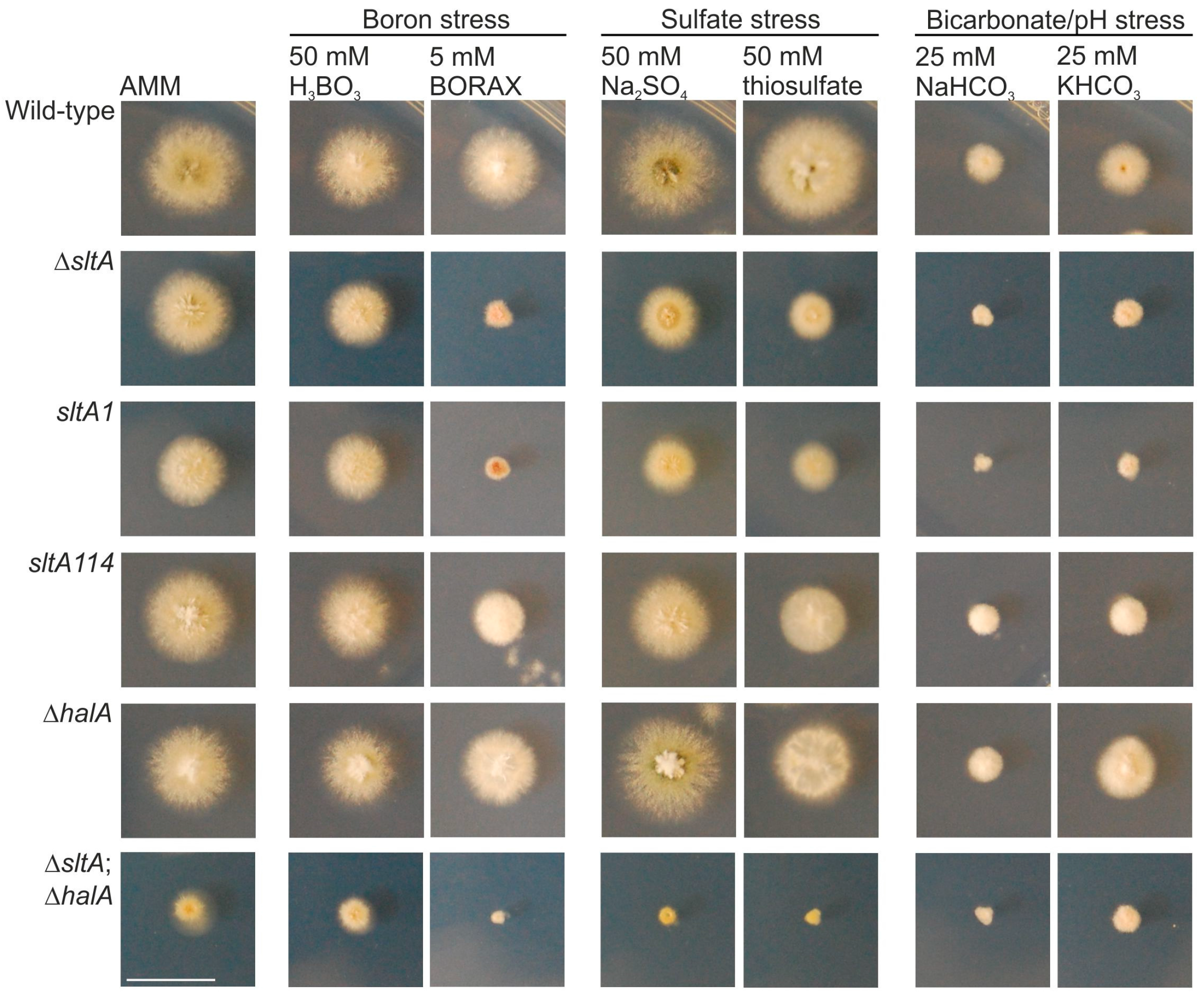
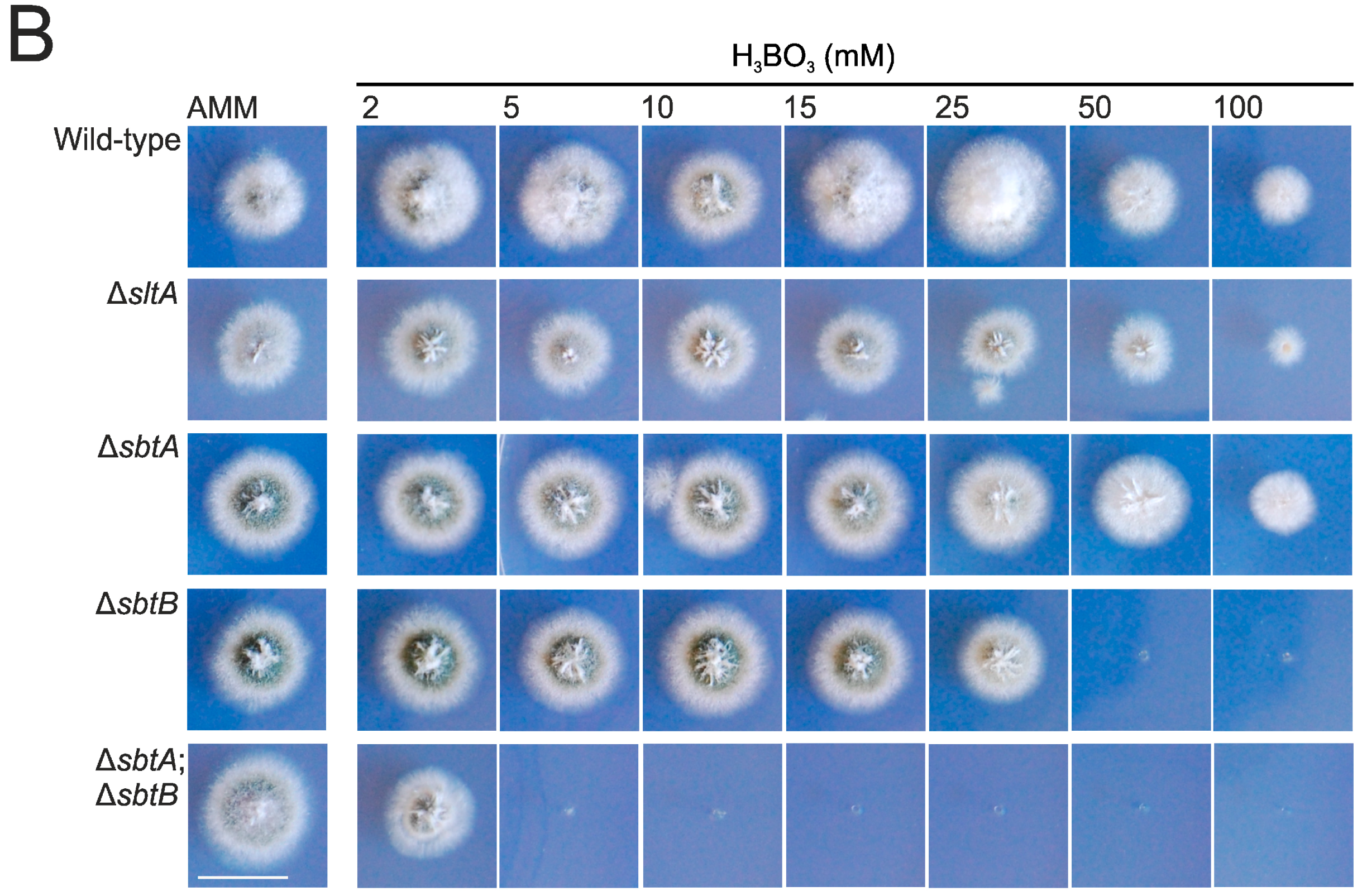
3.1. sltA Mutants Are Less Tolerant to Boron Stress
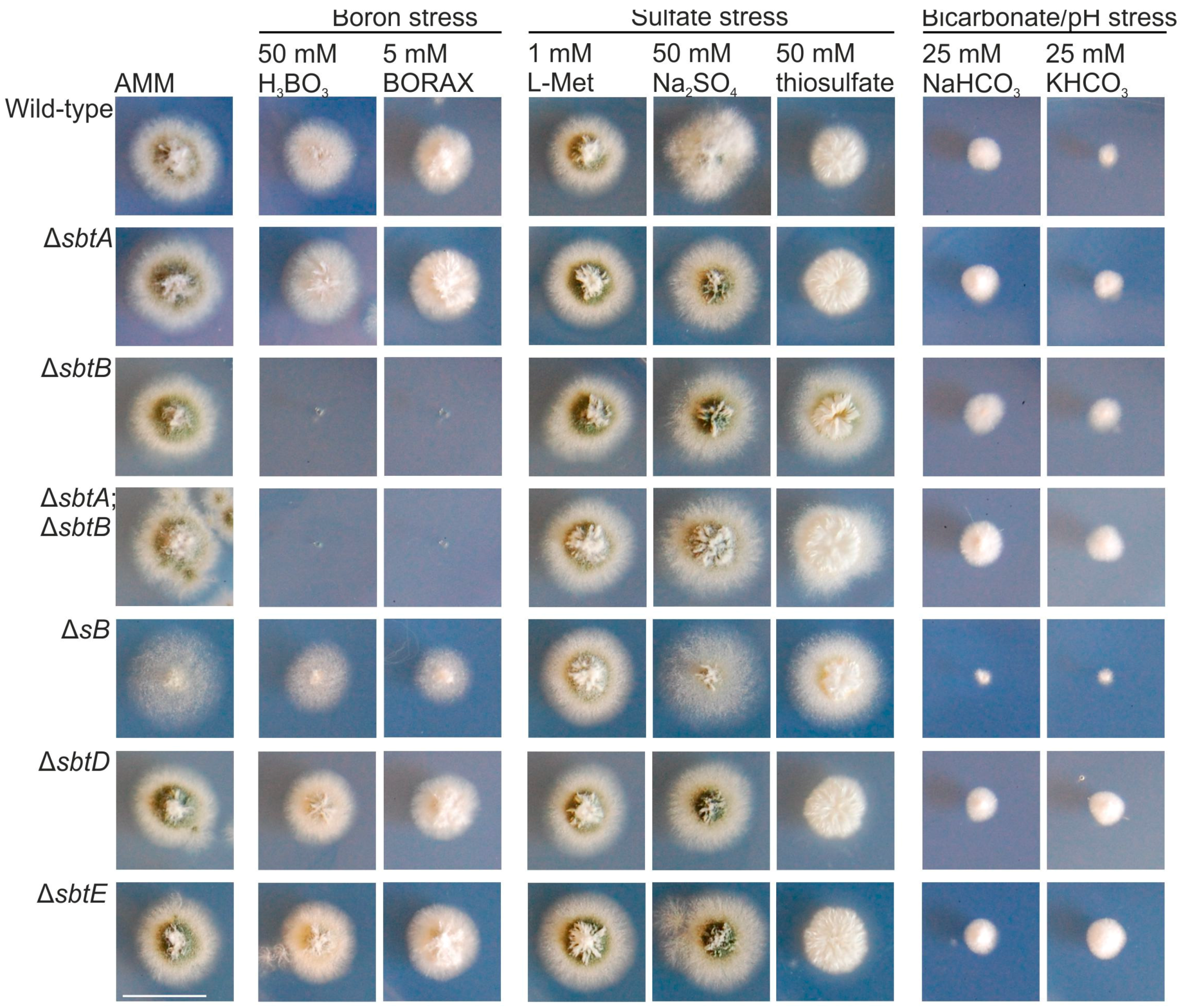
3.2. SbtA and SbtB Are Required for Tolerance to Borates
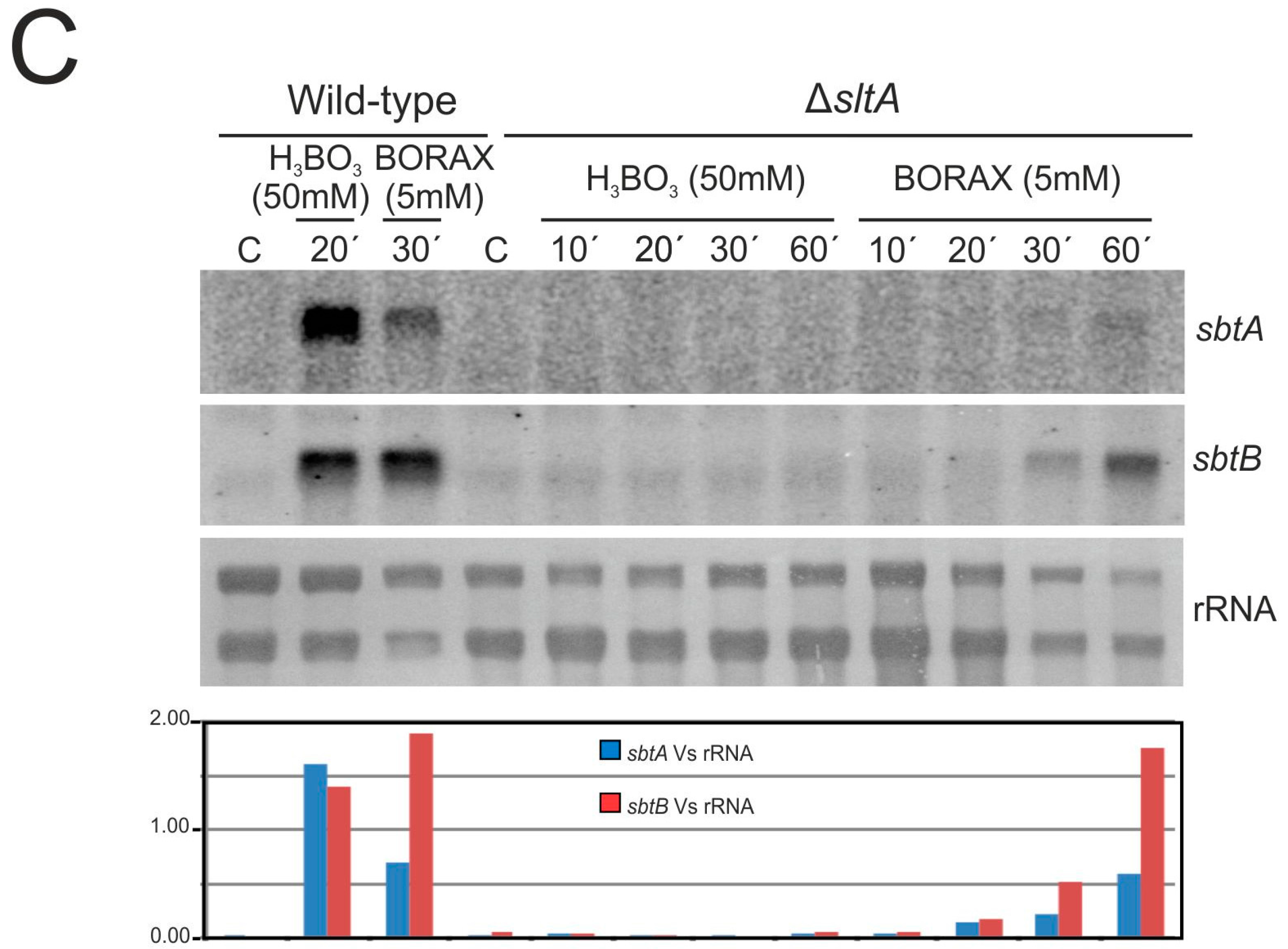
3.3. The Expression of sbtA and sbtB Is Induced Under Borate Stress and Depends on the Regulatory Activity of SltA
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Corrochano, L.M.; Kuo, A.; Marcet-Houben, M.; Polaino, S.; Salamov, A.; Villalobos-Escobedo, J.M.; Grimwood, J.; Álvarez, M.I.; Avalos, J.; Bauer, D.; et al. Expansion of Signal Transduction Pathways in Fungi by Extensive Genome Duplication. Curr. Biol. 2016, 26, 1577–1584. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Romero, J.; Hedtke, M.; Kastner, C.; Müller, S.; Fischer, R. Fungi, Hidden in Soil or Up in the Air: Light Makes a Difference. Annu. Rev. Microbiol. 2010, 64, 585–610. [Google Scholar] [CrossRef] [PubMed]
- Etxebeste, O.; Ugalde, U.; Espeso, E. Adaptative and developmental responses to stress in Aspergillus nidulans. Curr. Protein Pept. Sci. 2010, 11, 704–718. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Martínez, A.E.; Lara-Rojas, F.; Sánchez, O.; Aguirre, J. NapA Mediates a Redox Regulation of the Antioxidant Response, Carbon Utilization and Development in Aspergillus nidulans. Front. Microbiol. 2017, 8, 516. [Google Scholar] [CrossRef] [PubMed]
