The Landscape of Small Non-Coding RNAs in Triple-Negative Breast Cancer
Abstract
:1. Introduction
2. Methods
2.1. High-Throughput RNA Sequencing
2.2. Bioinformatics and Data Analyses
2.3. TNBC Subtyping
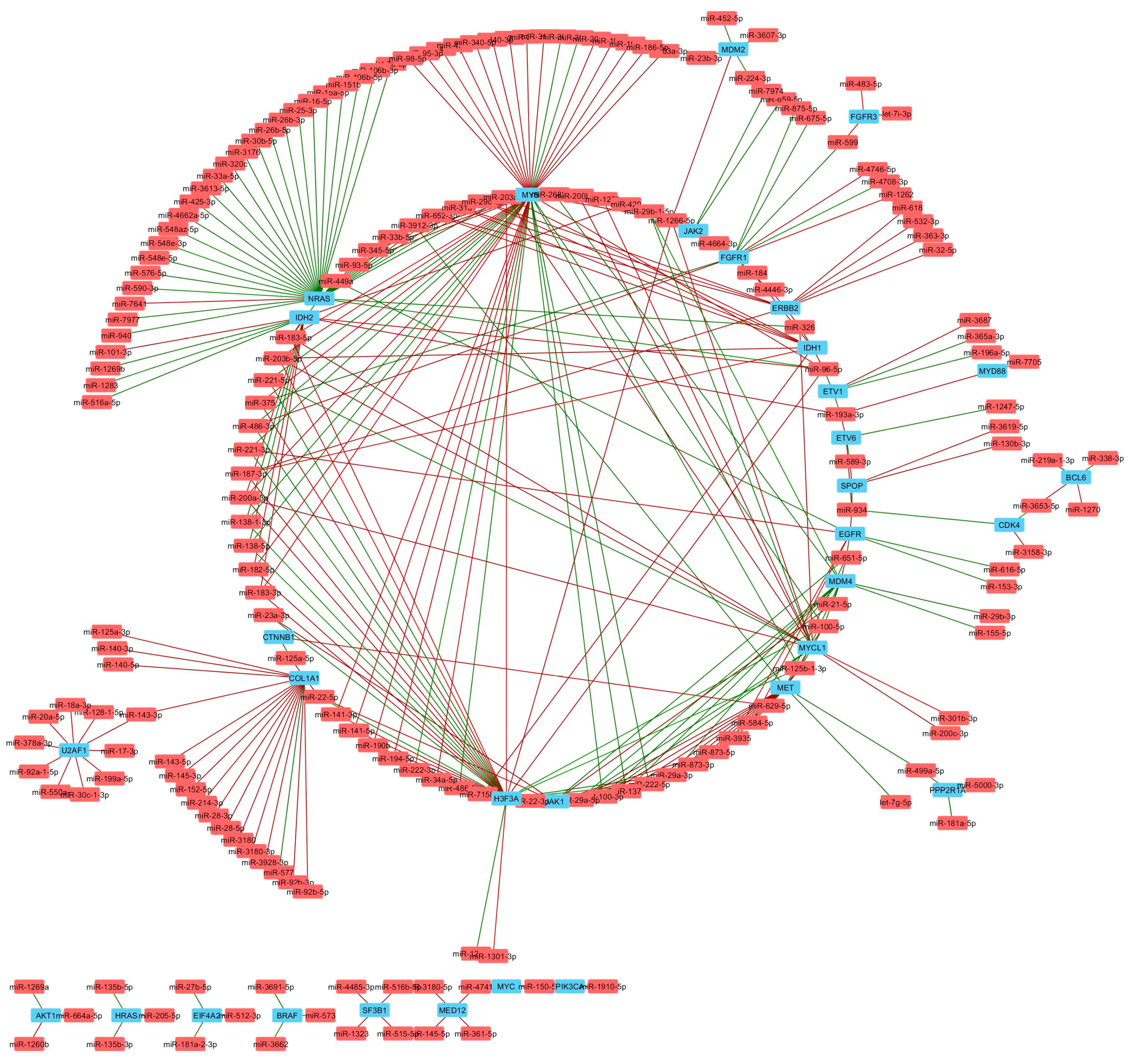
3. Results
4. Discussion
Supplementary Materials
Acknowledgments
Author contributions
Conflicts of Interest
References
- Shah, S.P.; Roth, A.; Goya, R.; Oloumi, A.; Ha, G.; Zhao, Y.J.; Turashvili, G.; Ding, J.R.; Tse, K.; Haffari, G.; et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 2012, 486, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, B.D.; Jovanovic, B.; Chen, X.; Estrada, M.V.; Johnson, K.N.; Shyr, Y.; Moses, H.L.; Sanders, M.E.; Pietenpol, J.A. Refinement of triple-negative breast cancer molecular subtypes: Implications for neoadjuvant chemotherapy selection. PLoS ONE 2016, 11, e0157368. [Google Scholar] [CrossRef] [PubMed]
- Jezequel, P.; Loussouarn, D.; Guerin-Charbonnel, C.; Campion, L.; Vanier, A.; Gouraud, W.; Lasla, H.; Guette, C.; Valo, I.; Verriele, V.; et al. Gene-expression molecular subtyping of triple-negative breast cancer tumours: Importance of immune response. Breast Cancer Res. 2015, 17, 43. [Google Scholar] [CrossRef] [PubMed]
- Burstein, M.D.; Tsimelzon, A.; Poage, G.M.; Coyington, K.R.; Contreras, A.; Fuqua, S.A.W.; Sayage, M.I.; Osborne, C.K.; Hilsenbeck, S.G.; Chang, J.C.; et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clinical Cancer Res. 2015, 21, 1688–1698. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The c. Elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef] [PubMed]
- Sai, S.; Ichikawa, D.; Tomita, H.; Ikoma, D.; Tani, N.; Ikoma, H.; Kikuchi, S.; Fujiwara, H.; Ueda, Y.; Otsuji, E. Quantification of plasma cell-free DNA in patients with gastric cancer. Anticancer Res. 2007, 27, 2747–2751. [Google Scholar] [PubMed]
