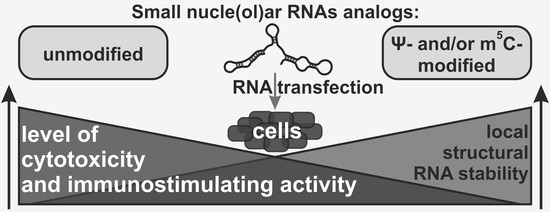
Nucleotide Modifications Decrease Innate Immune Response Induced by Synthetic Analogs of snRNAs and snoRNAs
Abstract
:1. Introduction
2. Materials and Methods
2.1. In Vitro Synthesis of Artificial RNAs
2.2. Enzymatic Digestion of RNAs and HPLC-UV Analysis
2.3. HPLC-MS/MS
2.4. Transfection of Human Cells with Synthetic RNAs
2.5. Differential Gene and Transcript Expression Analysis
2.6. qRT-PCR
2.7. Thermal Denaturation Experiments
2.8. MTT Assay
2.9. iCELLigence Assay
2.10. Statistical Analysis
3. Results
3.1. Synthesis of Modified ncRNA Analogs
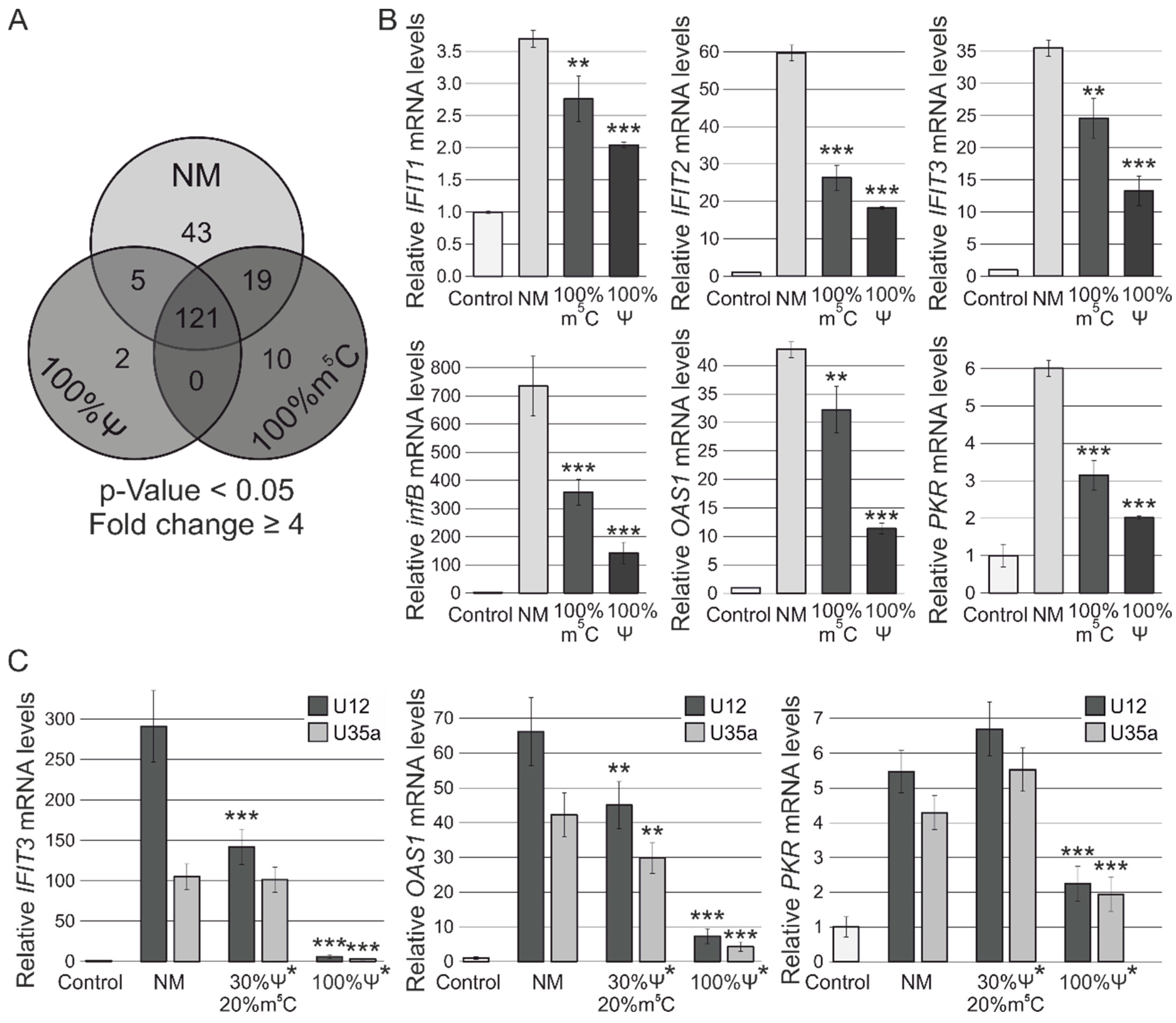
3.2. RNA-Seq Analysis of Human MCF-7 Cells Transfected with Ψ- and m5C-Modified RNAs
3.3. RT-PCR Verification of Gene Expression Changes
3.4. Analysis of Cytotoxicity and Antiproliferative Effects of Synthetic Modified ncRNAs
3.5. Thermal Denaturation Studies
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Valadkhan, S.; Mohammadi, A.; Jaladat, Y.; Geisler, S. Protein-free small nuclear RNAs catalyze a two-step splicing reaction. Proc. Natl. Acad. Sci. USA 2009, 106, 11901–11906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fica, S.M.; Tuttle, N.; Novak, T.; Li, N.S.; Lu, J.; Koodathingal, P.; Dai, Q.; Staley, J.P.; Piccirilli, J.A. RNA catalyses nuclear pre-mRNA splicing. Nature 2013, 503, 229–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, J.; Yu, Y.T. RNA pseudouridylation: New insights into an old modification. Trends Biochem. Sci. 2013, 38, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Dupuis-Sandoval, F.; Poirier, M.; Scott, M.S. The emerging landscape of small nucleolar RNAs in cell biology. Wiley Interdiscip. Rev. RNA 2015, 6, 381–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matera, A.G.; Terns, R.M.; Terns, M.P. Non-coding RNAs: Lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007, 8, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Cavaille, J.; Nicoloso, M.; Bachellerie, J.P. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature 1996, 383, 732–735. [Google Scholar] [CrossRef] [PubMed]
- Ganot, P.; Bortolin, M.L.; Kiss, T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 1997, 89, 799–809. [Google Scholar] [CrossRef]
- Baudin-baillieu, A.; Fabret, C.; Liang, X.H.; Piekna-Przybylska, D.; Fournier, M.J.; Rousset, J.P. Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy. Nucleic Acids Res. 2009, 37, 7665–7677. [Google Scholar] [CrossRef] [PubMed]
- Natchiar, S.K.; Myasnikov, A.G.; Kratzat, H.; Hazemann, I.; Klaholz, B.P. Visualization of chemical modifications in the human 80S ribosome structure. Nature 2017, 551, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Esguerra, J.; Warringer, J.; Blomberg, A. Functional importance of individual rRNA 2’-O-ribose methylation revealed by high-resolution phenotyping. RNA 2008, 14, 649–656. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N6 -methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2013, 505, 117–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reikine, S.; Nguyen, J.B.; Modis, Y. Pattern recognition and signaling mechanisms of RIG-I and MDA5. Front. Immunol. 2014, 5, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Schlee, M.; Hornung, V.; Hartmann, G. siRNA and isRNA: Two edges of one sword. Mol. Ther. 2006, 14, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Katze, M.G.; Fornek, J.L.; Palermo, R.E.; Walters, K.-A.; Korth, M.J. Innate immune modulation by RNA viruses: Emerging insights from functional genomics. Nat. Rev. Immunol. 2008, 8, 644–654. [Google Scholar] [CrossRef] [PubMed]
