Phosphate Deficiency Negatively Affects Early Steps of the Symbiosis between Common Bean and Rhizobia
Abstract
1. Introduction
2. Material and Methods
2.1. Plant Material
2.2. Bacterial Strains and Culture Conditions
2.3. Treatment
2.4. Quantification of Soluble Phosphate
2.5. Root Hair Deformation Analysis
2.6. Quantification of Number of Infection Threads
2.7. Gene Expression Analysis
2.8. Preparation of Messenger RNA-Seq Library and High-Throughput Sequencing
2.9. Mapping and Processing mRNA-Seq Reads
2.10. Quantification and Identification of Differentially Regulated Genes
2.11. Gene Functional Classification
2.12. Statistical Analyses
3. Results and Discussion
3.1. Rhizobia-Induced Root Hair Deformation and Infection Thread Formation is Affected in Pi-Deficient Common Bean Seedlings
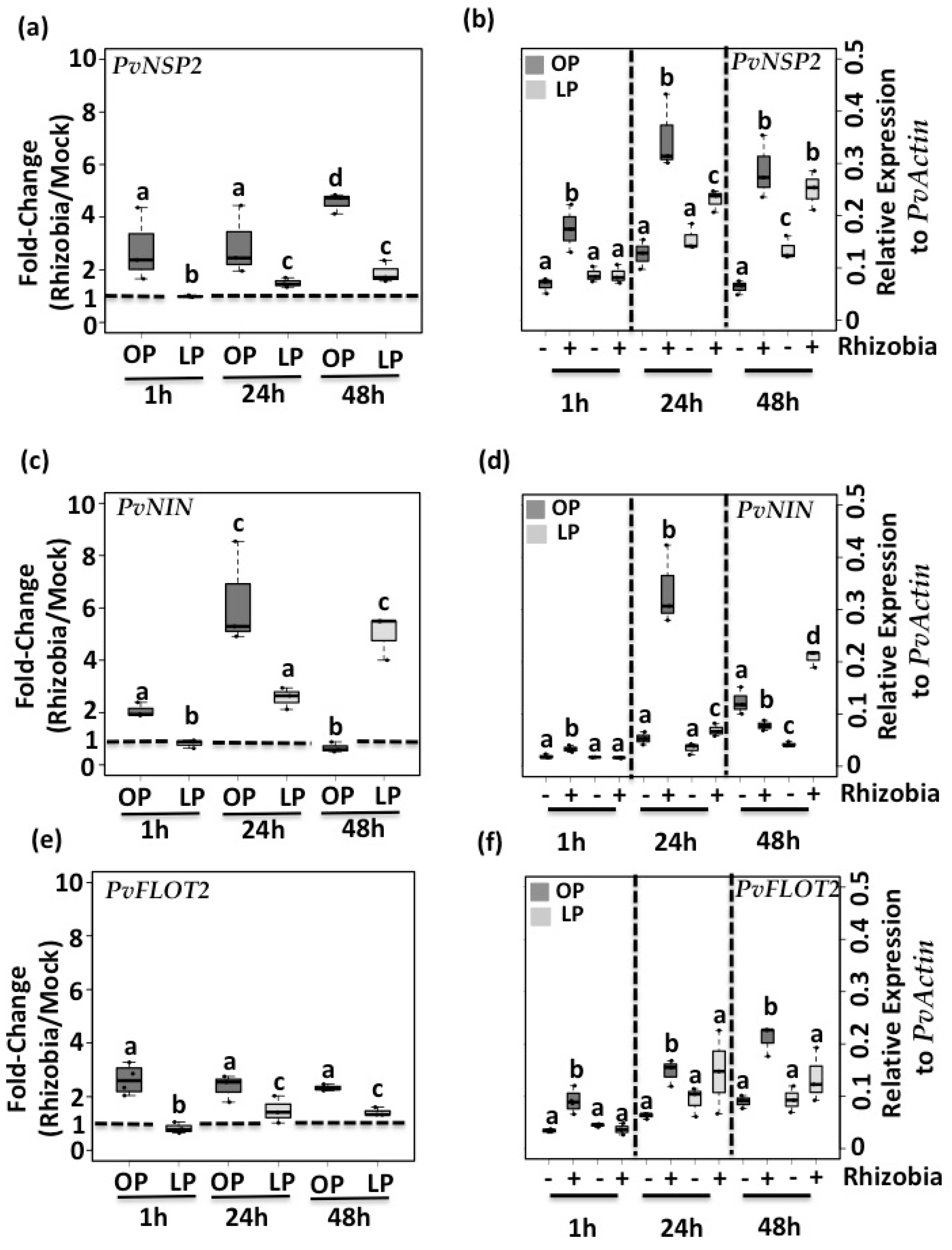
3.2. Pi Deficiency Decreases the Expression of Genes Required for Root Hair Curling and Rhizobia Infection
3.3. Pi Deficiency Increases the Expression of the PvRIC1 and PvRIC2 Genes in Common Bean Seedlings
3.4. mRNA-Seq Analysis
3.5. Global Transcriptional Responses of Pi-Deficient Common Bean Seedlings Interacting with Rhizobia
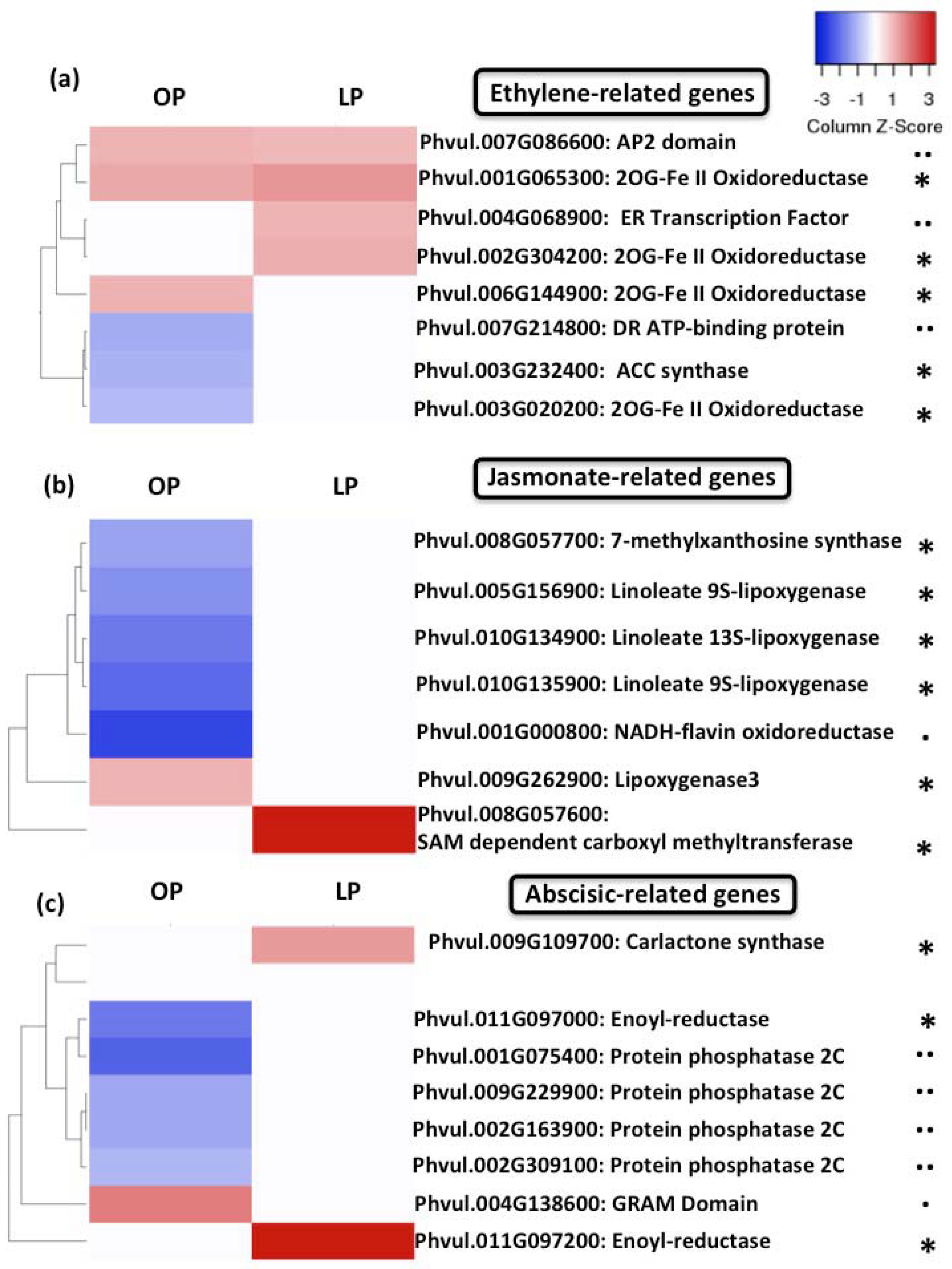
3.6. Expression of Hormone- and Signal-Transduction-Related Genes Is Compromised in Rhizobia Inoculated Pi-Deficient Common Bean Plants
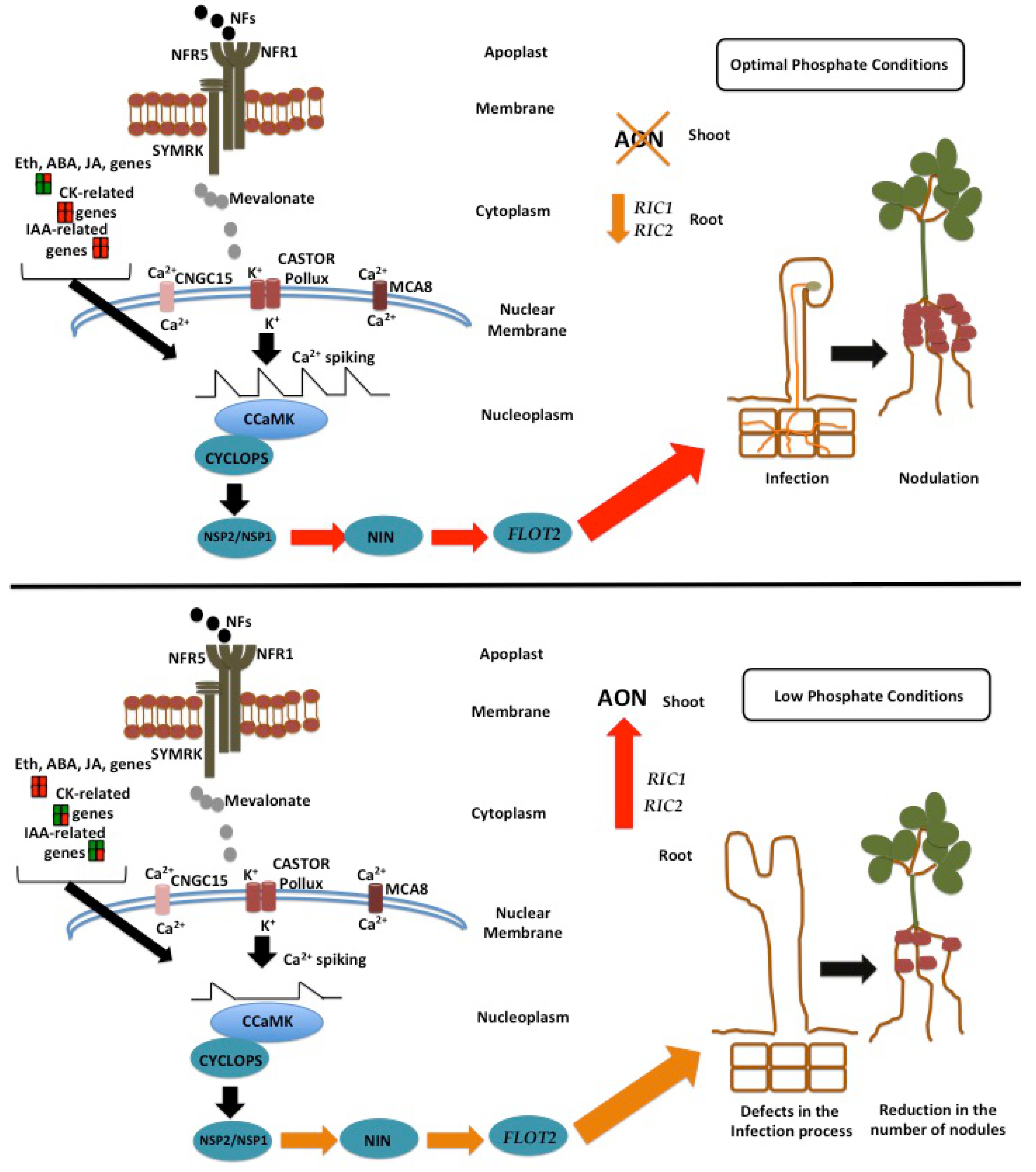
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Castro-Guerrero, N.A.; Isidra-Arellano, M.C.; Mendoza-Cozatl, D.M.; Valdés-López, O. Common bean: A legume model on the rise for unraveling adaptations to iron, zinc and phosphate deficiencies. Front. Plant Sci. 2016, 7, 600. [Google Scholar] [CrossRef] [PubMed]
- Blair, M.W.; Izquierdo, P.; Astudillo, C.; Grusak, M.A. A legume biofortification quandary: Variability and genetic control of seed coat micronutrient accumulation in common beans. Front. Plant Sci. 2013, 4, 275. [Google Scholar] [CrossRef] [PubMed]
- Jha, A.B.; Ashokkumar, K.; Diapari, M.; Ambrose, S.J.; Zhang, H.; Tar’an, B.; Bett, K.E.; Vandenberg, A.; Warkentin, T.D.; Purves, R.W. Genetic diversity of folate profiles in seeds of common bean, lentil, chickpea and pea. J. Food Compos. Anal. 2015, 42, 134–140. [Google Scholar] [CrossRef]
- Peters, N.K.; Frost, J.N.; Long, S.R. A plant flavonone, loteolin, induces expression of Rhizobium meliloti genes. Sciences 1996, 233, 977–980. [Google Scholar] [CrossRef]
- Liu, C.W.; Murray, J. The role of flavonoids in nodulation host-range specificity and update. Plants 2016, 5, 33. [Google Scholar] [CrossRef] [PubMed]
- Broghanmer, A.; Krusell, L.; Blaise, M.; Sauer, J.; Sullivan, J.T.; Maolanon, N.; Vinther, M.; Lorentzen, A.; Madsen, E.B.; Jensen, K.J.; et al. Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc. Natl. Acad. Sci. USA 2012, 109, 13859–13864. [Google Scholar] [CrossRef] [PubMed]