- Vinayanuwattikun, C.; Winayanuwattikun, P.; Chantranuwat, P.; Mutirangura, A.; Sriuranpong, V. The impact of non-tumor-derived circulating nucleic acids implicates the prognosis of non-small cell lung cancer. J. Cancer Res. Clin. Oncol. 2013, 139, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Shin, V.Y.; Siu, J.M.; Cheuk, I.; Ng, E.K.O.; Kwong, A. Circulating cell-free miRNAs as biomarker for triple-negative breast cancer. Br. J. Cancer 2015, 112, 1751–1759. [Google Scholar] [CrossRef] [PubMed]
- Mathe, A.; Scott, R.J.; Avery-Kiejda, K.A. MiRNAs and other epigenetic changes as biomarkers in triple negative breast cancer. Int. J. Mol. Sci. 2015, 16, 28347–28376. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.B.; Chen, X.; Zhao, Y.; Tian, T.; Jin, G.F.; Shu, Y.Q.; Chen, Y.J.; Xu, L.; Zen, K.; Zhang, C.Y.; et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J. Clin. Oncol. 2010, 28, 1721–1726. [Google Scholar] [CrossRef] [PubMed]
- Cloonan, N.; Wani, S.; Xu, Q.Y.; Gu, J.; Lea, K.; Heater, S.; Barbacioru, C.; Steptoe, A.L.; Martin, H.C.; Nourbakhsh, E.; et al. MicroRNAs and their isomirs function cooperatively to target common biological pathways. Genome Biol. 2011, 12, R126. [Google Scholar] [CrossRef] [PubMed]
- Tan, G.C.; Chan, E.; Molnar, A.; Sarkar, R.; Alexieva, D.; Isa, I.M.; Robinson, S.; Zhang, S.C.; Ellis, P.; Langford, C.F.; et al. 5′ isomir variation is of functional and evolutionary importance. Nucleic Acids Res. 2014, 42, 9424–9435. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Bosompem, A.; Mohan, S.; Erdogan, B.; Ye, F.; Vickers, K.C.; Sheng, Q.H.; Zhao, S.L.; Li, C.I.; Su, P.F.; et al. Transfer RNA detection by small RNA deep sequencing and disease association with myelodysplastic syndromes. BMC Genom. 2015, 16, 727. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Xiong, Y.; Sheng, Q.; Zhao, S.; Wattacheril, J.; Flynn, C.R. A micro-RNA expression signature for human nafld progression. J. Gastroenterol. 2016, 51, 1022–1030. [Google Scholar] [CrossRef] [PubMed]
- Vickers, K.C.; Roteta, L.A.; Hucheson-Dilks, H.; Han, L.; Guo, Y. Mining diverse small RNA species in the deep transcriptome. Trends Biochem. Sci. 2015, 40, 4–7. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Vickers, K.C.; Samuels, D.C.; Guo, Y. Alternative applications for distinct RNA sequencing strategies. Brief. Bioinform. 2014, 16, 629–639. [Google Scholar] [CrossRef] [PubMed]