- Peñalva, M.A.; Tilburn, J.; Bignell, E.; Arst, H.N. Ambient pH gene regulation in fungi: Making connections. Trends Microbiol. 2008, 16, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Mellado, L.; Arst, H.N.; Espeso, E.A. Proteolytic activation of both components of the cation stress-responsive Slt pathway in Aspergillus nidulans. Mol. Biol. Cell 2016, 27, 2598–2612. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Ortiz, P.; Espeso, E.A. Spatiotemporal dynamics of the calcineurin target CrzA. Cell Signal. 2017, 29, 168–180. [Google Scholar] [CrossRef] [PubMed]
- Mellado, L.; Calcagno-Pizarelli, A.M.; Lockington, R.A.; Cortese, M.S.; Kelly, J.M.; Arst, H.N., Jr.; Espeso, E.A. A second component of the SltA-dependent cation tolerance pathway in Aspergillus nidulans. Fungal Genet. Biol. 2015, 82, 116–128. [Google Scholar] [CrossRef] [PubMed]
- Spielvogel, A.; Findon, H.; Arst, H.N.; Araújo-Bazán, L.; Hernández-Ortíz, P.; Stahl, U.; Meyer, V.; Espeso, E.A. Two zinc finger transcription factors, CrzA and SltA, are involved in cation homoeostasis and detoxification in Aspergillus nidulans. Biochem. J. 2008, 414, 419–429. [Google Scholar] [CrossRef] [PubMed]
- O’Neil, J.D.; Bugno, M.; Stanley, M.S.; Barham-Morris, J.B.; Woodcock, N.A.; Clement, D.J.; Clipson, N.J. W.; Whitehead, M.P.; Fincham, D.A.; Hooley, P. Cloning of a novel gene encoding a C2H2 zinc finger protein that alleviates sensitivity to abiotic stresses in Aspergillus nidulans. Mycol. Res. 2002, 106, 491–498. [Google Scholar] [CrossRef]