- Der, S.D.; Zhou, A.; Williams, B.R.G.; Silverman, R.H. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 1998, 95, 15623–15628. [Google Scholar] [CrossRef] [PubMed]
- Trinchieri, G. Type I interferon: Friend or foe? J. Exp. Med. 2010, 207, 2053–2063. [Google Scholar] [CrossRef] [PubMed]
- Josset, L.; Tchitchek, N.; Gralinski, L.E.; Ferris, M.T.; Eisfeld, A.J.; Green, R.R.; Thomas, M.J.; Tisoncik-Go, J.; Schroth, G.P.; Kawaoka, Y.; et al. Annotation of long non-coding RNAs expressed in Collaborative Cross founder mice in response to respiratory virus infection reveals a new class of interferon-stimulated transcripts. RNA Biol. 2014, 11, 875–890. [Google Scholar] [CrossRef] [PubMed]
- Karikó, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 2005, 23, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Fortes, P.; Cuevas, Y.; Guan, F.; Liu, P.; Pentlicky, S.; Jung, S.P.; Martínez-Chantar, M.L.; Prieto, J.; Rowe, D.; Gunderson, S.I. Inhibiting expression of specific genes in mammalian cells with 5′ end-mutated U1 small nuclear RNAs targeted to terminal exons of pre-mRNA. Proc. Natl. Acad. Sci. USA 2003, 100, 8264–8269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knoepfel, S.A.; Abad, A.; Abad, X.; Fortes, P.; Berkhout, B. Design of modified U1i molecules against HIV-1 RNA. Antivir. Res. 2012, 94, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Blazquez, L.; Gonzalez-Rojas, S.J.; Abad, A.; Razquin, N.; Abad, X.; Fortes, P. Increased in vivo inhibition of gene expression by combining RNA interference and U1 inhibition. Nucleic Acids Res. 2012, 40, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Stepanov, G.A.; Semenov, D.V.; Kuligina, E.V.; Koval, O.A.; Rabinov, I.V.; Kit, Y.Y.; Richter, V.A. Analogues of Artificial Human Box C/D Small Nucleolar RNA As Regulators of Alternative Splicing of a pre-mRNA Target. Acta Nat. 2012, 4, 32–41. [Google Scholar]
- Stepanov, G.A.; Semenov, D.V.; Savelyeva, A.V.; Kuligina, E.V.; Koval, O.A.; Rabinov, I.V.; Richter, V.A. Artificial box C/D RNAs affect pre-mRNA maturation in human cells. Biomed. Res. Int. 2013, 2013, 656158. [Google Scholar] [CrossRef] [PubMed]
- Ono, M.; Yamada, K.; Avolio, F.; Afzal, V.; Bensaddek, D.; Lamond, A.I. Targeted knock-down of miR21 primary transcripts using snoMEN vectors induces apoptosis in human cancer cell lines. PLoS ONE 2015, 10, e0138668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ono, M.; Yamada, K.; Endo, A.; Avolio, F.; Lamond, A.I. Analysis of Human Protein Replacement Stable Cell Lines Established using snoMEN-PR Vector. PLoS ONE 2013, 8, e62305. [Google Scholar] [CrossRef] [PubMed]
- Holley, C.L.; Li, M.W.; Scruggs, B.S.; Matkovich, S.J.; Ory, D.S.; Schaffer, J.E. Cytosolic accumulation of small nucleolar RNAs (snoRNAs) is dynamically regulated by NADPH oxidase. J. Biol. Chem. 2015, 290, 11741–11748. [Google Scholar] [CrossRef] [PubMed]