- Morin, R.D.; O’Connor, M.D.; Griffith, M.; Kuchenbauer, F.; Delaney, A.; Prabhu, A.L.; Zhao, Y.J.; McDonald, H.; Zeng, T.; Hirst, M.; et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells (vol 18, pg 610, 2008). Genome Res. 2009, 19, 958. [Google Scholar]
- Guo, Y.; Strickland, S.A.; Mohan, S.; Li, S.; Bosompem, A.; Vickers, K.C.; Zhao, S.; Sheng, Q.; Kim, A.S. MicroRNAs and tRNA-derived fragments predict the transformation of myelodysplastic syndromes to acute myeloid leukemia. Leuk. Lymphoma 2017, 58, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Silva, M.R.; Cabrera-Cabrera, F.; Guida, M.C.; Cayota, A. Hints of tRNA-derived small RNAs role in RNA silencing mechanisms. Genes 2012, 3, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Feng, J.; Liu, Q.; Sun, F.; Tie, Y.; Zhu, J.; Xing, R.; Sun, Z.; Zheng, X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009, 583, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Sheng, Q.; Vickers, K.; Zhao, S.; Wang, J.; Samuels, D.C.; Koues, O.; Shyr, Y.; Guo, Y. Multi-perspective quality control of illumina RNA sequencing data analysis. Brief. Funct. Genom. 2016, 16, 194–204. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Zhao, S.; Sheng, Q.; Ye, F.; Li, J.; Lehmann, B.; Pietenpol, J.; Samuels, D.C.; Shyr, Y. Multi-perspective quality control of illumina exome sequencing data using QC3. Genomics 2014, 103, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Boil. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Berezikov, E.; Robine, N.; Samsonova, A.; Westholm, J.O.; Naqvi, A.; Hung, J.H.; Okamura, K.; Dai, Q.; Bortolamiol-Becet, D.; Martin, R.; et al. Deep annotation of drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res. 2011, 21, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, R.; Vaucheret, H.; Trejo, J.; Bartel, D.P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006, 20, 3407–3425. [Google Scholar] [CrossRef] [PubMed]