- Calcagno-Pizarelli, A.M.; Hervás-Aguilar, A.; Galindo, A.; Abenza, J.F.; Peñalva, M.A.; Arst, H.N. Rescue of Aspergillus nidulans severely debilitating null mutations in ESCRT-0, I, II and III genes by inactivation of a salt-tolerance pathway allows examination of ESCRT gene roles in pH signalling. J. Cell Sci. 2011, 124, 4064–4076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shantappa, S.; Dhingra, S.; Hernández-Ortiz, P.; Espeso, E.A.; Calvo, A.M. Role of the Zinc Finger Transcription Factor SltA in Morphogenesis and Sterigmatocystin Biosynthesis in the Fungus Aspergillus nidulans. PLoS ONE 2013, 8, e68492. [Google Scholar] [CrossRef] [PubMed]
- Saloheimo, A.; Aro, N.; Ilmén, M.; Penttilä, M. Isolation of the ace1 gene encoding a Cys(2)-His(2) transcription factor involved in regulation of activity of the cellulase promoter cbh1 of Trichoderma reesei. J. Biol. Chem. 2000, 275, 5817–5825. [Google Scholar] [CrossRef] [PubMed]
- Dubey, A.K.; Barad, S.; Luria, N.; Kumar, D.; Espeso, E.A.; Prusky, D.B. Cation-Stress-Responsive Transcription Factors SltA and CrzA Regulate Morphogenetic Processes and Pathogenicity of Colletotrichum gloeosporioides. PLoS ONE 2016, 11, e0168561. [Google Scholar] [CrossRef] [PubMed]
- Nozawa, A.; Takano, J.; Kobayashi, M.; von Wirén, N.; Fujiwara, T. Roles of BOR1, DUR3, and FPS1 in boron transport and tolerance in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 2006, 262, 216–222. [Google Scholar] [CrossRef] [PubMed]
- Piłsyk, S.; Natorff, R.; Sieńko, M.; Paszewski, A. Sulfate transport in Aspergillus nidulans: A novel gene encoding alternative sulfate transporter. Fungal Genet. Biol. 2007, 44, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Garzia, A.; Etxebeste, O.; Rodríguez-Romero, J.; Fischer, R.; Espeso, E.A.; Ugalde, U. Transcriptional