Satellite DNAs Unveil Clues about the Ancestry and Composition of B Chromosomes in Three Grasshopper Species
Abstract
1. Introduction
2. Material and Methods
2.1. Animal Sampling, Chromosome Preparations, and Genomic DNA Sequencing
2.2. SatDNAs Searching by Graph-Based Clustering Method
2.3. Amplification of SatDNAs through PCR, Probes and Fluorescence In Situ Hybridization
3. Results
3.1. Karyotypes, B Chromosomes, and Heterochromatin Distribution
3.2. In Silico SatDNA Analysis
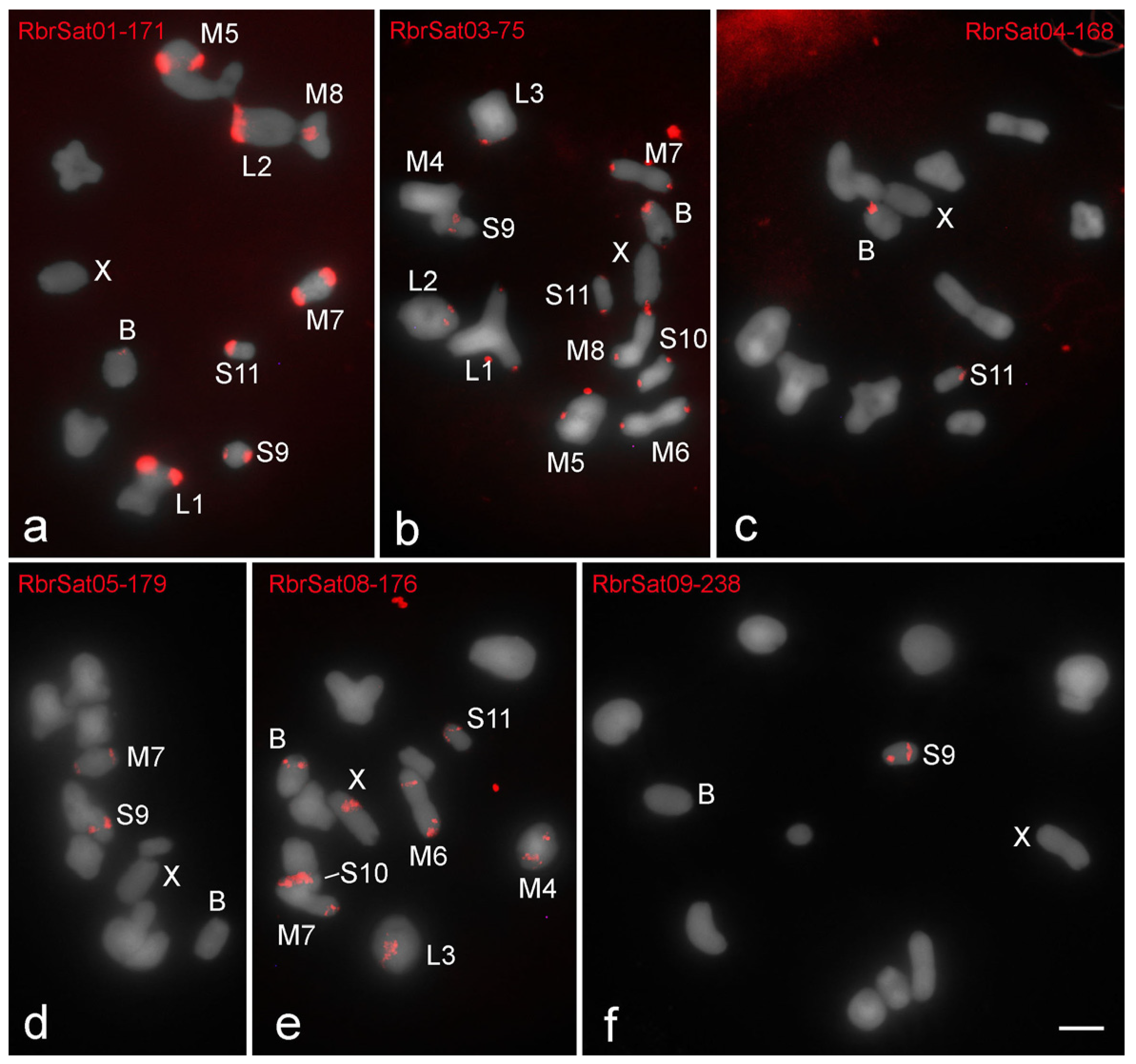
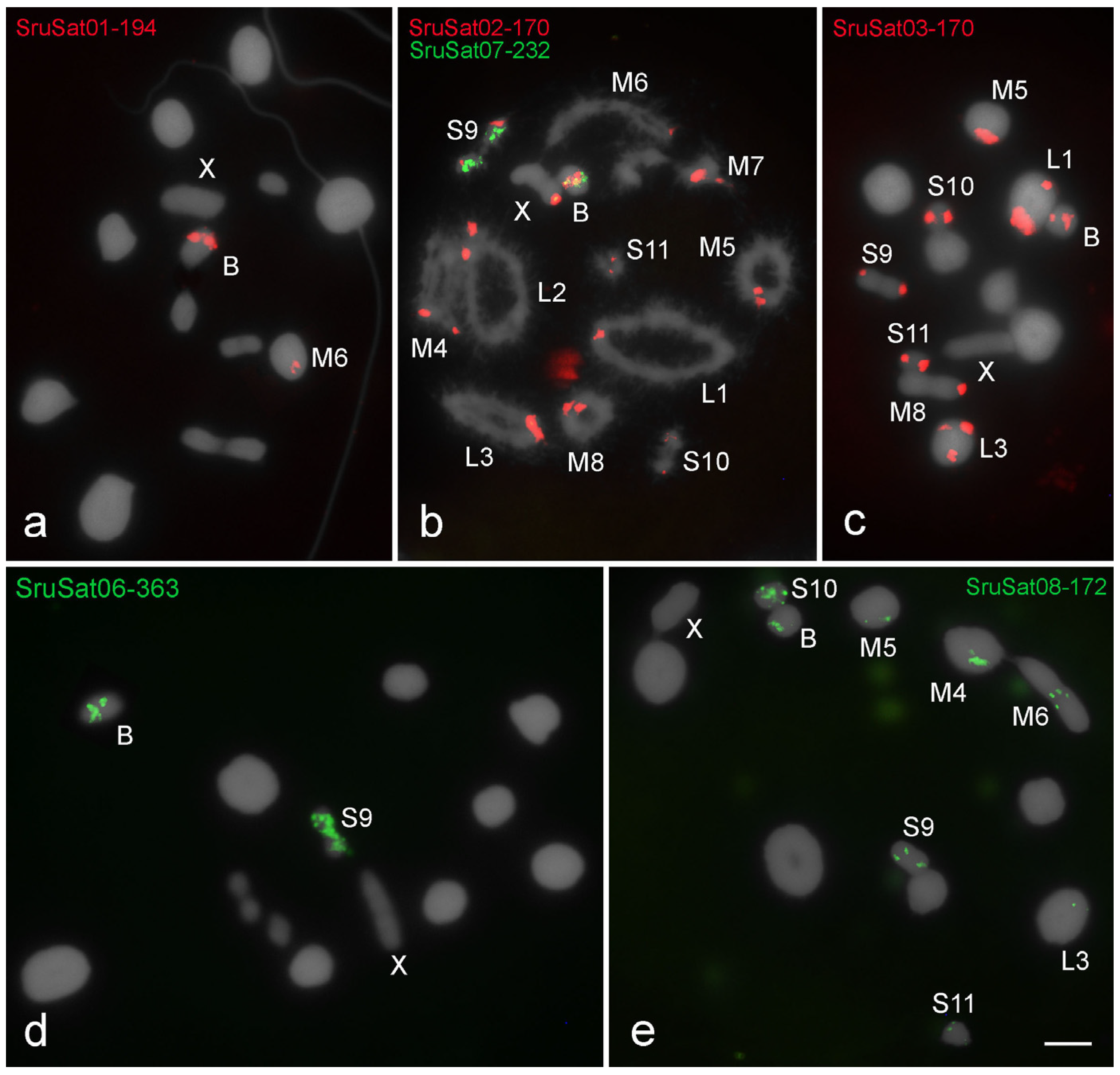
3.3. Chromosomal Location of SatDNAs
4. Discussion
SatDNAs Reveal Clues about B Chromosome Composition and Ancestry
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Charlesworth, B.; Sniegowski, P.; Stephan, W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 1994, 371, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Ramos, M.A. Satellite DNA: An evolving topic. Genes 2017, 8, 230. [Google Scholar] [CrossRef] [PubMed]
- Lower, S.S.; McGurk, M.P.; Clark, A.G.; Barbash, D.A. Satellite DNA evolution: Old ideas, new approaches. Curr. Opin. Genet. Dev. 2018, 49, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Camacho, J.P.M. B chromosomes. In The Evolution of the Genome; Gregory, T.R., Ed.; Elsevier: San Diego, CA, USA, 2005; pp. 223–286. [Google Scholar]
- Houben, A. B chromosomes—A matter of chromosome drive. Front. Plant Sci. 2017, 8, 210. [Google Scholar] [CrossRef] [PubMed]
- Wilson, E.B. The supernumerary chromosomes of Hemiptera. Science 1907, 26, 870–871. [Google Scholar]