- Michel, C.I.; Holley, C.L.; Scruggs, B.S.; Sidhu, R.; Brookheart, R.T.; Listenberger, L.L.; Behlke, M.A.; Ory, D.S.; Schaffer, J.E. Small nucleolar RNAs U32a, U33, and U35a are critical mediators of metabolic stress. Cell Metab. 2011, 14, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Malathi, K.; Dong, B.; Gale, M.; Silverman, R.H. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature 2007, 448, 816–819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, J.; Yu, L.; Mei, Y.; Guarnera, M.; Shen, J.; Li, R.; Liu, Z.; Jiang, F. Small nucleolar RNA signatures as biomarkers for non-small-cell lung cancer. Mol. Cancer 2010, 9, 198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savelyeva, A.V.; Kuligina, E.V.; Bariakin, D.N.; Kozlov, V.V.; Ryabchikova, E.I.; Richter, V.A.; Semenov, D.V. Variety of RNAs in Peripheral Blood Cells, Plasma, and Plasma Fractions. Biomed. Res. Int. 2017, 2017, 7404912. [Google Scholar] [CrossRef] [PubMed]
- Shender, V.O.; Pavlyukov, M.S.; Ziganshin, R.H.; Arapidi, G.P.; Kovalchuk, S.I.; Anikanov, N.A.; Altukhov, I.A.; Alexeev, D.G.; Butenko, I.O.; Shavarda, A.L.; et al. Proteome–Metabolome Profiling of Ovarian Cancer Ascites Reveals Novel Components Involved in Intercellular Communication. Mol. Cell. Proteom. 2014, 13, 3558–3571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steinbusch, M.M.F.; Fang, Y.; Milner, P.I.; Clegg, P.D.; Young, D.A.; Welting, T.J.M.; Peffers, M.J. Serum snoRNAs as biomarkers for joint ageing and post traumatic osteoarthritis. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yuan, T.; Tschannen, M.; Sun, Z.; Jacob, H.; Du, M.; Liang, M.; Dittmar, R.L.; Liu, Y.; Liang, M.; et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genom. 2013, 14, 319. [Google Scholar] [CrossRef] [PubMed]
- Warren, L.; Manos, P.D.; Ahfeldt, T.; Loh, Y.H.; Li, H.; Lau, F.; Ebina, W.; Mandal, P.K.; Smith, Z.D.; Meissner, A.; et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 2010, 7, 618–630. [Google Scholar] [CrossRef] [PubMed]
- Sioud, M.; Furset, G.; Cekaite, L. Suppression of immunostimulatory siRNA-driven innate immune activation by 2′-modified RNAs. Biochem. Biophys. Res. Commun. 2007, 361, 122–126. [Google Scholar] [CrossRef] [PubMed]
- Durbin, A.F.; Wang, C.; Marcotrigiano, J.; Gehrke, L. RNAs containing modified nucleotides fail to trigger RIG-I conformational changes for innate immune signaling. mBio 2016, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, R.A.P.; Suter, S.R.; Ball-Jones, A.A.; Ibarra-Soza, J.M.; Zheng, Y.; Beal, P.A. Base modification strategies to modulate immune stimulation by an siRNA. ChemBioChem 2014, 16, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Chappell, J.; Takahashi, M.K.; Meyer, S.; Loughrey, D.; Watters, K.E.; Lucks, J. The centrality of RNA for engineering gene expression. Biotechnol. J. 2013, 8, 1379–1395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stepanov, G.A.; Filippova, J.A.; Nushtaeva, A.A.; Kuligina, E.V.; Koval, O.A.; Richter, V.A.; Semenov, D.V. Artificial Analogues of Circulating Box C/D RNAs Induce Strong Innate Immune Response and MicroRNA