changes in the transition from vegetative cells to asexual development in the model fungus Aspergillus nidulans. Eukaryot. Cell 2013, 12, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Oiartzabal-Arano, E.; Garzia, A.; Gorostidi, A.; Ugalde, U.; Espeso, E.A.; Etxebeste, O. Beyond asexual development: Modifications in the gene expression profile caused by the absence of the Aspergillus nidulans transcription factor FlbB. Genetics 2015, 199, 1127–1142. [Google Scholar] [CrossRef] [PubMed]
- Coradetti, S.T.; Xiong, Y.; Glass, N.L. Analysis of a conserved cellulase transcriptional regulator reveals inducer-independent production of cellulolytic enzymes in Neurospora crassa. Microbiologyopen 2013, 2, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Ukil, L.; Osmani, A.; Nahm, F.; Davies, J.; De Souza, C.P.C.; Dou, X.; Perez-Balaguer, A.; Osmani, S.A. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot. Cell 2004, 3, 1359–1362. [Google Scholar] [CrossRef] [PubMed]
- Nayak, T.; Szewczyk, E.; Oakley, C.E.; Osmani, A.; Ukil, L.; Murray, S.L.; Hynes, M.J.; Osmani, S.A.; Oakley, B.R. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 2006, 172, 1557–1566. [Google Scholar] [CrossRef] [PubMed]
- Szewczyk, E.; Nayak, T.; Oakley, C.E.; Edgerton, H.; Xiong, Y.; Taheri-Talesh, N.; Osmani, S.A.; Oakley, B.R. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 2007, 1, 3111–3120. [Google Scholar] [CrossRef] [PubMed]