- Murakami, E.; Cheng, J.; Gysel, K.; Bozsoki, Z.; Kawaharada, Y.; Hjuler, C.T.; Sørensen, K.K.; Tao, K.; Kelly, S.; Venice, F.; et al. Epidermal LysM receptor ensure robust symbiotic signalling in Lotus japonicus. eLife 2018, 7, e33506. [Google Scholar] [CrossRef] [PubMed]
- Venkateshwaran, M.; Volkening, J.D.; Sussman, M.R.; Ané, J.M. Symbiosis and the social network of higher plants. Curr. Opin. Plant Biol. 2013, 16, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Lei, M.J.; Wang, Q.; Li, X.; Chen, A.; Luo, L.; Xie, Y.; Li, G.; Luo, D.; Mysore, K.S.; Wen, J.; et al. The small GTPase ROP10 of Medicago truncatula is required for both tip growth of root hairs and Nod Factor-induced root hair deformation. Plant Cell 2015, 27, 806–822. [Google Scholar] [CrossRef] [PubMed]
- Nadzieja, M.; Kelly, S.; Stougaard, J.; Reid, D. Epidermal auxin biosynthesis facilitates rhizobial infection in Lotus japonicus. Plant J. 2018, 95, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Fournier, J.; Timmers, A.C.J.; Sieberer, B.J.; Jauneau, A.; Chabaud, M.; Barker, D.G. Mechanism of infection thread elongation in root hairs of Medicago truncatula and dynamic interplay with associated rhizobial colonization. Plant Physiol. 2008, 148, 1985–1995. [Google Scholar] [CrossRef] [PubMed]
- Fournier, J.; Teillet, A.; Chabaud, M.; Ivanov, S.; Genre, A.; Limpens, E.; de Carvalho-Nievel, F.; Barker, D. Remodeling of the infection chamber before infection thread formation reveals a two-step mechanism for rhizobial entry into the host legume root hair. Plant Physiol. 2015, 167, 1233–1242. [Google Scholar] [CrossRef] [PubMed]
- Xiao, T.T.; Schilderink, S.; Moling, S.; Deinum, E.E.; Kondorosi, E.; Franssen, H.; Kulikova, O.; Niebel, A.; Bisseling, T. Fate map of Medicago truncatula root nodules. Development 2014, 141, 3517–3528. [Google Scholar] [CrossRef] [PubMed]
- Genre, A.; Russo, G. Does a common pathway transduce symbiotic signals in plant-microbe interactions? Front. Plant Sci. 2016, 7, 96. [Google Scholar] [CrossRef] [PubMed]
- Bersoult, A.; Camut, S.; Perhald, A.; Kereszt, A.; Kiss, G.B.; Cullimore, J.V. Expression of the Medicago truncatula DM12 gene suggests roles of the symbiotic nodulation receptor kinase in nodules and during early nodule development. Mol. Plant Microbe Interact. 2005, 18, 869–876. [Google Scholar] [CrossRef] [PubMed]