Activation in Human Adenocarcinoma Cells. Adv. Exp. Med. Biol. 2016, 924, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Nushtaeva, A.A.; Stepanov, G.A.; Semenov, D.V.; Juravlev, E.S.; Balahonova, E.A.; Gerasimov, A.V.; Sidorov, S.V.; Savelyev, E.I.; Kuligina, E.V.; Richter, V.A.; et al. Characterization of primary normal and malignant breast cancer cell and their response to chemotherapy and immunostimulatory agents. BMC Cancer 2018, 18, 728. [Google Scholar] [CrossRef] [PubMed]
- Youssef, O.A.; Safran, S.A.; Nakamura, T.; Nix, D.A.; Hotamisligil, G.S.; Bass, B.L. Potential role for snoRNAs in PKR activation during metabolic stress. Proc. Natl. Acad. Sci. USA 2015, 112, 5023–5028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernard, J.J.; Cowing-Zitron, C.; Nakatsuji, T.; Muehleisen, B.; Muto, J.; Borkowski, A.W.; Martinez, L.; Greidinger, E.L.; Yu, B.D.; Gallo, R.L. Ultraviolet radiation damages self noncoding RNA and is detected by TLR3. Nat. Med. 2012, 18, 1286–1290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, C.T.Y.; Dyavaiah, M.; DeMott, M.S.; Taghizadeh, K.; Dedon, P.C.; Begley, T.J. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 2010, 6, e1001247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thüring, K.; Schmid, K.; Keller, P.; Helm, M. Analysis of RNA modifications by liquid chromatography–tandem mass spectrometry. Methods 2016, 107, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2013, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, I.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3. [Google Scholar] [CrossRef] [Green Version]
- Lokhov, S.G.; Pyshnyi, D.V. Thermodynamic and spectral properties of DNA miniduplexes with the terminal GWA mispairs and 3P or 5P dangling bases. FEBS Lett. 1997, 420, 134–138. [Google Scholar] [CrossRef]
- Karijolich, J.; Yu, Y.-T. Spliceosomal snRNA modifications and their function. RNA Biol. 2010, 7, 192–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorenz, R.; Bernhart, S.H.; Höner, Z.; Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hudson, G.A.; Bloomingdale, R.J.; Znosko, B.M. Thermodynamic contribution and nearest-neighbor parameters of pseudouridine-adenosine base pairs in oligoribonucleotides. RNA 2013, 19, 1474–1482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kierzek, E.; Malgowska, M.; Lisowiec, J.; Turner, D.H.; Gdaniec, Z.; Kierzek, R. The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res. 2014, 42, 3492–3501. [Google Scholar] [CrossRef] [PubMed]
- Delorimier, E.; Hinman, M.N.; Copperman, J.; Datta, K.; Guenza, M.; Berglund, J.A. Pseudouridine modification inhibits muscleblind-like 1 (MBNL1) binding to CCUG repeats and minimally structured RNA through reduced RNA flexibility. J. Biol. Chem. 2017, 292, 4350–4357. [Google Scholar] [CrossRef] [PubMed]
- Kenworthy, R.; Lambert, D.; Yang, F.; Wang, N.; Chen, Z.; Zhu, H.; Zhu, F.; Liu, C.; Li, K.; Tang, H. Short-hairpin RNAs delivered by lentiviral vector transduction trigger RIG-I-mediated IFN activation. Nucleic Acids Res. 2009, 37, 6587–6599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pichlmair, A.; Schulz, O.; Tan, C.P.; Naslund, T.I.; Liljestrom, P.; Weber, F.; e Sousa, C.R. RIG-I-Mediated Antiviral Responses to Single-Stranded RNA Bearing 5′-Phosphates. Science 2006, 314, 997–1001. [Google Scholar] [CrossRef] [PubMed]