- Westholm, J.O.; Ladewig, E.; Okamura, K.; Robine, N.; Lai, E.C. Common and distinct patterns of terminal modifications to mirtrons and canonical microRNAs. RNA 2012, 18, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Larter, C.Z.; Yeh, M.M. Animal models of NASH: Getting both pathology and metabolic context right. J. Gastroenterol. Hepatol. 2008, 23, 1635–1648. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S. Mirbase: The microRNA sequence database. Methods Mol. Biol. 2006, 342, 129–138. [Google Scholar] [PubMed]
- Chan, P.P.; Lowe, T.M. GtRNAdb: A database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009, 37, D93–D97. [Google Scholar] [CrossRef] [PubMed]
- Flicek, P.; Amode, M.R.; Barrell, D.; Beal, K.; Billis, K.; Brent, S.; Carvalho-Silva, D.; Clapham, P.; Coates, G.; Fitzgerald, S.; et al. Ensembl 2014. Nucleic Acids Res. 2014, 42, D749–D755. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. Tophat: Discovering splice junctions with RNA-seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. Htseq-a python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Zhao, S.; Ye, F.; Sheng, Q.; Shyr, Y. Multirankseq: Multiperspective approach for RNAseq differential expression analysis and quality control. BioMed Res. Int. 2014, 2014, 248090. [Google Scholar] [CrossRef] [PubMed]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. Diana-mirpath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, J.; Gray, W.H.; Lehmann, B.D.; Bauer, J.A.; Shyr, Y.; Pietenpol, J.A. Tnbctype: A subtyping tool for triple-negative breast cancer. Cancer Inform. 2012, 11, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Diribarne, G.; Bensaude, O. 7SK RNA, a non-coding RNA regulating P-TEFB, a general transcription factor. RNA Biol. 2009, 6, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Christov, C.P.; Trivier, E.; Krude, T. Noncoding human Y RNAs are overexpressed in tumours and required for cell proliferation. Br. J. Cancer 2008, 98, 981–988. [Google Scholar] [CrossRef] [PubMed]