- Ruiz-Ruano, F.J.; Cabrero, J.; López-León, M.D.; Camacho, J.P.M. Satellite DNA content illuminates the ancestry of a supernumerary (B) chromosome. Chromosoma 2017, 126, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ruano, F.J.; Cabrero, J.; López-León, M.D.; Sánchez, A.; Camacho, J.P.M. Quantitative sequence characterization for repetitive DNA content in the supernumerary chromosome of the migratory locust. Chromosoma 2018, 127, 45–57. [Google Scholar] [CrossRef] [PubMed]
- McAllister, B.F.; Werren, J.H. Hybrid origin of a B chromosome (PSR) in the parasitic wasp Nasonia vitripennis. Chromosoma 1997, 106, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Silva, D.M.Z.A.; Utsunomia, R.; Ruiz-Ruano, F.J.; Daniel, S.N.; Porto-Foresti, F.; Hashimoto, D.T.; Oliveira, C.; Camacho, J.P.M.; Foresti, F. High-throughput analysis unveils a highly shared satellite DNA library among three species of fish genus Astyanax. Sci. Rep. 2017, 7, 12726. [Google Scholar] [CrossRef] [PubMed]
- Kumke, K.; Macas, J.; Fuchs, J.; Altschmied, L.; Kour, J.; Dhar, M.K.; Houben, A. Plantago lagopus B chromosome is enriched in 5S rDNA-derived satellite DNA. Cytogenet. Genome Res. 2016, 148, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Palestis, B.G.; Cabrero, J.; Trivers, R.; Camacho, J.P.M. Prevalence of B chromosomes in Orthoptera is associated with shape and number of A chromosomes. Genetica 2010, 138, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- López-León, M.D.; Neves, N.; Schwarzacher, T.; Heslop-Harrison, J.S.; Hewitt, G.M.; Camacho, J.P.M. Possible origin of a B chromosome deduced from its DNA composition using double FISH technique. Chromosom. Res. 1994, 2, 87–92. [Google Scholar] [CrossRef]