- Hogg, B.V.; Cullimore, J.V.; Ranjeva, R.; Bono, J.J. The DMI1 and DMI2 early symbiotic genes of Medicago truncatula are required for a high-affinity nodulation factor-binding site associate to a particulate fraction of roots. Plant Physiol. 2006, 140, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Imaizumi-Anraku, H.; Takeda, N.; Charpentier, M.; Perry, J.; Miwa, H.; Umehara, Y.; Kouchi, H.; Murakami, Y.; Mulder, L.; Vickers, K.; et al. Plastid proteins crucial for symbiotic fungal and bacterial entry into plant host. Nature 2005, 433, 527–531. [Google Scholar] [CrossRef] [PubMed]
- Kanamori, N.; Madsen, L.H.; Radatoiu, S.; Frantescu, M.; Quistgaard, E.M.; Miwa, H.; Downie, J.A.; James, E.K.; Felle, H.H.; Haaning, L.L.; et al. A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc. Natl. Acad. Sci. USA 2006, 103, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Yoshikawa, M.; Yano, K.; Miwa, H.; Uchida, H.; Asamizu, E.; Sato, S.; Tabata, S.; Imaizumi-Anraku, H.; Kouchi, H.; et al. NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell 2007, 19, 610–624. [Google Scholar] [CrossRef] [PubMed]
- Groth, M.; Takeda, N.; Perry, J.; Uchida, H.; Dräxl, S.; Brachmann, A.; Sato, S.; Tabata, S.; Kawaguchi, M.; Wang, T.L.; et al. NENA, a Lotus japonicus homolog of Sec13, is required for rhizodermal infection by arbuscular mycorrhiza fungi and rhizobia but dispensable for cortical endosymbiotic development. Plant Cell 2010, 22, 2509–2526. [Google Scholar] [CrossRef] [PubMed]
- Charpentier, M.; Sun, J.; Vaz Martins, T.; Radhakrishnan, G.V.; Findlay, K.; Soumpourou, E.; Thouin, J.; Véry, A.A.; Sanders, D.; Morris, R.J.; et al. Nuclear-localized cyclic nucleotide-gated channels mediate symbiotic calcium oscillations. Science 2016, 352, 1102–1105. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.B.; Pratap, A.; Miyahara, A.; Zhou, L.; Borneman, S.; Morris, R.J.; Oldroyd, G.E. Calcium/Calmodulin-dependent protein kinase is negatively and positively regulated by calcium, providing a mechanism for decoding calcium responses during symbiosis signaling. Plant Cell 2013, 25, 5053–5066. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Katzer, K.; Lambert, J.; Cerri, M.; Parniske, M. CYCLOPs, a DNA-binding transcriptional activator orchestrates symbiotic root nodule development. Cell Host Microbe 2014, 15, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, S.; Kim, J.; Muñoz, A.; Heckmann, A.B.; Downie, J.A.; Oldroyd, G.E.D. GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 2009, 21, 545–557. [Google Scholar] [CrossRef] [PubMed]
- Soyano, T.; Kouchi, H.; Hirota, A.; Hayashi, M. Nodule Inception directly targets NF-Y sub-unit genes to regulate essential processes of root nodule development in Lotus japonicus. PLoS Genet. 2013, 9, e1003352. [Google Scholar] [CrossRef] [PubMed]
- Haney, C.H.; Long, S.R. Plant flotillins are required for infection by nitrogen-fixing bacteria. Proc. Natl. Acad. Sci. USA 2010, 107, 478–483. [Google Scholar] [CrossRef] [PubMed]