- Christov, C.P.; Gardiner, T.J.; Szuts, D.; Krude, T. Functional requirement of noncoding Y RNAs for human chromosomal DNA replication. Mol. Cell. Biol. 2006, 26, 6993–7004. [Google Scholar] [CrossRef] [PubMed]
- Lerner, M.R.; Boyle, J.A.; Hardin, J.A.; Steitz, J.A. Two novel classes of small ribonucleoproteins detected by antibodies associated with lupus erythematosus. Science 1981, 211, 400–402. [Google Scholar] [CrossRef] [PubMed]
- Mamdani, M.; Williamson, V.; McMichael, G.O.; Blevins, T.; Aliev, F.; Adkins, A.; Hack, L.; Bigdeli, T.; van der Vaart, A.D.; Web, B.T.; et al. Integrating mRNA and miRNA weighted gene co-expression networks with eQTLs in the nucleus accumbens of subjects with alcohol dependence. PLoS ONE 2015, 10, e0137671. [Google Scholar] [CrossRef] [PubMed]
- Nunez, Y.O.; Truitt, J.M.; Gorini, G.; Ponomareva, O.N.; Blednov, Y.A.; Harris, R.A.; Mayfield, R.D. Positively correlated miRNA-mRNA regulatory networks in mouse frontal cortex during early stages of alcohol dependence. BMC Genom. 2013, 14, 725. [Google Scholar] [CrossRef] [PubMed]
- Tu, K.; Yu, H.; Hua, Y.J.; Li, Y.Y.; Liu, L.; Xie, L.; Li, Y.X. Combinatorial network of primary and secondary microRNA-driven regulatory mechanisms. Nucleic Acids Res. 2009, 37, 5969–5980. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, R.; Gonzalez-Angulo, A.M.; Myhre, S.; Carey, M.; Lee, J.S.; Overgaard, J.; Alsner, J.; Stemke-Hale, K.; Lluch, A.; Neve, R.M.; et al. Integrative analysis of cyclin protein levels identifies cyclin b1 as a classifier and predictor of outcomes in breast cancer. Clin. Cancer Res. 2009, 15, 3654–3662. [Google Scholar] [CrossRef] [PubMed]
- Stefansson, O.A.; Jonasson, J.G.; Olafsdottir, K.; Hilmarsdottir, H.; Olafsdottir, G.; Esteller, M.; Johannsson, O.T.; Eyfjord, J.E. CpG island hypermethylation of BRCA1 and loss of pRb as co-occurring events in basal/triple-negative breast cancer. Epigenetics 2011, 6, 638–649. [Google Scholar] [CrossRef] [PubMed]