- Tilburn, J.; Scazzocchio, C.; Taylor, G.G.; Zabicky-Zissman, J.H.; Lockington, R.A.; Davies, R.W. Transformation by integration in Aspergillus nidulans. Gene 1983, 26, 205–221. [Google Scholar] [CrossRef]
- Etxebeste, O.; Ni, M.; Garzia, A.; Kwon, N.J.; Fischer, R.; Yu, J.H.; Espeso, E.A.; Ugalde, U. Basic-zipper-type transcription factor FlbB controls asexual development in Aspergillus nidulans. Eukaryot. Cell 2008, 7, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Cove, D.J. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta Enzymol. Biol. Oxid. 1966, 113, 51–56. [Google Scholar] [CrossRef]
- Church, G.M.; Gilbert, W. Genomic sequencing. Biochemistry 1984, 81, 1991–1995. [Google Scholar] [CrossRef]
- Borokhov, O.; Schubert, D. Antimicrobial Properties of Boron Derivatives. In New Biocides Development: The Combined Approach of Chemistry and Microbiology; Zhu, P.C., Ed.; American Chemical Society: Washington, DC, USA, 2007; pp. 412–435. [Google Scholar]
- Findon, H.; Calcagno-Pizarelli, A.-M.; Martínez, J.L.; Spielvogel, A.; Markina-Iñarrairaegui, A.; Indrakumar, T.; Ramos, J.; Peñalva, M.A.; Espeso, E.A.; Arst, H.N. Analysis of a novel calcium auxotrophy in Aspergillus nidulans. Fungal Genet. Biol. 2010, 47, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Shibata, M.; Katoh, H.; Sonoda, M.; Ohkawa, H.; Shimoyama, M.; Fukuzawa, H.; Kaplan, A.; Ogawa, T. Genes essential to sodium-dependent bicarbonate transport in cyanobacteria: Function and phylogenetic analysis. J. Biol. Chem. 2002, 277, 18658–18664. [Google Scholar] [CrossRef] [PubMed]
- Aravind, L.; Koonin, E.V. The STAS domain—A link between anion transporters and antisigma-factor antagonists. Curr. Biol. 2000, 10, R53–R55. [Google Scholar] [CrossRef]