- van Zeijl, A.; Op den Camp, R.H.M.; Deinum, E.E.; Charnikhova, T.; Franssen, H.; Op den Camp, H.J.M.; Bouwmeester, H.; Kohlen, W.; Bisseling, T.; Geurts, R. Rhizobium lipo-chitooligosaccharide signaling triggers accumulation of cytokinins in Medicago truncatula roots. Mol. Plant 2015, 8, 1213–1226. [Google Scholar] [CrossRef] [PubMed]
- Reid, D.; Liu, H.; Kelly, S.; Kawaharada, Y.; Mun, T.; Andersen, S.U.; Desbrosses, G.; Stougaard, J. Dynamics of Ethylene production in response to compatible Nod Factor. Plant Physiol. 2018, 176, 1764–1772. [Google Scholar] [CrossRef] [PubMed]
- Penmetsa, V.R.; Cook, D.R. A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 1997, 275, 527–530. [Google Scholar] [CrossRef] [PubMed]
- Trichine, L.; Sandal, N.; Madsen, L.H.; Radutoiu, S.; Albrektsen, A.S.; Sato, S.; Asamizu, E.; Tabata, S.; Stougaard, J. A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 2007, 315, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, B.J.; Mens, C.; Hastwell, A.H.; Zhang, M.; Su, H.; Jones, C.H.; Chu, X.; Gresshoff, P.M. Legume nodulation: The host controls the party. Plant Cell Environ. 2018, 1, 11. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, B.J.; Dongxue, L.; Hastwell, A.H.; Reid, D.E.; Li, Y.; Jackson, S.A.; Gresshoff, P. The soybean (Glycine max) nodulation-suppressive CLE peptide, GmRIC1, functions interspecifically in common white bean (Phaseolus vulgaris), but not in a suppernodulating line mutated in the receptor PvNARK. Plant Biotechnol. J. 2014, 12, 1085–1097. [Google Scholar] [CrossRef] [PubMed]
- Krusell, L.; Madsen, L.H.; Sato, S.; Aubert, G.; Genua, A.; Szczyglowski, K.; Duc, G.; Kaneko, T.; Tabata, S.; de Bruijn, F.; et al. Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 2002, 420, 422–426. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, R.; Hayashi, M.; Wu, G.J.; Kouchi, H.; Imaizumi-Anraku, H.; Murakami, Y.; Kawasaki, S.; Akao, S.; Ohmori, M.; Nagasawa, M.; et al. HAR1 mediates systemic regulation of symbiotic organ development. Nature 2002, 420, 426–429. [Google Scholar] [CrossRef] [PubMed]
- Searle, I.R.; Men, A.E.; Laniya, T.S.; Buzas, D.M.; Iturbe-Ormaetxe, I.; Carroll, B.J.; Gresshoff, P.M. Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 2003, 299, 109–112. [Google Scholar] [CrossRef] [PubMed]
- Schnabel, E.; Journet, E.P.; de Carvalho-Niebel, F.; Duc, G.; Frugoli, J. The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol. Biol. 2005, 58, 809–822. [Google Scholar] [CrossRef] [PubMed]
- Broughton, W.J.; Hernandez, G.; Blair, M.; Beebe, S.; Gepts, P.; Vanderleyden, J. Beans (Phaseolus spp.)—Model food legumes. Plant Soil 2003, 252, 55–128. [Google Scholar] [CrossRef]