- Subhawong, A.P.; Subhawong, T.; Nassar, H.; Kouprina, N.; Begum, S.; Vang, R.; Westra, W.H.; Argani, P. Most basal-like breast carcinomas demonstrate the same Rb-/p16+immunophenotype as the HPV-related poorly differentiated squamous cell carcinomas which they resemble morphologically. Am. J. Surg. Pathol. 2009, 33, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.T.; Langley, A.R.; Christov, C.P.; Kheir, E.; Shafee, T.; Gardiner, T.J.; Krude, T. Dynamic interaction of y RNAs with chromatin and initiation proteins during human DNA replication. J. Cell Sci. 2011, 124, 2058–2069. [Google Scholar] [CrossRef] [PubMed]
- Bazeley, P.S.; Shepelev, V.; Talebizadeh, Z.; Butler, M.G.; Fedorova, L.; Filatov, V.; Fedorov, A. SnoTARGET shows that human orphan snoRNA targets locate close to alternative splice junctions. Gene 2008, 408, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.S.; Mao, X.H.; Shi, P.Y.; He, B.Y.; Xu, K.; Zhang, S.M.; Wang, J.M. MicroRNAs in the prognosis of triple-negative breast cancer a systematic review and meta-analysis. Medicine 2017, 96, e7085. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.L.; Zhang, Y.C.; Li, Q.F.; Li, J.F.; Ma, X.T.; Xing, J.F.; Rong, S.H.; Wu, Z.; Tian, Y.; Li, J.; et al. MiRNAs predict the prognosis of patients with triple negative breast cancer: A meta-analysis. PLoS ONE 2017, 12, e0170088. [Google Scholar] [CrossRef] [PubMed]
- Tuttle, T.R.; Mierzwa, M.L.; Wells, S.I.; Fox, S.R.; Ben-Jonathan, N. The cyclic GMP/protein kinase G pathway as a therapeutic target in head and neck squamous cell carcinoma. Cancer Lett. 2016, 370, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Swinnen, J.V.; Esquenet, M.; Goossens, K.; Heyns, W.; Verhoeven, G. Androgens stimulate fatty acid synthase in the human prostate cancer cell line LNCaP. Cancer Res. 1997, 57, 1086–1090. [Google Scholar] [PubMed]
- Itkonen, H.M.; Mills, I.G. N-linked glycosylation supports cross-talk between receptor tyrosine kinases and androgen receptor. PLoS ONE 2013, 8, e65016. [Google Scholar] [CrossRef] [PubMed]