- Oliveira, N.L.; Cabral-de-Mello, D.C.; Rocha, M.F.; Loreto, V.; Martins, C. Chromosomal mapping of rDNAs and H3 histone sequences in the grasshopper Rhammatocerus brasiliensis (acrididae, gomphocerinae): Extensive chromosomal dispersion and co-localization of 5S rDNA/H3 histone clusters in the A complement and B chromosome. Mol. Cytogenet. 2011, 4, 24. [Google Scholar] [CrossRef] [PubMed]
- Teruel, M.; Cabrero, J.; Perfectti, F.; Camacho, J.P.M. B chromosome ancestry revealed by histone genes in the migratory locust. Chromosoma 2010, 119, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Bueno, D.; Palacios-Gimenez, O.M.; Cabral-de-Mello, D.C. Chromosomal mapping of repetitive DNAs in Abracris flavolineata reveal possible ancestry for the B chromosome and surprisingly H3 histone spreading. PLoS ONE 2013, 8, e66532. [Google Scholar] [CrossRef] [PubMed]
- Montiel, E.E.; Cabrero, J.; Camacho, J.P.M.; López-león, M.D. Gypsy, RTE and Mariner transposable elements populate Eyprepocnemis plorans genome. Genetica 2012, 140, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Palacios-Gimenez, O.M.; Bueno, D.; Cabral-de-Mello, D.C. Chromosomal mapping of two Mariner-like elements in the grasshopper Abracris flavolineata (orthoptera: Acrididae) reveals enrichment in euchromatin. Eur. J. Entomol. 2014, 111, 329–334. [Google Scholar] [CrossRef]
- Milani, D.; Cabral-de-Mello, D.C. Microsatellite organization in the grasshopper Abracris flavolineata (Orthoptera: Acrididae) revealed by FISH mapping: Remarkable spreading in the A and B chromosomes. PLoS ONE 2014, 9, e97956. [Google Scholar] [CrossRef] [PubMed]