- Valdés-López, O.; Hernández, G. Transcriptional regulation and signaling in phosphorus starvation: What about legumes? J. Integr. Plant Biol. 2008, 50, 1213–1222. [Google Scholar] [CrossRef] [PubMed]
- Vance, C.P.; Uhde-Stone, C.; Allan, D.L. Phosphorus acquisition and use: Critical adaptations by plants for securing a nonrenewable resource. New Phytol. 2003, 157, 423–447. [Google Scholar] [CrossRef]
- Hernández, G.; Valdés-López, O.; Ramírez, M.; Goffard, N.; Weiller, G.; Aparicio-Fabre, R.; Fuentes, S.I.; Erban, A.; Kopka, J.; Udvardi, M.K.; et al. Global changes in the transcript and metabolic profiles during symbiotic nitrogen fixation in phosphorus-stressed common bean plants. Plant Physiol. 2009, 151, 1221–1238. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, G.; Devron, J.J. Influence of oxygen and acetylene during in situ open-flow assays of nitrogenase activity (C2H2 reduction) in Phaseolus vulgaris root nodule. J. Plant Physiol. 1991, 138, 587–590. [Google Scholar] [CrossRef]
- Vadez, V.; Rodier, F.; Payre, H.; Devon, J.J. Nodule conductance to O2 and nitrogenase-linked respiration in bean genotypes varying in the tolerance of N2 fixation to P deficiency. Plant Physiol. Biochem. 1996, 34, 871–878. [Google Scholar]
- Cabeza, R.A.; Liese, R.; Lingner, A.; von Steieglitz, I.; Neumann, J.; Salinas-Riester, G.; Pommerenke, C.; Dittert, K.; Schulze, J. RNA-seq transcriptome profiling reveals that Medicago trucatula nodules acclimate N2 fixation before emerging P deficiency reaches the nodules. J. Exp. Bot. 2014, 65, 6035–6048. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Cui, Q.; Liang, C.; Sun, L.; Tian, J.; Liao, H. Identification of differentially expressed proteins in soybean nodules under phosphorus deficiency through proteomics analysis. Proteomics 2011, 11, 4648–4659. [Google Scholar] [CrossRef] [PubMed]
- Suleiman, S.; Ha, C.V.; Schulze, J.; Tran, L.S.P. Growth and nodulation of symbiotic Medicago truncatula at different levels of phosphorus availability. J. Exp. Bot. 2013, 64, 2701–2712. [Google Scholar] [CrossRef] [PubMed]
- Nasr Esfahani, M.; Inoue, K.; Chu, H.D.; Nguyen, K.H.; Van Ha, C.; Watanabe, Y.; Burritt, D.J.; Herrera-Estrella, L.; Mochida, K.; Tran, L.P. Comparative transcriptome analysis of nodules of two Mesorhizobium-chickpea associations with differential symbiotic efficiency under phosphate deficiency. Plant J. 2017, 91, 911–926. [Google Scholar] [CrossRef] [PubMed]
- Sa, T.; Israel, D.W. Energy status and functioning of phosphorus-deficient soybean nodules. Plant Physiol. 1991, 97, 928–935. [Google Scholar] [CrossRef] [PubMed]
- Al-Niemi, T.S.; Kahn, M.L.; McDermott, T.R. P metabolism in the bean-Rhizobium tropicisymbiosis. Plant Physiol. 1997, 113, 1233–1242. [Google Scholar] [CrossRef] [PubMed]