- Weser, U.; Kaup, Y. Borate, an Effective Mummification Agent in Pharaonic Egypt. Z. Naturforsch. 2002, 57, 819–822. [Google Scholar]
- Scorei, R. Is Boron a Prebiotic Element? A Mini-review of the Essentiality of Boron for the Appearance of Life on Earth. Orig. Life Evol. Biosph. 2012, 42, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Loomis, W.D.; Durst, R.W. Chemistry and biology of boron. Biofactors 1992, 3, 229–239. [Google Scholar] [PubMed]
- Takano, J.; Kobayashi, M.; Noda, Y.; Fujiwara, T. Saccharomyces cerevisiae Bor1p is a boron exporter and a key determinant of boron tolerance. FEMS Microboil. Lett. 2006, 267, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Li, Q.; Shcheynikov, N.; Zeng, W.; Muallem, S. NaBC1 Is a Ubiquitous Electrogenic Na+-Coupled Borate Transporter Essential for Cellular Boron Homeostasis and Cell Growth and Proliferation. Mol. Cell 2004, 16, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Urra, A.B.; Jimenez, C.; Dueñas, M.; Ugalde, U. Bicarbonate gradients modulate growth and colony morphology in Aspergillus nidulans. FEMS Microbiol. Lett. 2009, 300, 216–221. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Mosa, K.A.; Chhikara, S.; Musante, C.; White, J.C.; Dhankher, O.P. Two rice plasma membrane intrinsic proteins, OsPIP2;4 and OsPIP2;7, are involved in transport and providing tolerance to boron toxicity. Planta 2014, 239, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Jennings, M.L.; Howren, T.R.; Cui, J.; Winters, M.; Hannigan, R. Transport and regulatory characteristics of the yeast bicarbonate transporter homolog Bor1p. Am. J. Physiol. Cell Physoil. 2007, 293, 468–476. [Google Scholar] [CrossRef] [PubMed]