- Milani, D.; Ramos, E.; Loreto, V.; Martí, D.A.; Cardoso, A.L.; Moraes, K.C.M.; Martins, C.; Cabral-de-Mello, D.C. The satellite DNA AflaSAT-1 in the A and B chromosomes of the grasshopper Abracris flavolineata. BMC Genet. 2017, 18, 81. [Google Scholar] [CrossRef] [PubMed]
- Novák, P.; Neumann, P.; Pech, J.; Steinhaisl, J.; Macas, J. RepeatExplorer: A Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next generation sequence reads. Bioinformatics 2013, 29, 792–793. [Google Scholar] [CrossRef] [PubMed]
- Utsunomia, R.; Silva, D.M.Z.A.; Ruiz-Ruano, F.J.; Araya-Jayme, C.; Pansonato-Alves, J.C.; Scacchetti, P.C.; Hashimoto, D.T.; Oliveira, C.; Trifonov, V.A.; Porto-Foresti, F.; et al. Uncovering the ancestry of B chromosomes in Moenkhausia sanctaefilomenae (Teleostei, Characidae). PLoS ONE 2016, 11, e0150573. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Mariño-Pérez, R.; Woller, D.A.; Cigliano, M.M. Evolution, diversification, and biogeography of grasshoppers (Orthoptera: Acrididae). Insect Sys. Div. 2018, 2, 1–25. [Google Scholar] [CrossRef]
- Song, H.; Foquet, B.; Mariño-Pérez, R.; Woller, D. Phylogeny of locusts and grasshoppers reveals complex evolution of density-dependent phenotypic plasticity. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Molecular Cloning, a Laboratory Manual, 3rd ed.; Cold Spring Harbor: New York, NY, USA, 2001. [Google Scholar]
- Sumner, A.T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell. Res. 1972, 75, 304–306. [Google Scholar] [CrossRef]
- FastQC; Version 0.10.1; A Quality Control Tool for High throughput Sequence Data; Babraham Bioinformatics: Cambridge, UK, 2012.