- Vadez, V.; Beck, D.P.; Lasso, J.H.; Devron, J.J. Utilization of the acetylene reduction assay to screen for tolerance of symbiotic N2 fixation to limiting P nutrition in common bean. Physiol. Plant 1997, 99, 227–232. [Google Scholar] [CrossRef]
- Tang, C.; Hinsinger, P.; Drevon, J.J.; Jaillard, B. Phosphorus deficiency impairs early nodule functioning and enhances proton release in roots of Medicago truncatula L. Ann. Bot. 2001, 88, 131–138. [Google Scholar] [CrossRef]
- Hogh-Jensen, H.; Schjoerring, J.K.; Soussana, J.F. The influence of phosphorus deficiency on growth and nitrogen fixation of white clover plants. Ann. Bot. 2002, 90, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Schulze, J.; Temple, G.; Temple, S.; Beschow, H.; Vance, C.P. Nitrogen fixation by white lupin under phosphorus deficiency. Ann. Bot. 2006, 98, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Catoira, R.; Galera, C.; de Billy, F.; Penmetsa, R.V.; Journet, E.P.; Maillet, F.; Rosenberg, C.; Cood, D.; Gough, C.; Dénarié, J. Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 2000, 12, 1647–1665. [Google Scholar] [CrossRef] [PubMed]
- Dalla Via, V.; Narduzzi, C.; Aguilar, O.M.; Zanetti, M.E.; Blanco, A. Changes in the common bean transcriptome in response to secreted and surface signal molecules of Rhizobium etli. Plant Physiol. 2015, 169, 1356–1370. [Google Scholar] [CrossRef] [PubMed]
- Taussky, H.H.; Shorr, E. A microcolorimetric method for determination of inorganic phosphorus. J. Biol. Chem. 1953, 202, 675–685. [Google Scholar] [PubMed]
- Reyero-Saavedra, M.D.R.; Qiao, Z.; Sánchez-Correa, M.D.S.; Díaz-Pineda, M.E.; Reyes, J.L.; Covarrubias, A.A.; Libault, M.; Valdés-López, O. Gene silencing of Argonaute5 negatively affects the establishment of the legume-rhizobia. Genes 2017, 8, 352. [Google Scholar] [CrossRef] [PubMed]
- Ramakers, C.; Ruijter, M.J.; Deprez, H.R.; Moorman, A.F. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Primer3. Available online: http://bioinfo.ut.ee/primer3-0.4.0/primer3/ (accessed on 10 July 2017).
- Schmutz, J.; McClean, P.E.; Mamidi, S.; Wu, G.A.; Cannon, S.B.; Grimwood, J.; Chavarro, C.; Torres-Torres, M.; Geffroy, V.; Moghaddam, S.M.; et al. A reference genome for common bean and geno-wide analysis of dual domestications. Nat. Genet. 2014, 46, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Phytozome. Available online: https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Pvulgaris (accessed on 8 June 2017).
- Barbraham Bionformatics. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 8 June 2017).
- Babraham Bioinformatics. Available online: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 8 June 2017).