- Nallagatla, S.R.; Hwang, J.; Toroney, R.; Zheng, X.; Cameron, C.E.; Bevilacqua, P.C. 5’-Triphosphate-Dependent Activation of PKR by RNAs with Short Stem-Loops. Science 2007, 318, 1455–1458. [Google Scholar] [CrossRef] [PubMed]
- Hornung, V.; Ellegast, J.; Kim, S.; Brzozka, K.; Jung, A.; Kato, H.; Poeck, H.; Akira, S.; Conzelmann, K.-K.; Schlee, M.; et al. 5’-Triphosphate RNA Is the Ligand for RIG-I. Science 2006, 314, 994–997. [Google Scholar] [CrossRef] [PubMed]
- Abbas, Y.M.; Pichlmair, A.; Górna, M.W.; Superti-Furga, G.; Nagar, B. Structural basis for viral 5′-PPP-RNA recognition by human IFIT proteins. Nature 2013, 494, 60–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pichlmair, A.; Lassnig, C.; Eberle, C.A.; Górna, M.W.; Baumann, C.L.; Burkard, T.R.; Búrckstúmmer, T.; Stefanovic, A.; Krieger, S.; Bennett, K.L.; et al. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat. Immunol. 2011, 12, 624–630. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Schwerd, T.; Hamm, W.; Hellmuth, J.C.; Cui, S.; Wenzel, M.; Hoffmann, F.S.; Michallet, M.-C.; Besch, R.; Hopfner, K.-P.; et al. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc. Natl. Acad. Sci. USA 2009, 106, 12067–12072. [Google Scholar] [CrossRef] [PubMed]
- Hull, C.M.; Bevilacqua, P.C. Discriminating Self and Non-Self by RNA: Roles for RNA Structure, Misfolding, and Modification in Regulating the Innate Immune Sensor PKR. Acc. Chem. Res. 2016, 49, 1242–1249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwartz, S.; Bernstein, D.A.; Mumbach, M.R.; Jovanovic, M.; Herbst, R.H.; León-Ricardo, B.X.; Engreitz, J.M.; Guttman, M.; Satija, R.; Lander, E.S.; et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 2014, 159, 148–162. [Google Scholar] [CrossRef] [PubMed]
- Carlile, T.M.; Rojas-Duran, M.F.; Zinshteyn, B.; Shin, H.; Bartoli, K.M.; Gilbert, W.V. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 2014, 515, 143–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filippova, J.A.; Semenov, D.V.; Juravlev, E.S.; Komissarov, A.B.; Richter, V.A.; Stepanov, G.A. Modern approaches for identification of modified nucleotides in RNA. Biochemistry 2017, 82, 1217–1233. [Google Scholar] [CrossRef] [PubMed]
- Squires, J.E.; Patel, H.R.; Nousch, M.; Sibbritt, T.; Humphreys, D.T.; Parker, B.J.; Suter, C.M.; Preiss, T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012, 40, 5023–5033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linder, B.; Grozhik, A.V.; Olarerin-George, A.O.; Meydan, C.; Mason, C.E.; Jaffrey, S.R. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 2015, 12, 767–772. [Google Scholar] [CrossRef] [PubMed]
- Slobodin, B.; Han, R.; Calderone, V.; Vrielink, J.A.F.O.; Loayza-Puch, F.; Elkon, R.; Agami, R. Transcription Impacts the Efficiency of mRNA Translation via Co-transcriptional N6-adenosine Methylation. Cell 2017, 169, 326–337. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Ashraf, S.; Wang, J.; Lilley, D.M. Control of box C/D snoRNP assembly by N6 -methylation of adenine. EMBO Rep. 2017, 18, 1631–1645. [Google Scholar] [CrossRef] [PubMed]