- Novák, P.; Neumann, P.; Macas, J. Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinform. 2010, 11, 378. [Google Scholar] [CrossRef] [PubMed]
- Geneious; Version 4.8.5; Biomatters Ltd.: Aukland, New Zealand, 2009.
- RepeatMasker Open; Version 4.0; Institute for Systems Biology: Seattle, WA, USA, 2013.
- Ruiz-Ruano, F.J.; López-León, M.D.; Cabrero, J.; Camacho, J.P.M. High-throughput analysis of the satellitome illuminates satellite DNA evolution. Sci. Rep. 2016, 6, 28333. [Google Scholar] [CrossRef] [PubMed]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers in Bioinformatics. Methods Mol. Biol 2000, 132, 365–386. [Google Scholar] [CrossRef] [PubMed]
- Pinkel, D.; Lanlegent, J.; Collins, C.; Fuscoe, J.; Segraves, R.; Lucas, J.; Gray, J. Fluorescence in situ hybridization with human chromosome-specific libraries: Detection of trisomy 21 and translocations of chromosome 4. Proc. Natl. Acad. Sci. USA 1986, 85, 9138–9142. [Google Scholar] [CrossRef]
- Camacho, J.P.M.; Cabrero, J.; López-León, M.D.; Cabral-de-Mello, D.C.; Ruiz-Ruano, F.J. Grasshoppers (Orthoptera). In Protocols for Cytogenetic Mapping of Arthropod Genomes, 1st ed.; Sharakhov, I.V., Ed.; CRC Press: Boca Raton, FL, USA, 2015; pp. 381–438. [Google Scholar]
- Loreto, V.; Cabrero, J.; López-León, M.D.; Camacho, J.P.M. Possible autosomal origin of macro B chromosomes in two grasshopper species. Chromosom. Res. 2008, 16, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Bernardino, A.C.S.; Cabral-de-Mello, D.C.; Machado, C.B.; Palacios-Gimenez, O.M.; Santos, N.; Loreto, V. B chromosome variants of the Grasshopper Xyleus discoideus angulatus are potentially derived from pericentromeric DNA. Cytogenet. Genome Res. 2017, 152, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Camacho, J.P.M.; Shaw, M.W.; Cabrero, J.; Bakkali, M.; Ruíz-Estévez, M.; Ruiz-Ruano, F.J.; Martín-Blázquez, R.; López-León, M.D. Transient microgeographic clines during B chromosome invasion. Am. Nat. 2015, 186, 675–681. [Google Scholar] [CrossRef] [PubMed]
- Souza, M.J.; Melo, N.F. Chromosome study in Schistocerca (Orthoptera-Acrididae-Cyrtacanthacridinae): Karyotypes and distribution patterns of constitutive heterochromatin and nucleolus organizer regions (NORs). Genet. Mol. Biol. 2007, 30, 54–59. [Google Scholar] [CrossRef]
- Ruiz-Ruano, F.J.; Castillo-Martínez, J.; Cabrero, J.; Gómez, R.; Camacho, J.P.M.; López-León, M.D. High-throughput analysis of satellite DNA in the grasshopper Pyrgomorpha conica reveals abundance of homologous and heterologous higher-order repeats. Chromosoma 2018, 127, 3. [Google Scholar] [CrossRef] [PubMed]