- Uluisik, I.; Kaya, A.; Fomenko, D.E.; Karakaya, H.C.; Carlson, B.A.; Gladyshev, V.N.; Koc, A.; Lustig, A.J. Boron Stress Activates the General Amino Acid Control Mechanism and Inhibits Protein Synthesis. PLoS ONE 2011, 6, e27772. [Google Scholar] [CrossRef] [PubMed]
- Bozdag, G.O.; Uluisik, I.; Gulculer, G.S.; Karakaya, H.C.; Koc, A. Roles of ATR1 paralogs YMR279c and YOR378w in boron stress tolerance. Biochem. Biophys. Res. Commun. 2011, 409, 748–751. [Google Scholar] [CrossRef] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villarino, M.; Etxebeste, O.; Mendizabal, G.; Garzia, A.; Ugalde, U.; Espeso, E.A. Boron Tolerance in Aspergillus nidulans Is Sustained by the SltA Pathway Through the SLC-Family Transporters SbtA and SbtB. Genes 2017, 8, 188. https://doi.org/10.3390/genes8070188
Villarino M, Etxebeste O, Mendizabal G, Garzia A, Ugalde U, Espeso EA. Boron Tolerance in Aspergillus nidulans Is Sustained by the SltA Pathway Through the SLC-Family Transporters SbtA and SbtB. Genes. 2017; 8(7):188. https://doi.org/10.3390/genes8070188
Chicago/Turabian StyleVillarino, María, Oier Etxebeste, Gorka Mendizabal, Aitor Garzia, Unai Ugalde, and Eduardo A. Espeso. 2017. "Boron Tolerance in Aspergillus nidulans Is Sustained by the SltA Pathway Through the SLC-Family Transporters SbtA and SbtB" Genes 8, no. 7: 188. https://doi.org/10.3390/genes8070188