- Munkley, J.; Vodak, D.; Livermore, K.E.; James, K.; Wilson, B.T.; Knight, B.; McCullagh, P.; Mcgrath, J.; Crundwell, M.; Harries, L.W.; et al. Glycosylation is an androgen-regulated process essential for prostate cancer cell viability. Ebiomedicine 2016, 8, 103–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Truong, H.H.; Xiong, J.L.; Ghotra, V.P.S.; Nirmala, E.; Haazen, L.; Le Devedec, S.E.; Balcioglu, H.E.; He, S.N.; Snaar-Jagalska, B.E.; Vreugdenhil, E.; et al. Beta(1) integrin inhibition elicits a prometastatic switch through the TGF beta-miR-200-ZEB network in E-cadherin-positive triple-negative breast cancer. Sci. Signal 2014, 7, ra15. [Google Scholar] [CrossRef] [PubMed]
- Trahan, C.; Dragon, F. Dyskeratosis congenita mutations in the H/ACA domain of human telomerase RNA affect its assembly into a pre-RNP. RNA 2009, 15, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Traina, T.A.; Miller, K.; Yardley, D.A.; O’Shaughnessy, J.; Cortes, J.; Awada, A.; Kelly, C.M.; Trudeau, M.E.; Schmid, P.; Gianni, L.; et al. Results from a phase 2 study of enzalutamide (ENZA), an androgen receptor (AR) inhibitor, in advanced ar plus triplenegative breast cancer (TNBC). J. Clin. Oncol. 2015, 33, 1003. [Google Scholar]
- Tosar, J.P.; Gambaro, F.; Sanguinetti, J.; Bonilla, B.; Witwer, K.W.; Cayota, A. Assessment of small RNA sorting into different extracellular fractions revealed by high-throughput sequencing of breast cell lines. Nucleic Acids Res. 2015, 43, 5601–5616. [Google Scholar] [CrossRef] [PubMed]
- Dhahbi, J.M.; Spindler, S.R.; Atamna, H.; Boffelli, D.; Martin, D.I. Deep sequencing of serum small RNAs identifies patterns of 5′ tRNA half and yRNA fragment expression associated with breast cancer. Biomark. Cancer 2014, 6, 37–47. [Google Scholar] [CrossRef] [PubMed]


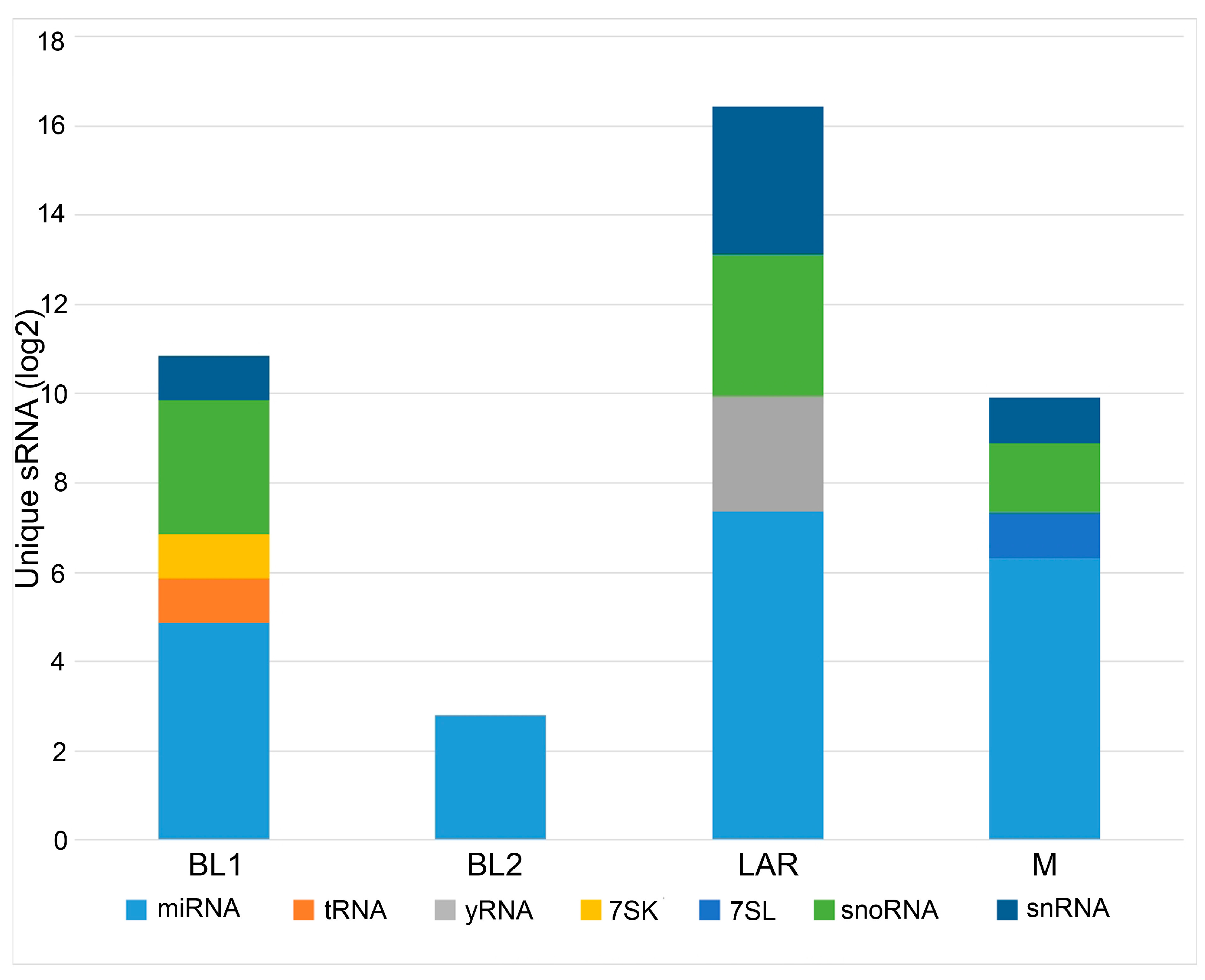
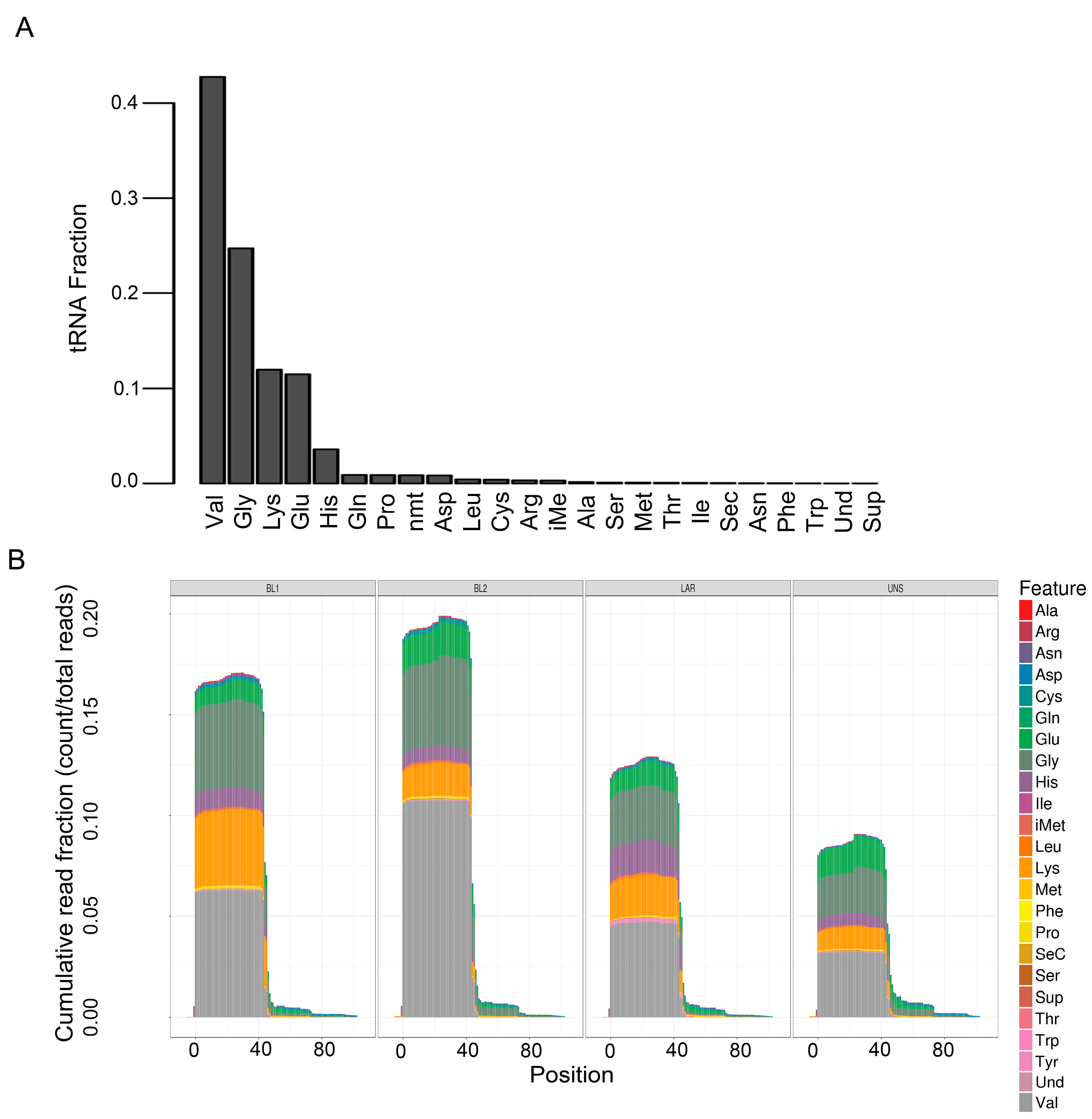
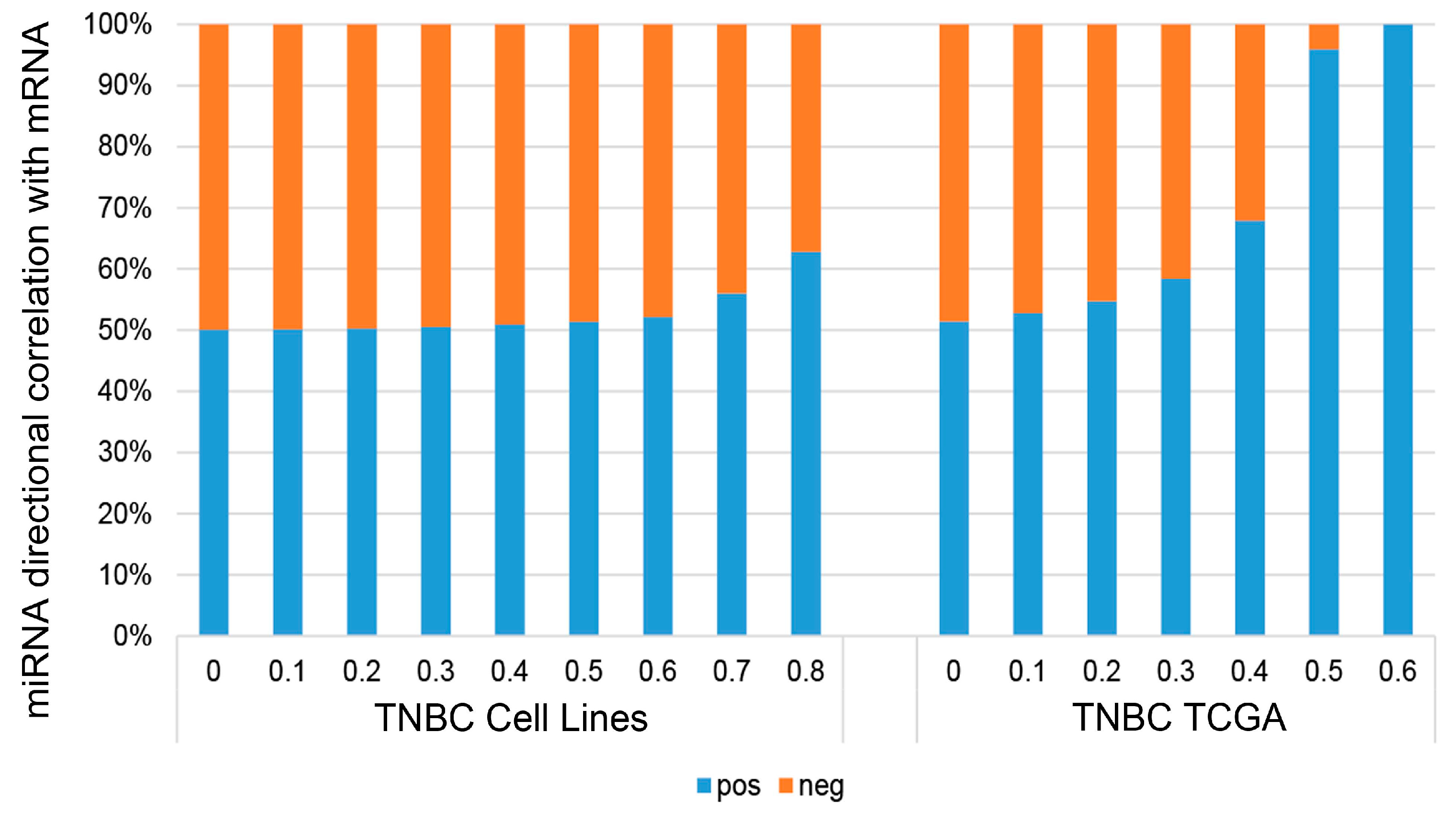
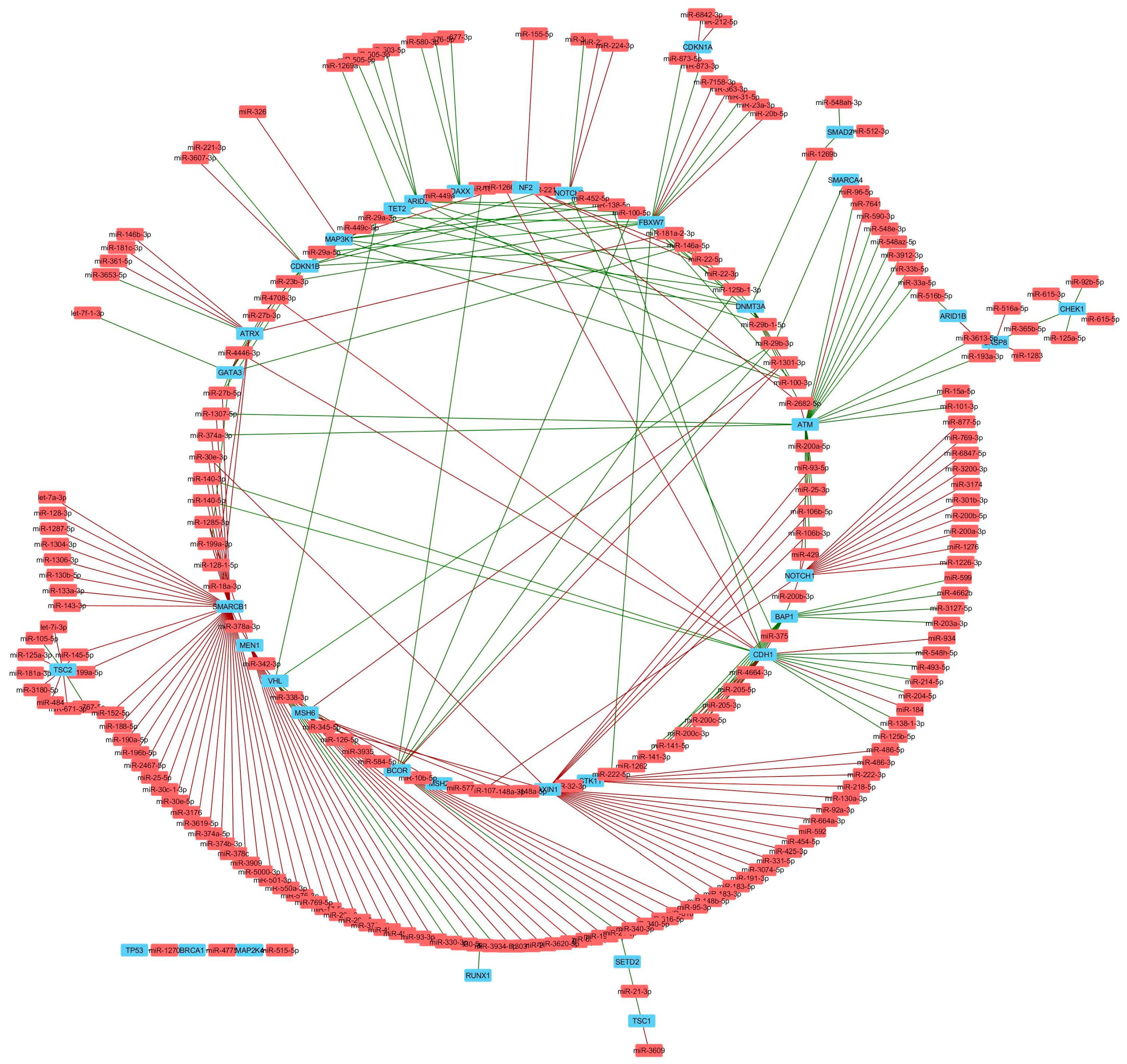

| Comparison | miRNA | isomiR | tRNA | snRNA | snoRNA | yRNA | 7SK | 7SL | Total |