- Palacios-Gimenez, O.M.; Dias, G.B.; de Lima, L.G.; Kuhn, G.C.E.S.; Ramos, E.; Martins, C.; Cabral-de-Mello, D.C. High-throughput analysis of the satellitome revealed enormous diversity of satellite DNAs in the neo-Y chromosome of the cricket Eneoptera surinamensis. Sci. Rep. 2017, 7, 6422. [Google Scholar] [CrossRef] [PubMed]
- Palacios-Gimenez, O.M.; Milani, D.; Lemos, B.; Castillo, E.R.; Martí, D.A.; Ramos, E.; Martins, C.; Cabral-de-Mello, D.C. Uncovering the evolutionary history of neo-XY sex chromosomes in the grasshopper Ronderosia bergii (Orthoptera, Melanoplinae) through satellite DNA analysis. BMC Evol. Biol. 2018, 18, 2. [Google Scholar] [CrossRef] [PubMed]
- Palacios-Gimenez, O.M.; Bardella, V.B.; Lemos, B.; Cabral-de-Mello, D.C. Satellite DNAs are conserved and differentially transcribed among Gryllus cricket species. DNA Res. 2018, 25, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Marques, A.; Klemme, S.; Guerra, M.; Houben, A. Cytomolecular characterization of de novo formed rye B chromosome variants. Mol. Cytogenet. 2012, 5, 34. [Google Scholar] [CrossRef] [PubMed]
- Cabrero, J.; López-León, M.D.; Ruíz-Estévez, M.; Gómez, R.; Petitpierre, E.; Rufas, J.S.; Massa, B.; Kamel Ben Halima, M.; Camacho, J.P.M. B1 was the ancestor B chromosome variant in the western Mediterranean area in the grasshopper Eyprepocnemis plorans. Cytogenet. Genome Res. 2014, 142, 54–58. [Google Scholar] [CrossRef] [PubMed]
- Melo, A.S.; Moura, R.C. (Universidade de Pernambuco, Recife, Pernambuco, BR). Personal communication, 2018.
- Martis, M.M.; Klemme, S.; Banaei-Moghaddam, A.M.; Blattner, F.R.; Macas, J.; Schmutzer, T.; Scholz, U.; Gundlach, H.; Wicker, T.; Simkova, H.; et al. Selfish supernumerary chromosome reveals its origin as a mosaic of host genome and organellar sequences. Proc. Natl. Acad. Sci. USA 2012, 109, 13343–13346. [Google Scholar] [CrossRef] [PubMed]
- Teruel, M.; Ruiz-Ruano, F.J.; Marchal, J.A.; Sánchez, A.; Cabrero, J.; Camacho, J.P.M.; Perfectti, F. Disparate molecular evolution of two types of repetitive DNAs in the genome of the grasshopper Eyprepocnemis plorans. Heredity 2014, 112, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, G.M. The integration of supernumerary chromosomes into the orthopteran genome. Cold Spring Harb. Symp. Quant. Biol. 1974, 38, 183–194. [Google Scholar] [CrossRef] [PubMed]



| Species | SatDNA Superfamily | SatDNA Family | Monomer Size (nt) | A+T (%) | Abundance (%) | Divergence (%) |
|---|---|---|---|---|---|---|
| R. brasiliensis | SF1 | RbrSat01-171 | 171 | 59.3 | 0.766 | 4.98 |
| - | RbrSat02-410 | 410 | 47.9 | 0.224 | 8.94 | |
| - | RbrSat03-36 | 36 | 44.5 | 0.126 | 8.45 | |
| SF1 | RbrSat04-168 | 168 | 59.4 | 0.105 | 18.14 | |
| - | RbrSat05-179 | 179 | 59.1 | 0.061 | 4.96 | |
| - | RbrSat06-165 | 165 | 59.2 | 0.056 | 1.23 | |