- Falaleeva, M.; Welden, J.R.; Duncan, M.J.; Stamm, S. C/D-box snoRNAs form methylating and non-methylating ribonucleoprotein complexes: Old dogs show new tricks. BioEssays 2017, 39, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Van Hoof, A.; Lennertz, P.; Parker, R.; Parker, R.O.Y. Yeast Exosome Mutants Accumulate 3′-Extended Polyadenylated Forms of U4 Small Nuclear RNA and Small Nucleolar RNAs. Mol. Cell. Biol. 2000, 20, 441–452. [Google Scholar] [CrossRef] [PubMed]
- LaCava, J.; Houseley, J.; Saveanu, C.; Petfalski, E.; Thompson, E.; Jacquier, A.; Tollervey, D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 2005, 121, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Slomovic, S.; Laufer, D.; Geiger, D.; Schuster, G. Polyadenylation of ribosomal RNA in human cells. Nucleic Acids Res. 2006, 34, 2966–2975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van, D.N.; Roberts, C.F.; Marion, J.D.; Lepine, S.; Harikumar, K.B.; Schreiter, J.; Dumur, C.I.; Fang, X.; Spiegel, S.; Bell, J.K. Innate immune agonist, dsRNA, induces apoptosis in ovarian cancer cells and enhances the potency of cytotoxic chemotherapeutics. FASEB J. 2012, 26, 3188–3198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kabilova, T.O.; Meschaninova, M.I.; Venyaminova, A.G.; Nikolin, V.P.; Zenkova, M.A.; Vlassov, V.V.; Chernolovskaya, E.L. Short Double-Stranded RNA with Immunostimulatory Activity: Sequence Dependence. Nucleic Acid Ther. 2012, 22, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Poeck, H.; Besch, R.; Maihoefer, C.; Renn, M.; Tormo, D.; Morskaya, S.S.; Kirschnek, S.; Gaffal, E.; Landsberg, J.; Hellmuth, J.; et al. 5′-triphosphate-siRNA: Turning gene silencing and Rig-I activation against melanoma. Nat. Med. 2008, 14, 1256–1263. [Google Scholar] [CrossRef] [PubMed]
- Putzbach, W.; Gao, Q.Q.; Patel, M.; Van Dongen, S.; Haluck-Kangas, A.; Sarshad, A.A.; Bartom, E.T.; Kim, K.Y.A.; Scholtens, D.M.; Hafner, M.; et al. Many si/shRNAs can kill cancer cells by targeting multiple survival genes through an off-target mechanism. eLife 2017, 6, 1–43. [Google Scholar] [CrossRef] [PubMed]





© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stepanov, G.; Zhuravlev, E.; Shender, V.; Nushtaeva, A.; Balakhonova, E.; Mozhaeva, E.; Kasakin, M.; Koval, V.; Lomzov, A.; Pavlyukov, M.; et al. Nucleotide Modifications Decrease Innate Immune Response Induced by Synthetic Analogs of snRNAs and snoRNAs. Genes 2018, 9, 531. https://doi.org/10.3390/genes9110531
Stepanov G, Zhuravlev E, Shender V, Nushtaeva A, Balakhonova E, Mozhaeva E, Kasakin M, Koval V, Lomzov A, Pavlyukov M, et al. Nucleotide Modifications Decrease Innate Immune Response Induced by Synthetic Analogs of snRNAs and snoRNAs. Genes. 2018; 9(11):531. https://doi.org/10.3390/genes9110531
Chicago/Turabian StyleStepanov, Grigory, Evgenii Zhuravlev, Victoria Shender, Anna Nushtaeva, Evgenia Balakhonova, Elena Mozhaeva, Marat Kasakin, Vladimir Koval, Alexander Lomzov, Marat Pavlyukov, and et al. 2018. "Nucleotide Modifications Decrease Innate Immune Response Induced by Synthetic Analogs of snRNAs and snoRNAs" Genes 9, no. 11: 531. https://doi.org/10.3390/genes9110531