|---|---|---|---|---|---|---|---|---|---|
| BL1 vs. BL2 | 29 | 33 | 1 | 1 | 6 | 0 | 1 | 0 | 71 |
| LAR vs. BL2 | 82 | 74 | 0 | 9 | 2 | 0 | 0 | 0 | 167 |
| LAR vs. BL1 | 49 | 40 | 0 | 9 | 10 | 2 | 1 | 0 | 111 |
| M vs. BL2 | 26 | 16 | 0 | 2 | 6 | 0 | 0 | 0 | 50 |
| M vs. BL1 | 71 | 84 | 2 | 3 | 15 | 0 | 4 | 2 | 181 |
| M vs. LAR | 105 | 105 | 0 | 5 | 19 | 5 | 0 | 2 | 241 |
| Total | 362 | 352 | 3 | 29 | 58 | 7 | 6 | 4 | 821 |
| Comparison | miRNA | isomiR | tRNA | snRNA | snoRNA | yRNA | 7SK | 7SL | Total |
|---|---|---|---|---|---|---|---|---|---|
| BL1 vs. others | 18 | 0 | 2 | 2 | 8 | 0 | 2 | 0 | 32 |
| BL2 vs. others | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 |
| M vs. others | 41 | 0 | 0 | 2 | 3 | 0 | 0 | 2 | 48 |
| LAR vs. others | 82 | 0 | 0 | 10 | 6 | 6 | 0 | 0 | 104 |
| Subtype | Gene Class | Decreased miRNA Target | Increased miRNA |
|---|---|---|---|
| BL1 | Oncogene | H3F3A | HRAS, MDM4 |
| Tumor Suppressor | DNMT3A | CDH1, BCOR, BAP1, FBXW7 | |
| BL2 | Oncogene | EIF4A2, PPP2R1A | |
| Tumor Suppressor | FBXW7 | ||
| M | Oncogene | MDM4, HRAS | CDK4, EIF4A2, FGFR1, H3F3A, MDM4, MYB, MYCL1, NRAS, |
| Tumor Suppressor | FBXW7, CDH1 | NOTCH2, SMAD2 | |
| LAR | Oncogene | EIF4A2, HF3F3A, MDM2, MDM4, MYCL1 | CDK4, EIF4A2, ETV1, FGFR1, IDH2, MDM2, MDM4 |
| Tumor Suppressor | GATA3, VHL | FBXW7, NOTCH2 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Y.; Yu, H.; Wang, J.; Sheng, Q.; Zhao, S.; Zhao, Y.-Y.; Lehmann, B.D. The Landscape of Small Non-Coding RNAs in Triple-Negative Breast Cancer. Genes 2018, 9, 29. https://doi.org/10.3390/genes9010029
Guo Y, Yu H, Wang J, Sheng Q, Zhao S, Zhao Y-Y, Lehmann BD. The Landscape of Small Non-Coding RNAs in Triple-Negative Breast Cancer. Genes. 2018; 9(1):29. https://doi.org/10.3390/genes9010029
Chicago/Turabian StyleGuo, Yan, Hui Yu, Jing Wang, Quanhu Sheng, Shilin Zhao, Ying-Yong Zhao, and Brian D. Lehmann. 2018. "The Landscape of Small Non-Coding RNAs in Triple-Negative Breast Cancer" Genes 9, no. 1: 29. https://doi.org/10.3390/genes9010029