| - | RbrSat07-240 | 240 | 60.4 | 0.047 | 11.56 | |
| - | RbrSat08-176 | 176 | 58.0 | 0.042 | 6.27 | |
| - | RbrSat09-238 | 238 | 63.9 | 0.025 | 8.75 | |
| - | RbrSat10-268 | 268 | 62.7 | 0.021 | 7.79 | |
| - | RbrSat11-233 | 233 | 58.0 | 0.016 | 7.85 | |
| - | RbrSat12-180 | 180 | 57.8 | 0.010 | 2.06 | |
| Total | 1.499 | |||||
| S. rubiginosa | - | SruSat01-194 | 194 | 58.3 | 0.730 | 4.30 |
| - | SruSat02-170 | 170 | 54.4 | 0.476 | 4.64 | |
| - | SruSat03-170 | 170 | 59.9 | 0.287 | 7.79 | |
| - | SruSat04-301 | 301 | 48.9 | 0.244 | 4.77 | |
| - | SruSat05-441 | 441 | 50.6 | 0.135 | 23.18 | |
| - | SruSat06-363 | 363 | 57.1 | 0.126 | 9.61 | |
| - | SruSat07-232 | 232 | 61.2 | 0.116 | 9.23 | |
| - | SruSat08-172 | 172 | 58.2 | 0.032 | 16.24 | |
| - | SruSat09-107 | 107 | 61.7 | 0.026 | 10.85 | |
| Total | 2.172 | |||||
| X. d. angulatus | - | XanSat01-8 | 8 | 62.5 | 0.627 | 4.62 |
| - | XanSat02-21 | 21 | 28.6 | 0.586 | 4.41 | |
| - | XanSat03-10 | 10 | 60.0 | 0.464 | 9.38 | |
| - | XanSat04-10 | 10 | 60.0 | 0.228 | 5.22 | |
| SF1 | XanSat05-267 | 267 | 56.7 | 0.087 | 5.50 | |
| - | XanSat06-168 | 168 | 64.3 | 0.069 | 4.53 | |
| SF1 | XanSat07-279 | 279 | 60.2 | 0.053 | 11.08 | |
| - | XanSat08-16 | 16 | 56.2 | 0.033 | 4.38 | |
| SF2 | XanSat09-130 | 130 | 63.1 | 0.024 | 10.61 | |
| - | XanSat10-289 | 289 | 60.2 | 0.022 | 7.27 | |
| - | XanSat11-51 | 51 | 47.1 | 0.019 | 3.97 | |
| - | XanSat12-246 | 246 | 59.2 | 0.018 | 11.81 | |
| - | XanSat13-281 | 281 | 56.3 | 0.018 | 4.61 | |
| SF2 | XanSat14-128 | 128 | 62.5 | 0.017 | 14.90 | |
| - | XanSat15-228 | 228 | 59.7 | 0.017 | 5.18 | |
| - | XanSat16-21 | 21 | 42.9 | 0.014 | 9.79 | |
| - | XanSat17-15 | 15 | 53.4 | 0.013 | 11.56 | |
| - | XanSat18-21 | 21 | 76.2 | 0.013 | 6.33 | |
| Total | 2.322 |
| Species | SatDNA Family | Chromosome Location | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | X | B | ||
| Rhammatocerus brasiliensis | RbrSat01-171 | p | p | p | p | p | p | p | p | |||||
| RbrSat02-410 | nc | |||||||||||||
| RbrSat03-36 | p | p | p | p | p | p | p | p | p | p | p | p | p | |
| RbrSat04-168 | p | p | ||||||||||||
| RbrSat05-179 | p | p | ||||||||||||
| RbrSat06-165 | nc | |||||||||||||
| RbrSat07-240 | nc | |||||||||||||
| RbrSat08-176 | p | p | p,i | p | p | p | i | p | ||||||
| RbrSat09-238 | i | |||||||||||||
| RbrSat10-268 | nc | |||||||||||||
| RbrSat11-233 | nc | |||||||||||||
| RbrSat12-180 | nc | |||||||||||||
| Total | 2 | 2 | 2 | 2 | 2 | 2 | 4 | 2 | 4 | 2 | 4 | 2 | 4 | |
| shared with B | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 4 | 2 | ||
| Schistocerca rubiginosa | SruSat01-194 | d | i | |||||||||||
| SruSat02-170 | p | p | p | p | p | p | p | p | p | p | p | p | i | |
| SruSat03-170 | p,d | p,d | p | p | p | p | p | i | ||||||