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Pachter, L.; Sazlberg, S.L. TopHat: Discovering splice junctions with RNA-seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, C.; Salzberg, S.L.; Rinn, J.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protocols 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Zhou, X.; Liang, Y.; Zhang, Z.; Su, Z. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010, 38, W64–W70. [Google Scholar] [CrossRef] [PubMed]
- AGRIGO. Available online: http://systemsbiology.cau.edu.cn/agriGOv2/ (accessed on 20 August 2017).
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO summarizes and visualizes long list of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed]
- REVIGO. Available online: http://revigo.irb.hr (accessed on 20 August 2017).
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef] [PubMed]
- Usadel, B.; Poree, F.; Nagel, A.; Lohse, M.; Czedik-Eysenberg, A.; Stitt, M. A guide to using MapMan to visualize and compare Omics data in plants: A case study in the crop species, maize. Plant Cell Environ. 2009, 32, 1211–1229. [Google Scholar] [CrossRef] [PubMed]
- MapMan. Available online: https://mapman.gabipd.org (accessed on 15 April 2018).
- Qiu, L.; Lin, J.S.; Xu, J.; Sato, S.; Parniske, M.; Wang, J.; Xie, F. SCARN a novel class of SCAR protein that is required for root-hair infection during legume nodulation. PLoS Genet. 2015, 11, e1005623. [Google Scholar] [CrossRef] [PubMed]
- Cerri, M.R.; Frances, L.; Laloum, T.; Auriac, M.C.; Niebel, A.; Oldroyd, G.E.; Barke, D.G.; Fournier, J.; de Carvalho-Niebel, F. Medicago truncatula ERN transcription factors: Regulatory interplay with NSP1/NSP2 GRAS factors and expression dynamics throughout rhizobia infection. Plant Physiol. 2012, 160, 2155–2172. [Google Scholar] [CrossRef] [PubMed]
- Howieson, J.G.; Robson, A.D.; Ewing, M.A. External phosphate and calcium concentrations and pH, but not the products of rhizobial nodulation genes, affects the attachment of Rhizobium meliloti to roots of annual medics. Soil Biol. Biochem. 1993, 25, 567–573. [Google Scholar] [CrossRef]
- Smit, P.; Raedts, J.; Portyanko, V.; Debellé, F.; Gough, C.; Bisseling, T.; Geurts, R. NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 2005, 308, 1789–1791. [Google Scholar] [CrossRef] [PubMed]
- Marsh, J.F.; Rakocevic, A.; Mitra, R.M.; Brocard, L.; Sun, J.; Eschstruth, A.; Long, S.; Schultze, M.; Ratet, P.; Oldroyd, G.E.D. Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 2007, 144, 324–335. [Google Scholar] [CrossRef] [PubMed]
- Oldroyd, G.E.; Long, S.R. Identification and characterization of nodulation-signalling pathway 2, a gene of Medicago truncatula involved in Nod Factor signaling. Plant Physiol. 2003, 131, 1027–1032. [Google Scholar] [CrossRef] [PubMed]
- Reid, D.E.; Ferguson, B.J.; Gresshoff, P.M. Inoculation- and nitrate-induced CLE peptides of soybean control NARK-dependent Nodule formation. Mol. Plant Microbe Interact. 2011, 24, 606–618. [Google Scholar] [CrossRef] [PubMed]
- Funayama-Noguchi, S.; Noguchi, K.; Yoshida, C.; Kawaguchi, M. Two CLE genes are induced by phosphate in roots of Lotus japonicus. J. Plant Res. 2011, 124, 155–163. [Google Scholar] [CrossRef] [PubMed]
- de Bang, T.C.; Lundquist, P.K.; Dai, X.; Boschiero, C.; Zhuang, Z.; Pant, P.; Torres-Jerez, I.; Roy, S.; Nogales, J.; Veerappan, V.; et al. Genome-Wide identification of Medicago peptides involved in macronutrient responses and nodulation. Plant Physiol. 2017, 175, 1669–1689. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Alanis, D.; Yong, L.; Jiménez, P.; Alatorre-Cobos, F.; Oropeza-Aburto, A.; Mora-Macías, J.; Sánchez-Rodríguez, F.; Cruz-Ramirez, A.; Herrera-Estrella, L. Phosphate Starvation-Dependent Iron Mobilization Induces CLE14 Expression to Trigger Root Meristem Differentiation through CLV2/PEPR2 Signaling. Dev. Cell 2017, 41, 555–570. [Google Scholar] [CrossRef] [PubMed]
- GEO. Available online: https://www.ncbi.nlm.nih.gov/geo/ (accessed on 23 August 2018).