| SruSat04-301 | nc | |||||||||||||
| SruSat05-441 | nc | |||||||||||||
| SruSat06-363 | i,d | 2i | ||||||||||||
| SruSat07-232 | i,d | i | ||||||||||||
| SruSat08-172 | p | i | p | p | p | p,i | p | p | ||||||
| SruSat09-207 | nc | |||||||||||||
| Total | 2 | 1 | 3 | 2 | 3 | 3 | 1 | 2 | 5 | 3 | 3 | 1 | 6 | |
| shared with B | 2 | 1 | 3 | 2 | 3 | 3 | 1 | 2 | 5 | 3 | 3 | 1 | ||
| Xylleus discoideus angulatus | XanSat01-8 | i | i | d | i | d | i | d | d | d | d | d | 2i | |
| XanSat02-21 | p | p | p | p | p | p | p | p | p | p,d | p | p | ||
| XanSat03-10 | p | p | p | p | p | p | p | p | p | p | p | p | p,i,d | |
| XanSat04-10 | p | p | p | |||||||||||
| XanSat05-267 | p | p,i | 2i | |||||||||||
| XanSat06-168 | p | i | ||||||||||||
| XanSat07-279 | i | d | p | |||||||||||
| XanSat08-16 | d | |||||||||||||
| XanSat09-130 | nc | |||||||||||||
| XanSat10-289 | nc | |||||||||||||
| XanSat11-51 | d | |||||||||||||
| XanSat12-246 | p | p | p | p | p | p | p | p | p | p | ||||
| XanSat13-281 | p | d | i | i,d | ||||||||||
| XanSat14-128 | nc | |||||||||||||
| XanSat15-228 | nc | |||||||||||||
| XanSat16-21 | nc | |||||||||||||
| XanSat17-15 | nc | |||||||||||||
| XanSat18-21 | nc | |||||||||||||
| Total | 5 | 6 | 5 | 4 | 5 | 9 | 6 | 3 | 5 | 6 | 4 | 3 | 3 | |
| shared with B | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 2 | 3 | 1 | 1 | ||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Milani, D.; Bardella, V.B.; Ferretti, A.B.S.M.; Palacios-Gimenez, O.M.; Melo, A.d.S.; Moura, R.C.; Loreto, V.; Song, H.; Cabral-de-Mello, D.C. Satellite DNAs Unveil Clues about the Ancestry and Composition of B Chromosomes in Three Grasshopper Species. Genes 2018, 9, 523. https://doi.org/10.3390/genes9110523
Milani D, Bardella VB, Ferretti ABSM, Palacios-Gimenez OM, Melo AdS, Moura RC, Loreto V, Song H, Cabral-de-Mello DC. Satellite DNAs Unveil Clues about the Ancestry and Composition of B Chromosomes in Three Grasshopper Species. Genes. 2018; 9(11):523. https://doi.org/10.3390/genes9110523
Chicago/Turabian StyleMilani, Diogo, Vanessa B. Bardella, Ana B. S. M. Ferretti, Octavio M. Palacios-Gimenez, Adriana de S. Melo, Rita C. Moura, Vilma Loreto, Hojun Song, and Diogo C. Cabral-de-Mello. 2018. "Satellite DNAs Unveil Clues about the Ancestry and Composition of B Chromosomes in Three Grasshopper Species" Genes 9, no. 11: 523. https://doi.org/10.3390/genes9110523
APA StyleMilani, D., Bardella, V. B., Ferretti, A. B. S. M., Palacios-Gimenez, O. M., Melo, A. d. S., Moura, R. C., Loreto, V., Song, H., & Cabral-de-Mello, D. C. (2018). Satellite DNAs Unveil Clues about the Ancestry and Composition of B Chromosomes in Three Grasshopper Species. Genes, 9(11), 523. https://doi.org/10.3390/genes9110523