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, B.J.; Mathesius, U. Phytohormone regulation of legume-rhizobia interactions. J. Chem. Ecol. 2014, 40, 770–790. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, A.M.; Bhuvaneswari, T.V.; Torrey, J.G.; Bisseling, T. Early nodulin genes are induced in alfalfa roots outgrowths elicited by auxin transport inhibitors. Proc. Natl. Acad. Sci. USA 1989, 86, 1244–1248. [Google Scholar] [CrossRef] [PubMed]
- Ng, J.L.; Hassan, S.; Truong, T.T.; Hocart, C.H.; Laffont, C.; Frugier, F.; Mathesius, U. Flavonoids and auxin transport inhibitors rescue symbiotic nodulation in Medicago truncatula cytokinin perception cre1. Plant Cell 2015, 27, 2210–2226. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, C.; Yang, J.; Yu, N.; Wang, E. Hormone modulation of legume-rhizobial symbiosis. J. Integr. Plant Biol. 2018, 60, 632–648. [Google Scholar] [CrossRef] [PubMed]
- Rouached, H.; Arpat, A.B.; Poirier, Y. Regulation of phosphate starvation responses in plants: Signaling players and cross-talks. Mol. Plant 2010, 3, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Oldroyd, G.E.D.; Engstrom, E.M.; Long, S.R. Ethylene inhibits the Nod factor signaling transduction pathway of Medicago truncatula. Plant Cell 2001, 13, 1835–1849. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Cardoza, V.; Mitchell, D.M.; Brigth, L.; Oldroyd, G.; Harris, J.M. Crosstalk between jasmonic acid, ethylene and Nod factor signaling allows integrations of diverse inputs for regulation of nodulation. Plant J. 2006, 46, 961–970. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Kalo, P.; Yendrek, C.; Sun, J.; Liang, Y.; Marsh, J.F.; Harris, J.M.; Oldroyd, G.E. Abscisic acid coordinates Nod factors and cytokinins signaling during the regulation of nodulation in Medicago truncatula. Plant Cell 2008, 20, 2681–2695. [Google Scholar] [CrossRef] [PubMed]
- Madsen, E.B.; Madsen, L.H.; Radutoiu, S.; Olbryt, M.; Rakwalska, M.; Szczyglowski, K.; Sato, S.; Kaneko, T.; Tabata, S.; Sandal, N.; et al. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 2003, 425, 637–640. [Google Scholar] [CrossRef] [PubMed]
- Radutoiu, S.; Madsen, L.H.; Madsen, E.B.; Felle, H.H.; Umehara, Y.; Sato, S.; Nakamura, Y.; Tabata, S.; Sandal, N.; Stougaard, J. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 2003, 425, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Kwaharada, Y.; Kelly, S.; Nielsen, M.W.; Hjuler, C.T.; Gysel, K.; Muszynski, A.; Carlson, R.W.; Thygesen, M.B.; Sandal, N.; Asmussen, M.H.; et al. Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 2015, 523, 308–312. [Google Scholar] [CrossRef] [PubMed]
- Rose, C.M.; Venkateshwaran, M.; Volkening, J.D.; Grimsrud, P.A.; Maeda, J.; Park, K.; Howes-Podoll, M.; den Os, D.; Yeun, L.H.; Westphall, M.S.; et al. Rapid phosphoproteomic and transcriptomic changes in the rhizobia-legume symbiosis. Mol. Cell. Proteomics 2012, 11, 724–744. [Google Scholar] [CrossRef] [PubMed]
- Vankateshwaran, M.; Jayaramar, D.; Chabaud, M.; Genre, A.; Ballon, A.J.; Maeda, J.; Forshey, K.; den Os, D.; Kwiecien, N.W.; Coon, J.J.; et al. A role for the mevalonate pathway in early symbiotic signaling. Proc. Natl. Acad. Sci. USA 2015, 112, 9781–9786. [Google Scholar] [CrossRef] [PubMed]







© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Isidra-Arellano, M.C.; Reyero-Saavedra, M.D.R.; Sánchez-Correa, M.D.S.; Pingault, L.; Sen, S.; Joshi, T.; Girard, L.; Castro-Guerrero, N.A.; Mendoza-Cozatl, D.G.; Libault, M.; et al. Phosphate Deficiency Negatively Affects Early Steps of the Symbiosis between Common Bean and Rhizobia. Genes 2018, 9, 498. https://doi.org/10.3390/genes9100498
Isidra-Arellano MC, Reyero-Saavedra MDR, Sánchez-Correa MDS, Pingault L, Sen S, Joshi T, Girard L, Castro-Guerrero NA, Mendoza-Cozatl DG, Libault M, et al. Phosphate Deficiency Negatively Affects Early Steps of the Symbiosis between Common Bean and Rhizobia. Genes. 2018; 9(10):498. https://doi.org/10.3390/genes9100498
Chicago/Turabian StyleIsidra-Arellano, Mariel C., María Del Rocio Reyero-Saavedra, Maria Del Socorro Sánchez-Correa, Lise Pingault, Sidharth Sen, Trupti Joshi, Lourdes Girard, Norma A. Castro-Guerrero, David G. Mendoza-Cozatl, Marc Libault, and et al. 2018. "Phosphate Deficiency Negatively Affects Early Steps of the Symbiosis between Common Bean and Rhizobia" Genes 9, no. 10: 498. https://doi.org/10.3390/genes9100498
APA StyleIsidra-Arellano, M. C., Reyero-Saavedra, M. D. R., Sánchez-Correa, M. D. S., Pingault, L., Sen, S., Joshi, T., Girard, L., Castro-Guerrero, N. A., Mendoza-Cozatl, D. G., Libault, M., & Valdés-López, O. (2018). Phosphate Deficiency Negatively Affects Early Steps of the Symbiosis between Common Bean and Rhizobia. Genes, 9(10), 498. https://doi.org/10.3390/genes9100498





